Abstract
Nucleosomes and their histone components have generally been recognized to act negatively on transcription. However, purified upstream activating factor (UAF), a transcription initiation factor required for RNA polymerase (Pol) I transcription in Saccharomyces cerevisiae, contains histones H3 and H4 and four nonhistone protein subunits. Other studies have shown that histones H3 and H4 are associated with actively transcribed rRNA genes. To examine their functional role in Pol I transcription, we constructed yeast strains in which synthesis of H3 is achieved from the glucose-repressible GAL10 promoter. We found that partial depletion of H3 (≈50% depletion) resulted in a strong inhibition (>80%) of Pol I transcription. A combination of biochemical analysis and electron microscopic (EM) analysis of Miller chromatin spreads indicated that initiation and elongation steps and rRNA processing were compromised upon histone depletion. A clear decrease in relative amounts of UAF, presumably caused by reduced stability, was also observed under the conditions of H3 depletion. Therefore, the observed inhibition of initiation can be explained, in part, by the decrease in UAF concentration. In addition, the EM results suggested that the defects in rRNA transcript elongation and processing may be a result of loss of histones from rRNA genes rather than (or in addition to) an indirect consequence of effects of histone depletion on expression of other genes. Thus, these results show functional importance of histones associated with actively transcribed rRNA genes.
Keywords: histones H3 and H4, upstream activating factor
In eukaryotic cells, many genes are packaged into nucleosomes and are not transcriptionally active. Nucleosomes and their histone components have generally been recognized to act negatively on transcription at both initiation and elongation steps. For this reason, extensive studies have been carried out on ways to overcome the negative effects of nucleosomal chromatin structures (such as alterations of chromatin structure by histone modifications or by chromatin remodeling factors) and on their significance in regulation of gene expression (reviewed in, e.g., refs. 1 and 2). Most of these previous studies focused on gene expression by RNA polymerase (Pol) II. Here, we focus on the role of histone H3 (and H4) in rRNA gene transcription by Pol I in the yeast Saccharomyces cerevisiae (called “yeast” in this paper).
Genes for rRNA in eukaryotic cells are usually present in many tandemly repeated copies, only a fraction of which are normally transcribed, whereas the remaining genes are associated with typical nucleosomal structures and kept inactive (3, 4). Although recent studies have revealed considerable information on the question of how modifications of histones and chromatin structures at Pol I promoter regions are involved in regulation of rRNA genes (e.g., see refs. 5 and 6) questions regarding the presence of histones in the actively transcribed rRNA coding region, which was controversial in the past (see ref. 7), and their possible functional significance have not been specifically addressed. If histones and nucleosomes play only negative roles, decreasing the association of histones with rRNA genes might lead to activation of inactive rRNA genes kept in the nucleosome structure. In addition, even active rRNA genes might undergo some stimulation if the actively transcribed coding region is associated with histones, which would likely inhibit Pol I elongation. Earlier work with a histone H4 depletion system was relevant to such questions and concluded that, in contrast to activation of some Pol II genes such as PHO5 (ref. 8; see also ref. 9), there was no evidence for alteration in Pol I transcription of rRNA genes upon histone H4 depletion (10). Thus, the results were consistent with the view that histones are either not associated or only weakly associated with actively transcribed rRNA genes and do not play any functionally significant role in Pol I transcription.
In our studies on Pol I transcription of yeast rRNA genes, we initially characterized essential transcription initiation factors and their protein components (for reviews, see refs. 11 and 12). One of them, upstream activating factor (UAF), which tightly binds to the upstream promoter element and is essential for Pol I transcription (13), was found to contain H3 and H4 in addition to four other protein subunits, Rrn5p, Rrn9p, Rrn10p, and Uaf30p (14). This study was initiated to examine a possible functional significance for the histone components in UAF functions. We have found that, contrary to the expectation from the earlier work (10), depletion of histone H3 (or H4) leads to strong inhibition of Pol I transcription of rRNA genes, and that the inhibition takes place not only at the initiation of transcription, but also at later steps during elongation of the transcripts.
Materials and Methods
Media and Strains. Yeast extract/peptone (YEP)-galactose, YEP-glucose, synthetic galactose (SG) and synthetic glucose (SD) complete media, were described in refs. 15 and 16. For [methyl-3H]methionine incorporation experiments, methionine was omitted from SD complete medium. In all of the experiments, cells were grown at 30°C.
The yeast strains and plasmids used are described in Table 1. To construct H3 depletion strain NOY2022 and control strain NOY2025, the starting strain was a segregant from the cross between NOY844 (14) and MX1-4C (17) (see Table 1) that was Leu- (i.e., RRN5) and Ura+ (i.e., HHT1-HHF1 present on pMS329) and was additionally sensitive to 5-FOA, indicating the presence of chromosomal deletions of both the HHT1-HHF1 and the HHT2-HHF2 loci. Plasmid pNOY684 (CEN6 TRP1 PGAL10-HHT2 HHF2) described below or pNOY435 (CEN6 TRP1 HHT2 HHF2) (14) was transformed into this strain, followed by plating onto 5-FOA to eliminate pMS329 (URA3) and transformation with pRS316 (URA3) to generate NOY2022 and NOY2025, respectively. pNOY684 was constructed by PCR amplification of a fragment carrying PGal10-HHT2 from pRM102 (18) and replacing the HHT2 gene in pNOY435 (CEN6 TRP1 HHT2 HHF2, ref. 14) with PGal10-HHT2.
Table 1. Yeast strains and plasmids used.
| Strain | Description |
|---|---|
| MX1-4C | MATα ura3-52 leu2-3, 112 trp1 his 3 Δ(hht1-hhf1) Δ(hht2-hhf2), pMS329 [CEN4 URA3 SUP11 HHT1-HHF1] (17) |
| NOY844 | MATa ade2-1 ura3-1 his3-11, 15 trp1-1 leu2-3, 112 can1-100 rrn5Δ::LEU2, pNOY434 [CEN6 HIS3 RRN5-(HA)3-(His)6] (14) |
| NOY848 | MATα ura3 his3 trp1 leu2-3, 112, rrn5Δ::LEU2 Δ(hht1-hhf1) Δ(hht2-hhf2), pNOY434 [CEN6 HIS3 RRN5-(HA)3 (His)6] (14), pNOY439 [CEN6 TRP1 myc-HHT2-HHF2] |
| NOY2020 | Same as NOY848 but carries pNOY682 [CEN6 TRP1 PGAL10-myc-HHT2 HHF2] instead of pNOY439 |
| NOY2022 | MAT? ade2-1 ura3 his3 trp1 leu2-3, 112 Δ(hht1 hhf1) Δ(hht2 hhf2), pNOY684 [CEN6 TRP1 PGAL10-HHT2 HHF2] and pRS316 [CEN6 URA3] |
| NOY2025 | Same as NOY2022 but carries pNOY435 [CEN6 TRP1 HHT2 HHF2] (14) instead of pNOY684 |
Another pair of the strains, H3 depletion strain NOY2020 and control strain NOY848, carried the genes for H3 and H4 on plasmid pNOY682 and pNOY439, respectively. Plasmid pNOY439 carrying HHT2 with a myc-tag sequence (SEQKLI-SEEDL) inserted between the first and the second codons of HHT2 was constructed by a method similar to that used for pNOY436 carrying myc-tagged HHF2 (14). For construction of pNOY682, the nontagged HHF2 gene and HHT2 with a myc tag to the N terminus were first constructed from yeast genomic DNA by standard PCR methods. The PCR product containing HHF2 was ligated into the SmaI and EagI sites in pRS314 creating plasmid PEC612 (HHF2). The PCR product containing myc-tagged HHT2 was inserted between NcoI and XhoI sites in pSJ1 (2μ LEU2 PGal10; a gift from B. Schwer, Weil Medical College of Cornell University, New York) to generate the plasmid carrying PGal10-myc-HHT2. This plasmid was digested with EcoRI and XhoI, generating a fragment containing PGal10-myc-HHT2, which was ligated into the EcoRI and XhoI sites of PEC612 (HHF2), resulting in pNOY682.
Biochemical Analyses. Analyses of rRNA synthesis by labeling RNA with [3H]uridine or [methyl-3H]methionine were carried out as described in refs. 16 and 19). Fractions used for determination of the amount of (His)6- and (HA)3-tagged Rrn5p or for UAF activity assay in vitro were prepared according to UAF purification methods described in refs. 14 and 20 (see Fig. 2 A). The in vitro Pol I transcription assay with purified components was also described in refs. 14 and 20.
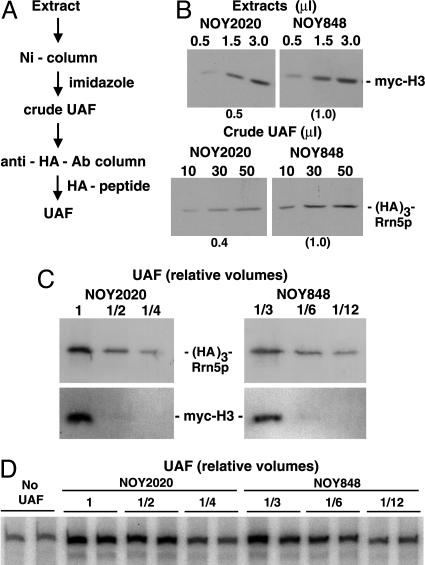
Fig. 2.
Analysis of UAF in H3-depleted cells. (A) Purification steps used to make UAF fractions. (B) H3 depletion strain NOY2020 and control strain NOY848 were grown in YEP-galactose. Cells at a midlog (A600 ≈ 0.2) were harvested and resuspended in YEP-glucose and incubated for 4 h. Cells were then collected, and extracts were prepared. After total protein concentrations of the two extracts were adjusted to be equal, portions were used to determine the amount of myc-tagged H3 by Western blotting with anti-myc Ab (Upper). Remaining portions were used to obtain “crude UAF” fractions by Ni-column followed by elution with imidazole. The amounts of Rrn5p were then determined by Western blotting with anti-HA Ab (Lower). (C) Extracts were prepared from H3-depleted cells (NOY2020) and control cells (NOY848) as described in B. During the 4-h depletion of H3 in YEP-glucose, NOY2020 cell mass increased 2.0-fold and NOY848 cell mass increased 2.5-fold. After total protein concentrations of the two extracts were adjusted to the same values, equal volumes of the two extracts were used to purify UAF as described in A. Series of 2-fold diluted samples as indicated were subjected to gel electrophoresis. The gel was cut into two parts: Upper was used to determine the amount of HA-tagged Rrn5p by using anti-HA Ab, and Lower was used to determine myc-tagged H3 by using anti-myc Ab. Note that each of the control samples shown was 3-fold more diluted than each of the samples from H3-depleted cells. If the relative synthesis rate of UAF (per cell density) in glucose is similar to that in galactose, the theoretical maximum decrease in UAF concentration when H3-synthesis is halted upon transfer to glucose would be 2-fold. The observed 3-fold decrease strongly suggests an instability of UAF in the H3-depleted cells. (D) The purified UAF preparations used in the experiment shown in C were examined for their ability to stimulate Pol I transcription in vitro. Series of 2-fold diluted samples were analyzed. An autoradiogram of transcription products is shown. As in C, 3-fold differences in dilution factors were used to make comparison of H3-depleted samples and control samples easier. The UAF purified from NOY2020 and NOY848 activated transcription 2.7-fold and 2.8-fold compared with control without UAF, respectively, at the highest concentration.
Electron Microscopic (EM) Miller Chromatin Spread Analysis. Yeast were grown in SG complete medium containing 1 M sorbitol, and H3 was depleted by the addition of glucose to a final concentration of 2%, followed by incubation for 4 h at 30°C. Miller chromatin spread and EM analysis were performed as described in ref. 21. As in the previous study, chromatin spreads from multiple cultures of each strain were examined. For quantitative analysis, entire EM grids were scanned, and all rRNA genes were photographed and analyzed. The polymerase number per gene was determined by counting the number of RNA polymerases (or nascent rRNA transcripts) on all rRNA genes that could be unambiguously followed from 5′ to 3′ end.
Results
Effects of Histone H3 (or H4) Depletion on rRNA Transcription. We constructed yeast strains that allow H3 (or H4) depletion to be achieved by a galactose to glucose medium shift similar to that used in ref. 10. In initial experiments, we compared rRNA synthesis rate measured by [32P]orthophosphate labeling at 4 h after the shift to glucose (depleted) with that of the same strain growing in galactose medium (nondepleted). In contrast to the reported results, a large inhibition (≈90%) of rRNA synthesis was observed for H4 (or H3)-depleted cells relative to nondepleted cells (data not shown). The reason for the discrepancy between these results and previously reported results is unknown. In these initial experiments, rRNA transcription rates were compared in the same strain growing on two different carbon sources. For subsequent experiments to compare effects of histone depletion in cells growing on the same carbon source, a control strain was constructed that grows on glucose without histone depletion, and we focused on effects of histone H3 depletion on Pol I transcription.
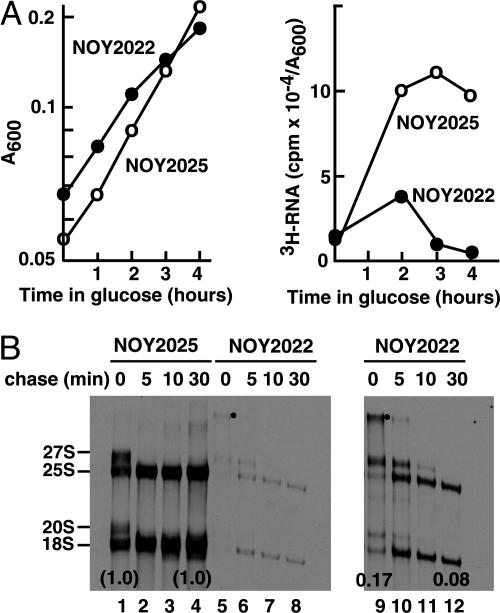
Two strains were used in this experiment: NOY2022 as the H3 depletion strain and NOY2025 as the control strain. Cells were grown in SG complete medium, and histone depletion was achieved (in NOY2022) by the addition of glucose directly to the cultures. Total RNA synthesis rates were first measured by 5 min of [3H]uridine pulse labeling. Two hours after the addition of glucose, when effects of H3 depletion on cell growth were minimal, the RNA synthesis rate in H3-depleted cells was inhibited 60% relative to control cells. Thus, inhibition of RNA synthesis cannot be explained by inhibitory effects of H3 depletion on cell growth. At 3 and 4 h after addition of glucose, inhibition by H3 depletion was ≈90% (Fig. 1A). Inhibition of synthesis of individual large rRNAs, 5S RNA, and tRNAs was also confirmed by gel electrophoresis (data not shown). Next, pulse-chase experiments were done by using [methyl-3H]methionine at 4 h after the addition of glucose. The pulse was terminated at 4 min by the addition of excess nonradioactive methionine, and the amounts of the 3H-labeled methyl group of methionine in the total trichloroacetic acid-precipitable fraction (mostly protein) and rRNAs separated by gel electrophoresis were measured at times indicated in Fig. 1B. Only a weak decrease (8% and 17% decrease compared with controls before and after the 30-min chase, respectively) was observed in the incorporation into protein (data not shown). This result confirms that effects of H3 depletion on cell growth were small in this experiment. In addition, the result also indicates that H3 depletion does not cause a significant inhibition of methionine uptake. In contrast, incorporation into rRNAs and their precursors (per unit amount of cell mass) was greatly inhibited (83% and 94% before and after the 30-min chase, Fig. 1B). The results also clearly indicated that processing of rRNA precursors to mature 18S and 25S rRNAs is slowed by H3 depletion. For example, at the end of 4 min pulse labeling, a significant fraction (≈30%) of pulse-labeled rRNAs exist as 35S pre-rRNA (Fig. 1B, lanes 5 and 9; bands with a dot) in H3-depleted cells, whereas a very small amount, if any, is detectable in control cells (lane 1).
Fig. 1.
Effects of H3 depletion on cell growth and rRNA synthesis. (A) NOY2022 and NOY2025 were grown in SG complete medium to a cell density of A600 0.06-0.08, glucose was then added to a final concentration of 2%, and incubation was continued for 4 h. Cell density increase was followed (Left). At indicated times, aliquots were taken, cells were pulse-labeled with [3H]uridine (5 μCi/ml; 39 Ci/mmol; 1 Ci = 37 GBq) for 5 min, and incorporation into a total trichloroacetic acid-insoluble RNA fraction was determined (Right). (B) In another experiment similar to above, cells were pulse-labeled with [methyl-3H]methionine (33 μCi/ml; 83 mCi/μmol) for 4 min at 4 h after the addition of glucose. Nonradioactive methionine (500 μg/ml) was then added (time 0) to chase [methyl-3H]methionine. At indicated times, samples were taken for measurements of incorporation of 3H-label into total trichloroacetic acid-insoluble fraction (mostly protein together with small amounts of RNA; data not shown, but mentioned in Results). 3H-labeled RNA was also isolated, and samples (derived from equal volumes of the original cultures) were subjected to a polyacrylamide/agarose composite gel electrophoresis followed by autoradiography (lanes 1-8). Lanes 9-12 are the same as lanes 5-8 but with a longer exposure time. The amount of 3H in radioactive rRNA bands (precursor 35S, 27S, 20S, and mature 25S and 18S) were quantified, and the sums of these values were used to calculate rRNA synthesized per unit amount of cell mass (A600 unit). The values obtained for samples before and after chase were compared between the two strains and indicated below the 18S bands.
Effects of H3 Depletion on UAF. To examine whether the observed decrease in rRNA synthesis in H3 (or H4)-depleted cells could be explained by a decrease in UAF containing these histones, we measured the amounts of Rrn5p, an essential component of UAF. We used H3 depletion strain NOY2020, which carries a myc-tagged HHT2 gene fused to the GAL10 promoter, and control strain NOY848, in which the myc-tagged HHT2 is transcribed from the native promoter. Both strains also carry (His)6- and (HA)3-tagged Rrn5p. The two strains were grown in YEP-galactose and then shifted to YEP-glucose. At 4 h after the shift, the amounts of HA-tagged Rrn5p (and myc-tagged H3) associated with UAF were measured. In addition, the total amount of H3 in cell extracts was also analyzed. After 4 h of depletion, the total amount of H3 was ≈50% of that in the control strain NOY848, as determined by Western blot with anti-myc Ab (Fig. 2B Upper). The amount of Rrn5p in “crude UAF” (see Fig. 2 A) in H3 depleted cells was then analyzed by Western blot with anti-HA Ab and found to be ≈40% the total of control cells (Fig. 2B Lower). Thus, H3 depletion caused a decrease in the amount of Rrn5p (per total protein), and the degree of decrease was roughly comparable with that of H3 in cell extracts. Similar observations were also made with respect to H4 depletion (unpublished experiments). In other experiments, UAF was further purified by immunoaffinity purification by using an anti-HA Ab column (see Fig. 2 A), and the amounts of Rrn5p and H3 were analyzed simultaneously by Western blot with anti-HA and anti-myc Abs, respectively. The amount of Rrn5p in UAF recovered from the H3 depleted cells was ≈3-fold less than that of the control strain. However, Rrn5p was associated with H3 to approximately the same extent as that observed for Rrn5p from the control cells (Fig. 2C). We also examined the activity of UAF in stimulating Pol I transcription in vitro (20) by using a series of dilutions of the UAF preparations (Fig. 2D). We found that, when the same amounts of UAF (as judged by the amounts of Rrn5p) were used, there was no significant difference in stimulatory activity between UAF from H3-depleted cells and that from control cells. Thus, we conclude that H3 depletion leads to a 2- to 3-fold decrease in cellular amounts of UAF, but that the remaining UAF is still associated with H3 (to the same extent as the control UAF) and other essential subunits and retains its activity in Pol I transcription as assayed in vitro.
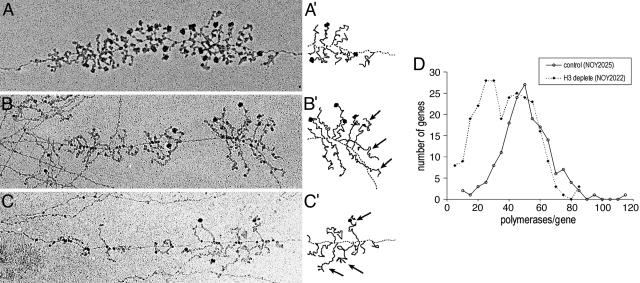
Transcription of rRNA Genes in H3-Depleted Cells Analyzed by the EM Chromatin Spread Method. We examined transcription patterns for individual rRNA genes by using an EM chromatin spread method. NOY2022 for H3 depletion and NOY2025 as a control were used, and H3 depletion was carried out by the direct addition of glucose to cultures in SG complete medium containing 1 M sorbitol and incubating for 4 h. Examples of rRNA genes for each strain are shown in Fig. 3 A-C, and the summary of quantification of polymerase density is shown in Fig. 3D and Table 2.
Fig. 3.
Representative electron micrographs of 35S rRNA genes from H3-depleted cells. (A) Control NOY2025 cells. A gene with ≈51 polymerases (close to the average polymerase density for this control strain) is shown. (B) H3-depleted NOY2022 cells. A gene with ≈34 polymerases (close to the average polymerase density for H3-depleted cells) is shown. (C) H3-depleted NOY2022 cells. A gene with 26 polymerases is shown. Schematic representations of the ends of the genes in A, B, and C are in A′, B′, and C′, respectively. Arrows indicate the longer uncleaved rRNA transcripts. (D) Decrease in polymerase density per gene in H3-depleted NOY2022 relative to control NOY2025 cells (at 4 h after the shift to glucose). The number of polymerases per gene was counted for 261 genes from H3-depleted cells and for 170 genes from control cells. Distribution of polymerases among transcribed genes is plotted by grouping with an increment of 5.
Table 2. Summary of the EM analysis of the effects of H3 depletion on the transcription of the 355 rRNA genes and comparison with biochemical analysis of rRNA synthesis.
| Polymerase density (EM)
|
Inactive genes
|
Relative rRNA synthesis rate, %
|
|||
|---|---|---|---|---|---|
| Strains | Avg. (%) | n | Inactive/active | % of total | |
| NOY2025 | 49 ± 16 (100) | 170 | 23/93 | 20 | 100 |
| NOY2022 | 35 ± 17 (71) | 261 | 10/43 | 19 | 15 |
Average polymerase density was calculated from the data shown in Fig. 3D. Counting the number of inactive genes relative to active genes is explained in the text. [Methyl-3H]methionine incorporation experiments were carried out by using the same conditions as used for the EM analysis. The values (NOY2022/NOY2025) obtained in two independent experiments were 0.12 and 0.17, and the average value is shown.
We found that although H3-depleted cells showed a clear decrease in polymerase density in many individual rRNA genes, as is evident from Fig. 3D, the average polymerase density was 71% relative to the control. Because this decrease was significantly less than that determined by biochemical analyses, we repeated the biochemical experiments by using cells grown in the sorbitol-containing medium used for the EM experiments. By 4 min pulse labeling with [methyl-3H]methionine, we again found that rRNA synthesis in H3-depleted cells was only ≈15% of that in control cells, the value similar to that (17%, Fig. 1C) obtained in the absence of sorbitol (Table 2, sixth column). To determine whether fewer genes were active in H3-depleted cells, we examined linear stretches of rRNA genes that could be followed at least 3 repeat lengths and counted the number of inactive genes relative to active genes, as in our previous study (21). Although inactive genes without adjacent recognizable active genes may be missed in this assay, the number of inactive genes counted in this way did not show any significant difference between H3-depleted cells and control cells (Table 2, fifth column). Thus, H3 depletion apparently did not cause any significant decrease in the number of active genes (and, presumably, did not cause derepression of inactive genes). It is clear that the large decrease in rRNA synthesis rate observed upon H3 depletion cannot be explained by the observed small decrease in average polymerase density if the rate of elongation of polymerase (and its processivity) remained the same. Thus, these data indicate that Pol I engaged in rRNA gene transcription in H3-depleted cells is not functioning efficiently, i.e., overall transcript elongation is inefficient relative to the control cells. Consistent with elongation defects, many of the genes in H3-depleted cells have a polymerase distribution that could arise from periodic impediments to polymerase movement down the gene, namely clumps of densely packed polymerases separated by polymerase-free gaps (Fig. 3 B and C). Alternatively, this feature may be a result of bursts of initiation events, as if UAF were temporarily associated and then dissociated from the promoter. However, even if this situation were the case, overall efficiency of elongation must be decreased in H3-depleted cells because the decrease in the average polymerase density was much smaller than that in overall rRNA synthesis rate. Genes from control cells (Fig. 3A) typically have more uniform polymerase distributions.
The EM analysis showed that the control strain had a polymerase density distribution very similar to other control strains (21, 22), with a normal distribution and a mean of 49 polymerases per gene (n = 170). There was, however, considerable heterogeneity in polymerase density in rRNA genes after H3 depletion (mean of 35; n = 261 genes from 23 cells), with some genes showing a large reduction and others being similar to controls (Fig. 3D), indicating heterogeneity in the degree or type of inhibition. This finding cannot entirely be explained by heterogeneity in the response of individual cells to H3 depletion. By examining the distribution of polymerase density in individual cells, we found that only a small fraction of cells [4 cells, or 17% of total number of cells, whose rRNA genes were analyzed] showed “high polymerase density” (defined as >35 polymerases per gene) in all of the observable rRNA genes. A major fraction of cells (12 cells or 52% of cells analyzed) showed both high polymerase density genes and “low polymerase density genes” (defined as genes with <35 polymerases per gene). The remaining cells analyzed (7 cells or 30% of cells analyzed) showed mostly low polymerase density genes. Our interpretation of these results is that H3 depletion decreases both initiation and elongation steps stochastically. If the frequency of initiation is affected much more strongly than elongation rate is decreased, polymerase density is expected to decrease strongly relative to rRNA genes in control cells. If the elongation rate decreases strongly, together with decreased initiation frequency, there will be little or no change in polymerase density. Because the decrease in the amount of bulk H3 in the H3-depleted cells is only partial (≈50% relative to control; compare Fig. 2B), the exact nature of defects in elongation or initiation by H3 depletion might be different among some ≈80 or so active genes within a single cell, thus explaining the greater heterogeneity of polymerase density than is normally observed among individual rRNA genes within a single cell. This characteristic feature makes it more difficult to explain Pol I transcription defects as being solely due to indirect effects of H3 depletion on expression of some other genes (see Discussion).
Examination of EM pictures of H3-depleted cells also revealed defects in pre-rRNA processing. For example, large terminal knobs at the 5′-end of transcripts (small subunit processomes, ref. 23) form somewhat later in the depletion strain relative to the control strain. In addition, cotranscriptional cleavage in ITS1 (23) is less frequent in the depletion strain, in that longer uncleaved transcripts can be seen at or near the end of genes more often than the control strain (arrows in Fig. 3 B′, C′ and results not shown). These observations are consistent with the conclusion obtained by pulse-chase experiments, namely that histone depletion slows rRNA processing (Fig. 1B).
Discussion
We have observed that the partial depletion of histone H3 (or H4) leads to a large decrease in rRNA synthesis rates. No derepression of inactive rRNA genes was evident. Our observations are most consistent with the model that histones (H3 and H4) are associated with actively transcribed genes and play functionally important roles not only in initiation but also in other steps, including elongation, in Pol I transcription. In separate studies, we have directly demonstrated the association of histones H3 and H4 with actively transcribed rRNA-encoding DNA copies, not only at the promoter but also at the coding region, by chromatin immunoprecipitation with a yeast strain in which all rRNA genes are actively transcribed during exponential growth (unpublished data).
H3 (or H4) Depletion Causes a Decrease in Pol I Transcription and in the Relative Amount of UAF. Studies on global gene expression by using DNA microarrays showed that ≈10% and ≈15% of yeast protein coding genes showed reduced and increased expression, respectively, upon H4 depletion (9). Thus, the large decrease in Pol I transcription of rRNA genes observed at 4 h of H3 or H4 depletion could be a consequence of indirect effects through alterations in the expression of other Pol II-transcribed genes. However, the decrease in Pol I transcription must be at least partly due to the observed decrease in UAF, an essential Pol I initiation factor. The decrease in the amount of UAF relative to the total amount of protein may be due to instability of UAF under conditions of histone depletion. Because H4 depletion was found not to cause any significant changes in the amount of mRNAs for the components of UAF (Rrn5p, Rrn9p, Rrn10p, and Uaf30p) relative to the control strain (9), and total protein synthesis rates are only slightly decreased under such conditions (see Results), it is unlikely that synthesis of UAF subunits is significantly decreased by H4 or H3 depletion. In addition, we found that the remaining UAF is associated with H3 to the same extent as the control UAF, that is, we failed to observe any significant increase in the amount of Rrn5p that is not associated with H3 in H3-depleted cells. This observation is also consistent with the hypothesis that UAF and/or UAF subunits are unstable in the absence of associated H3 and H4. Therefore, we suggest that UAF can stably exist in vivo only in the form containing both H3 and H4, presumably bound to the upstream element of the Pol I promoter. Thus, decrease in the synthesis of H3 (or H4) may cause a decrease in the stable form of UAF, which, in turn, leads to a decrease in the initiation of Pol I transcription.
Although H3 and H4 are known UAF components (14), their functional significance in the purified UAF is unknown. Without suitable histone-free preparations, it has not been possible to compare the degree of stimulation of Pol I transcription between purified or reconstituted UAF preparations with and without these histones in vitro. The results of histone depletion experiments now suggest that the association of H3 and H4 with the other UAF components is important at least for the stability of UAF in vivo and, hence, for rRNA transcription by Pol I.
In addition to an inhibition expected by the decrease in the stable form of UAF, there may be an additional mechanism(s) that inhibits transcription initiation either directly or indirectly as a result of histone depletion. We observed that H3 depletion also caused inhibition of Pol I elongation and processivity either directly or indirectly (see below). We note that the extreme heterogeneity in polymerase density among individual rRNA genes within a single cell observed by EM analysis indicates that individual genes are affected differently by histone depletion. If inhibition is caused entirely by an indirect mechanism, e.g., an induction of a regulatory mechanism that inhibits an initiation and/or some other step(s), one would expect more equal effects on all of the active genes within a single cell as was observed previously for the EM analysis of rapamycin-treated cells (16). The heterogeneous distribution of polymerase density observed for H3 depleted cells is clearly different from that for control cells (or previously analyzed rapamycin-treated cells). Therefore we favor the model that random loss of histones from rRNA-encoding DNA “directly” affects different genes in different ways and leads to the increased heterogeneity of polymerase density in H3-depleted cells.
The heterogeneous polymerase density observed for H3-depleted cells as shown in Fig. 3D might suggest the presence of two distinct populations. Possibly, these populations correspond to genes with and without UAF bound to promoter regions. Further studies are clearly needed to test the validity of such a possibility.
Histone H3 Depletion Leads to Defects in Elongation of Pol I Transcription and rRNA Processing. As can be seen clearly in Table 2, the decrease in the average polymerase density is much smaller than the decrease in the synthesis rate of rRNA. In addition, no significant decrease in the number of active rRNA genes was observed. Therefore, the overall rate of elongation must have decreased relative to the control strain upon the depletion of H3 for 4 h. A formal calculation from the value shown in Table 1 (polymerase density 71% and synthesis rate 15% relative to control) indicates that the elongation rates have decreased, on average, to ≈20% of control (0.71 × 0.2 = 0.142 = ≈0.15). In addition, pulse-chase experiments indicate that processing of rRNA precursors is also impaired under the conditions of H3 depletion. The longer transcripts observed near the end of the rRNA gene in the EM analysis also supports this conclusion.
What is the mechanism that leads to defects in transcription elongation and rRNA processing? As discussed above, we favor direct roles of histones in efficient elongation of transcripts. Thus, just as UAF, with its tightly associated H3 and H4, plays an important role in the initiation of Pol I transcription, specific histone containing chromatin structures may recruit some (undiscovered) protein factors required for efficient Pol I elongation, or factors/machineries required for cotranscriptional events, such as the cotranscriptional cleavage of rRNA transcripts at the A2 site (23). Perhaps efficient cotranscriptional rRNA cleavage/processing might be important for achieving efficient elongation in vivo. Compared with the mechanism of transcription initiation and its regulation, regulation of elongation steps in Pol I transcription is poorly studied. Thus, regardless of the question of direct vs. indirect effects of histone H3 depletion on elongation, the mechanisms involved deserve further study in connection with possible regulation of Pol I transcription at elongation steps.
Finally, it should be noted that histones H2A and H2B are not present in UAF (14). However, these histones may also be associated with actively transcribed rRNA genes together with H3 and H4, as was shown for extrachromosomal rRNA genes from Physarum polycephalum (24). Thus, it is possible that H2A (and H2B) may also play important roles in rRNA gene transcription that are independent of UAF functions. Thus, analysis of effects of H2A (or H2B) depletion on rRNA transcription may deserve future pursuit.
Acknowledgments
We thank Dominique Lalo for construction of some of the plasmids and yeast strains used in this study; Kristilyn Eliason, Cathleen A. Josaitis, and Jonathan A. Dodd for participation in this project at various stages; and Drs. M. Grunstein (Geffen School of Medicine at the University of California, Los Angeles), M. Mitchell Smith (University of Virginia), and B. Schwer for gifts of yeast strains and plasmids. The work was supported by Public Health Service Grants GM-35940 (to M.N.) and GM-63952 (to A.L.B.) and by a postdoctoral research fellowship from Jane Coffin Childs Memorial Fund for Medical Research (to D.A.S.).
Author contributions: M.N. designed research; P.T., S.L.F., M.L.O., L.V., and D.A.S. performed research; L.V. contributed new reagents/analytic tools; P.T., S.L.F., A.L.B., and M.N. analyzed data; S.L.F., D.A.S., A.L.B., and M.N. wrote the paper; and A.L.B. supervised EM experiments.
Abbreviations: EM, electron microscopic; Pol, RNA polymerase; SG, synthetic galactose; UAF, upstream activating factor; YEP, yeast extract/peptone.
References
- 1.Fischle, W., Wang, Y. & Allis, C. D. (2003) Curr. Opin. Cell Biol. 15, 172-183. [DOI] [PubMed] [Google Scholar]
- 2.Khorasanizadeh, S. (2004) Cell 116, 259-272. [DOI] [PubMed] [Google Scholar]
- 3.Conconi, A., Widmer, R. M., Koller, T. & Sogo, J. M. (1989) Cell 57, 753-761. [DOI] [PubMed] [Google Scholar]
- 4.Dammann, R., Lucchini, R., Koller, T. & Sogo, J. M. (1993) Nucleic Acids Res. 21, 2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santoro, R. & Grummt, I. (2005) Mol. Cell. Biol. 25, 2539-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawrence, R. J., Earley, K., Pontes, O., Silva, M., Chen, Z. J., Neves, N., Viegas, W. & Pikaard, C. S. (2004) Mol. Cell 13, 599-609. [DOI] [PubMed] [Google Scholar]
- 7.Lucchini, R. & Sogo, J. M. (1998) in Transcription of Ribosomal RNA Genes by Eukaryotic RNA Polymerase I, ed. Paule, M. (R.G. Landes, Austin, TX), pp. 255-276.
- 8.Han, M., Kim, U. J., Kayne, P. & Grunstein, M. (1988) EMBO J. 7, 2221-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyrick, J. J., Holstege, F. C., Jennings, E. G., Causton, H. C., Shore, D., Grunstein, M., Lander, E. S. & Young, R. A. (1999) Nature 402, 418-421. [DOI] [PubMed] [Google Scholar]
- 10.Kim, U. J., Han, M., Kayne, P. & Grunstein, M. (1988) EMBO J. 7, 2211-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nomura, M. (2001) Cold Spring Harbor Symp. Quant. Biol. 66, 555-565. [DOI] [PubMed] [Google Scholar]
- 12.Nomura, M., Nogi, Y. & Oakes, M. (2004) in The Nucleolus, ed. Olson, M. O. J. (R.G. Landes, Austin, TX), pp. 128-153.
- 13.Keys, D. A., Lee, B. S., Dodd, J. A., Nguyen, T. T., Vu, L., Fantino, E., Burson, L. M., Nogi, Y. & Nomura, M. (1996) Genes Dev. 10, 887-903. [DOI] [PubMed] [Google Scholar]
- 14.Keener, J., Dodd, J. A., Lalo, D. & Nomura, M. (1997) Proc. Natl. Acad. Sci. USA 94, 13458-13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherman, F., Fink, G. R. & Hicks, J. B. (1986) Laboratory Course Manual for Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 16.Claypool, J. A., French, S. L., Johzuka, K., Eliason, K., Vu, L., Dodd, J. A., Beyer, A. L. & Nomura, M. (2004) Mol. Biol. Cell 15, 946-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan, B. A., Mittman, B. A. & Smith, M. M. (1991) Mol. Cell. Biol. 11, 4111-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mann, R. K. & Grunstein, M. (1992) EMBO J. 11, 3297-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warner, J. R. (1991) Methods Enzymol. 194, 423-428. [DOI] [PubMed] [Google Scholar]
- 20.Keener, J., Josaitis, C. A., Dodd, J. A. & Nomura, M. (1998) J. Biol. Chem. 273, 33795-33802. [DOI] [PubMed] [Google Scholar]
- 21.French, S. L., Osheim, Y. N., Cioci, F., Nomura, M. & Beyer, A. L. (2003) Mol. Cell. Biol. 23, 1558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandmeier, J. J., French, S., Osheim, Y., Cheung, W. L., Gallo, C. M., Beyer, A. L. & Smith, J. S. (2002) EMBO J. 21, 4959-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osheim, Y. N., French, S. L., Keck, K. M., Champion, E. A., Spasov, K., Dragon, F., Baserga, S. J. & Beyer, A. L. (2004) Mol. Cell 16, 943-954. [DOI] [PubMed] [Google Scholar]
- 24.Prior, C. P., Cantor, C. R., Johnson, E. M., Littau, V. C. & Allfrey, V. G. (1983) Cell 34, 1033-1042. [DOI] [PubMed] [Google Scholar]