Abstract
Telomerase deficiency leads to a progressive loss of telomeric DNA that eventually triggers cell apoptosis in human primary cells during prolonged growth in culture. Rare survivors can maintain telomere length through either activation of telomerase or recombination-based telomere lengthening, and thus proliferate indefinitely. We have explored the possibility that telomeres may be maintained through telomere sister chromatid exchange (T-SCE) in murine telomere reverse transcriptase-deficient (mTert-/-) splenocytes and ES cells. Because telomerase deficiency leads to gradual loss of telomeric DNA in mTert-/- splenocytes and ES cells and eventually to chromosomes with telomere signal-free ends (SFEs), we examined these cell types for evidence of sister chromatid exchange at telomeres, and observed an increase in T-SCEs only in a subset of mTert-/- splenocytes or ES cells that possessed multiple SFEs. Furthermore, T-SCEs were more often detected in ES cells than in splenocytes that harbored a similar frequency of SFEs. In mTert heterozygous (mTert+/-) ES cells or splenocytes, which are known to exhibit a decrease in average telomere length but no SFEs, no increase in T-SCE was observed. In addition to T-SCE, other genomic rearrangements (i.e., SCE) were also significantly increased in mTert-/- ES cells possessing critically short telomeres, but not in splenocytes. Our results suggest that animals and cell culture differ in their ability to carry out genomic rearrangements as a means of maintaining telomere integrity when telomeres become critically shortened.
Keywords: murine telomerase reverse transcriptase, telomere signal-free end
Telomeres are unique DNA-protein structures that contain noncoding TTAGGG repeats and telomere-binding or associated proteins. Telomeres allow cells to distinguish natural chromosome ends from damaged DNA and to protect chromosomes against degradation and fusion (1). Telomere integrity in cells thus plays an essential role in the control of genomic stability. A large body of evidence suggests that cells respond to dysfunctional telomeres by undergoing senescence, cell death, or genomic instability (1-4). Thus, telomere integrity depends on the ability to maintain telomere length and/or the ability to mask telomeres from being recognized as damaged DNA.
Telomerase contains two core components, telomerase reverse transcriptase and telomerase RNA, the latter serving as an integral template for synthesis of telomeric DNA. In mice, disruption of either component abolishes telomerase activity, leading to progressive telomere shortening accompanied by cell apoptosis in highly proliferative organs, and male testicular atrophy and infertility in late generations (5-12). The infertility or cell apoptosis correlates with the presence of critically shortened chromosome ends (11-13). In culture, murine ES cells deficient in telomerase RNA (Terc) undergo telomere shortening and an increase in chromosome end-to-end fusions (14, 15) and show growth retardation after 450 cell divisions, although surviving clones regain growth potential and maintain telomere length through a telomerase-independent telomere lengthening characterized by the presence of both telomeric and nontelomeric sequences at most chromosome ends (16). A gradual loss of telomere integrity is also observed in other eukaryotic models that possess a telomerase defect. In yeast, critically short telomeres induce a DNA damage response, loss of growth potential, and eventually a recombination-dependent or -independent process that leads to telomere elongation in the few surviving cells that regain growth potential (17-25). Similarly in plants, critically short telomeres lead to chromosome end-to-end fusions and sterility (26, 27).
Recent studies have shown that uncapped telomeres, either by loss of function of telomere-binding proteins or by loss of telomeric repeats, directly associate with many DNA damage response proteins and induce a response similar to that observed with DNA breaks (28-40). Several proteins known to play a role in the response to DNA damage (Ku, Mre11, Rad50, etc.) are also integral telomere-associated components, and mounting evidence suggests a dual role of those DNA damage response proteins in the protection of chromosome ends and the ability to herald cell-cycle arrest in response to dysfunctional telomeres (41-44).
A subset of human tumors and immortal cell lines utilizes a telomerase-independent mechanism to maintain telomeric DNA, termed alternative lengthening of telomeres (ALT) (45-48). ALT cells are typified by extreme heterogeneity of telomere length and the presence of ALT-associated promyelocytic leukemia bodies that contain extrachromosomal telomeric DNA, telomere-specific binding proteins, and proteins involved in DNA recombination and replication (45, 49-51). ALT is thought to occur as a recombination-mediated lengthening of telomeres (48, 52-56). Recent reports have demonstrated that frequent recombination at telomeres between sister chromatids occurred in ALT cells and may extend the proliferative life of telomerase-negative cells (54-56). Bailey et al. (54) proposed that clonal senescence may be delayed by unequal telomeric DNA exchanges that allow those cells to gain telomeric DNA and further proliferate without the need of telomerase-mediated telomeric DNA extension. Bechter et al. (55) reported that telomere sister chromatid exchanges (T-SCEs) were present in a variety of different human ALT cell lines but universally missing in human telomerase-positive cells. In addition, the ALT-like telomere elongation event was associated with the presence of T-SCEs in a human cancer cell line after inhibition of telomerase. Furthermore, reactivation of telomerase completely abolished the T-SCE pathway in these cells, suggesting that T-SCE is a special type of ALT that does not coexist with telomerase (55).
In this study, we explored T-SCE and other genomic rearrangements as possible mechanisms in the protection or maintenance of critically eroded telomeres of murine splenocytes and long-term culture ES cells. Our results indicate that murine splenocytes and long-term culture ES cells may respond to critically short telomeres differently. Although mTert-/- ES cells and splenocytes are “permissive” for T-SCEs when telomere integrity is compromised, T-SCEs are more readily detected in long-term culture ES cells than in splenocytes. Furthermore, other types of genomic rearrangements (i.e., SCE and nonhomologous end joining) were significantly increased in mTert-/- ES cells possessing critically short telomeres, but not in splenocytes. In addition, we did not find any evidence of T-SCEs in ES cells or splenocytes heterozygous for mTert, despite the presence of average short telomere lengths.
Materials and Methods
Generation of mTert Heterozygous or Null Mice and ES Cells. The generation of mTert+/- mice (C57BL/6) and ES cells and the mating strategy for different generations of mTert+/- and mTert-/- mice have been described (10, 12). In brief, mTert+/- founder mice (C57BL/6/R129J) were mated to wild-type C57BL/6 mice (The Jackson Laboratory), and the mTert+/- progeny were successively backcrossed to wild-type C57BL/6 mice for up to 11 generations (BC11). The first generation (G1) of mTert-/- mice was obtained through intercrossing of mTert+/- BC6 mice. Subsequent generations (G2 to G4) were obtained by mating mTert-/- mice. mTert-/- ES cell clones were generated from G418-resistant mTert+/- ES cell clones by incubation at an increased G418 concentration (4 mg/ml) (10, 15). ES cell culture was carried out as described (57). Cells were split 1:4 every 3 days, which counted as one passage (p) (15).
Activation of Murine Splenocytes. Single-cell suspensions were prepared from spleens of 3-month-old mice. T cell activation by anti-CD3 antibody (PharMingen, 145-2C11) and interleukin 2 (Biosource, PMC0024) have been described (58). Activation lasted 24 or 48 h, which allowed T cells to proliferate for one or two population doublings.
Telomeric FISH. Metaphase spread preparation and telomeric FISH were performed as described (5, 59). The Cy-3-labeled (CCCTAA)3 PNA (Applied Biosystems) was used as a probe. Metaphases were examined with a Zeiss axiophot fluorescence microscope. Approximately 100 metaphases from each sample were analyzed for chromosome abnormalities, including chromosome end-to-end fusion and telomere signal-free ends (SFEs).
Chromosome Orientation FISH (CO-FISH) and SCE. CO-FISH and SCE have been described in detail (54-56), and similar protocols were used here with some modification. Briefly, ES cells or splenocytes were subcultured in medium containing a 3:1 ratio of BrdUrd/BrdC (Sigma) at a final concentration of 1 × 10-5 M and collected at either 12 (ES cells) or 24 (splenocytes) hours for detecting T-SCE or two doubling times for detecting SCE or interspersed G+C tracts. Colcemid (0.1 μg/ml) was added for the final 4 h. Metaphase spreads were then stained with Hoechst dye 33258, exposed to UV light, and digested with exonuclease III to remove newly synthesized DNA strands as performed in the previous protocols. Hybridization and wash conditions were identical to those described for telomeric FISH (5, 59). A chromosome with more than two telomeric DNA signals was scored as T-SCE positive. A SCE was scored each time a color switch between dark or light sister chromatids occurred. Approximately 100 metaphases from each sample were analyzed for T-SCE or SCE, and ≈50 metaphases from each sample were analyzed for interspersed G+C tracts.
Statistical analysis was performed by using a χ2 test with 1 df. Resulting P values were compared to an α of 0.05. Only those comparisons that showed an increase over the control were evaluated for significance with a one-sided hypothesis test.
Results and Discussion
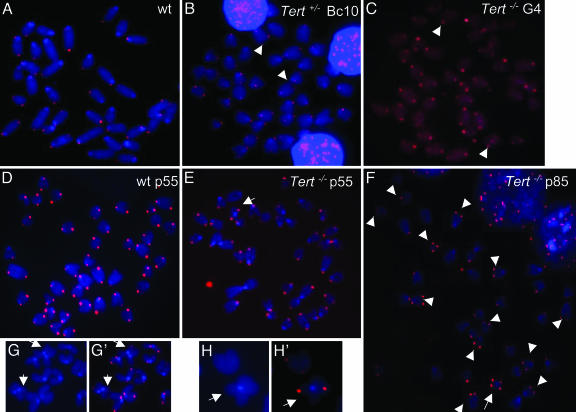
Multiple SFEs, Not Few SFEs, Are More Permissive for T-SCE in mTert-/- ES Cells but Not in mTert-/- Mice. Recent studies have indicated that a specific type of telomere recombination, called T-SCE, is involved in telomere length maintenance in telomerase negative cells (54-56). To understand the mechanism of T-SCE in telomere length maintenance in different experimental systems, we examined the frequencies of T-SCEs in mTert-/- mice and in vitro cultured ES cells with critically short telomeres. To determine the frequency of T-SCEs in mice, we examined splenocytes from wild-type and different generations of mTert-/- mice. We found no detectable increase in T-SCE in wild-type or early generations of mTert-/- mice, but only a slight increase in T-SCEs in G4 mTert-/- mice that possessed multiple SFEs (Table 1 and Fig. 1). Although this result may accurately reflect the occurrence of T-SCEs in vivo, it is possible that the frequencies of T-SCEs is underestimated because of undetectable telomeric DNA signal at very short telomeres. The latter possibility seems unlikely, as one splenocyte from a G4 mTert-/- mouse possessed extremely weak telomeric fluorescent signals and multiple SFEs, yet showed several chromosome ends with T-SCEs (Fig. 2 and Table 4, which are published as supporting information on the PNAS web site). It is noteworthy that data compiled from 54 of 100 G4 mTert-/-, 40 of 100 G2 mTert-/-, or 38 of 100 wild-type splenocytes showed a low rate of T-SCEs (less than two chromosome ends are positive for T-SCEs) (Table 4). Thus, an increase in overall frequency of T-SCEs in G4 mTert-/- splenocytes was not remarkably significant compared to wild-type splenocytes (Tables 1 and 4).
Table 1. Chromosome abnormalities and T-SCEs in murine splenocytes derived from wild-type and early or late generations of mTert heterozygous and null mice.
| Cell type | End-to-end fusions per cell | % (no. of SFEs/chromosomes) | % (no. of T-SCEs/chromosomes) |
|---|---|---|---|
| Wild type | 0 | 0.1 (4/3,999)* | 1.1 (44/3,995) |
| mTert+/– | |||
| Bc10 | 0 | 0 (1/4,001) | 1.1 (45/3,990) |
| Bc11 | 0 | 0.1 (4/3,988) | 1.2 (48/3,998) |
| mTert-/- | |||
| G2 | 0 | 0.4 (16/3,993) | 1.3 (52/4,001) |
| G3 | 0 | ND | 1.3 (51/3,990) |
| G4 | 0 | 17.9 (700/3,920) | 1.9 (74/3,960) |
ND, not determined.
Each chromosome was counted for two ends
Fig. 1.
CO-FISH analysis of representative metaphase spreads of murine splenocytes (A-C) and different passages of ES cells (D-H). A wild-type (wt) murine splenocyte (A) showed normal CO-FISH pattern with two typical telomeric DNA signals. No T-SCE was found in this cell. One mTert+/- backcross generation 10 (Tert+/- BC10) (B) and one G4 mTert-/- (Tert-/- G4) splenocyte (C) showed a low rate of T-SCEs. At passage 55, a wild-type (D) or mTert-/- (E) ES cell showed a normal CO-FISH pattern. At passage 85, an mTert-/- ES cell (F) had multiple SFEs and T-SCEs. T-SCE-positive chromosome ends had stronger telomeric DNA signals than T-SCE negative chromosome ends in this ES cell. (G and H) Enlarged versions of telomere association and end-to-end fusion. Arrowhead, chromosomes with T-SCEs (more than two telomeric signals/chromosome); arrow, end-to-end fusion. mTert-/- G4 splenocyte and ES cells at p85 were overexposed to intensify the weak telomeric fluorescent signals.
We showed previously that mTert-/- ES cells undergo progressive telomere loss and an increase in SFEs during prolonged growth in culture, whereas wild-type ES cells, unlike other murine primary cells, gradually gain telomeric DNA (15). Thus, we examined the frequency of T-SCEs in wild-type and mTert-/- ES cells at early and late passages. We found no detectable increase in T-SCEs in wild type, early passages of mTert-/- ES cells, or mTert-/- ES cells at the earliest passages in which few SFE and end-to-end fusions were observed (Table 2 and Fig. 1E). However a significant increase (P < 0.01) in T-SCEs was observed in late passage of mTert-/- ES cells (p85, 8.7%) that harbored multiple SFEs in comparison with early passage of mTert-/- ES cells (p5, 1.7%), and T-SCEs often appeared at multiple chromosome ends (Table 2 and Fig. 1F). Furthermore, cell populations with a high frequency of T-SCEs increased dramatically in late passage (p85: 18%) in comparison with early passage (p5; 3.1%) of mTert-/- ES cells (Table 4). Interestingly, the chromosome ends with T-SCEs often had robust telomeric DNA signal intensity (Fig. 1F). No growth arrest was yet observed in these cells.
Table 2. Frequencies of chromosome end-to-end fusions, SFEs, and T-SCEs in early or late passages of wild-type, mTert heterozygous, and null ES cells.
| Cell type | End-to-end fusions per cell | % (no. of SFEs/chromosomes)* | % (no. of T-SCEs/chromosomes) |
|---|---|---|---|
| Wild type | |||
| p5 | 0 | 0 | 1.6 (66/4,022) |
| p55 | 0 | 0 | 2.0 (79/3,854) |
| p85 | 0 | 0 | 1 (38/3,846) |
| mTert+/– | |||
| p5 | 0 | 0 | 2.2 (76/3,512) |
| p55 | 0 | 0 | ND |
| p85 | 0 | 0 | 0.9 (32/3,699) |
| p150 | 0 | 0 | 0.9 (34/3,908) |
| mTert-/- | |||
| p5 | 0 | 0 | 1.7 (63/3,638) |
| p55 | 0.5 | 7.4 (290/3,896) | 2.1 (83/3,896) |
| p85 | 2.9 | 20.2 (802/3,964) | 8.7 (346/3,964)† |
ND, not determined.
The frequencies of SFEs was adopted from published work (15), except data from mTert+/– p150 and mTert+/– p55 and p85
The rates of T-SCEs between mTert-/- p85 and p5 are statistically different (P < 0.01)
Our data suggest that T-SCE does not appear to be an immediate by-product of SFEs, because earlier generations of mTert-/- mice and the earliest passages of mTert-/- ES cells that possess SFEs do not possess a significant increase in T-SCE events. Although nearly every G4 mTert-/- splenocyte or mTert-/- ES cell at passage 85 had a roughly similar frequency of multiple SFEs (18% vs. 20%), these cells had very different T-SCEs (1.9% vs. 8.7%) (Table 1 and 2). These data clearly indicate that T-SCEs do not follow automatically from SFEs. Furthermore, an increase in T-SCEs was only observed in a subset of cells and/or at chromosome ends with SFEs (Figs. 1F and 2 and Table 4). We propose that the delayed and incomplete penetrance of T-SCEs relative to the appearance of SFEs is not merely due to culling of T-SCE positive cells from the population at earlier generations/passages, because T-SCEs become readily detected in G4 mTert-/- mice or in late passages of mTert-/- ES cells.
T-SCEs were more frequent in late passage of mTert-/- ES cells (Table 2, 8.7%) than in G4 mTert-/- mice (Table 1, 1.9%). Furthermore, a high rate of T-SCEs (multiple chromosome ends are positive for T-SCEs) was found in 18% of mTert-/- ES cells (p85), but in only 1% of G4 mTert-/- splenocytes (Table 4). Thus, critically short telomeres may be more permissive for T-SCE in long-term cultured ES cells than in mice, or there may be cell-type specificity in the propensity to undergo T-SCE in response to SFE.
Our observations support previously proposed models and observations that multiple critically short telomeres may activate telomerase-independent telomere length maintenance in human or yeast cells during prolonged growth in culture (18, 54, 60-62). In line with this notion, we demonstrated that the chromosome ends with T-SCE indeed retained telomeric DNA signals (Figs. 1F and 2), indicating that telomere length may be maintained through T-SCE in these instances. Alternatively, critically short telomeres could be maintained via a different mechanism through which T-SCE occurs. Recent data suggested that frequent t-loop deletion by homologous recombination could promote rolling-circle replication of telomeres in the absence of telomerase in human ALT cells (38). It is possible that, in these subpopulations of cells, critically short telomeres are selectively elongated via a rolling circle, which then allows for T-SCE (38).
Telomere Length Maintenance in mTert Heterozygous Splenocytes or ES Cells Is T-SCE-Independent. mTert heterozygous splenocytes or ES cells exhibit a decrease in average length of telomeres, but no increase in SFEs or end-to-end fusions (12, 15). We also examined the frequency of T-SCEs in these cell types and observed no detectable increase in T-SCEs in splenocytes of successively backcrossed mTert+/- mice at generations 10 and 11 (BC10 and BC11) or in early or late passage of mTert+/- ES cells (Tables 1 and 2 and Fig. 1B).
Our results suggest that T-SCE does not participate in the telomere length maintenance of mTert+/- mice or ES cells with average short telomeres. One functional allele of mTert may be directly responsible for the inhibition of T-SCE or its ability to maintain telomere integrity at critically short telomeres in mTert+/- mice and ES cells may inhibit T-SCE. This notion is supported indirectly by Bechter et al. (55), who report that a high frequency of T-SCEs arises in a subpopulation of human colon cancer cells upon telomerase inhibition. T-SCE was completely abolished when cells escaped from crisis due to the induction of telomerase (55).
An Increase in Other Genomic Rearrangements in mTert-/- ES Cells, but Not in Mice, May Help to Maintain or Protect Eroded Telomeres of Long-Term Cultured Cells. In addition to T-SCE, other genomic rearrangements, e.g., SCE or nonhomologous end joining (including chromosome end-to-end fusion), may also help to mask unprotected chromosome ends with eroded telomeres. Some of these genomic rearrangements could account for an increase in T-SCEs observed in G4 mTert-/- splenocytes or late passage of mTert-/- ES cells. For example, multiple SFEs in these cell types may represent DNA damage and trigger genomic SCE, which could be detected by CO-FISH as T-SCE. Furthermore, extrachromosomal G-rich telomeric DNA fragments may serve as substrates for nonhomologous end joining, resulting in telomeric repeat tracts being ligated to unprotected chromosome ends. This event would create a C-rich strand with interspersed G-rich or C-rich telomeric DNA (interspersed G+C), which can be detected by CO-FISH using a C-rich telomeric DNA probe (56, 63). Thus, T-SCE could be a by-product of SCE or nonhomologous end joining of extrachromosomal telomeric fragments, especially in mTert-/- ES cells or mice with multiple SFEs.
Therefore, we analyzed the frequency of SCEs and interspersed G+C tracts in wild-type and G4 mTert-/- splenocytes, and in early and late passages of mTert-/- ES cells, after two rounds of BrdUrd/BrdC incorporation (Table 3 and Fig. 3, which is published as supporting information on the PNAS web site). A slight but not significant increase in SCEs or interspersed G+C was detected in G4 mTert-/- splenocytes (Table 3). Thus, these genomic rearrangements would be unlikely to contribute to the frequencies of T-SCEs observed in G4 mTert-/- splenocytes. A slight increase in SCEs was also reported in late generations of telomerase RNA deficient murine splenocytes (64). However, an increase in SCEs or interspersed G+C was readily detected in late passage of mTert-/- ES cells (p85) that possessed multiple SFEs (Tables 2 and 3). Taking a perhaps unsupported presumption, we subtracted the events of both SCEs and interspersed G+C tracts from the events of T-SCEs; after this subtraction, the occurrence of T-SCEs remained significantly higher in late passage of mTert-/- ES cells (p85) than early passage of mTert-/- ES cells (p5) (Table 3, P < 0.01).
Table 3. Frequencies of T-SCEs, SCEs, and interspersed G + C tracts in early and late generations or passages of mTert-/- splenocytes and ES cells.
| Cell type | % (no. of T-SCEs/chromosomes) | % (no. of SCEs/chromosomes) | % (no. of interspersed G + C tracts/chromosomes) |
|---|---|---|---|
| Splenocytes | |||
| BL6 | 1.1 (44/3,995) | 28.1 (1,127/4,010) | 0.2 (4/2,004) |
| G4 | 1.9 (74/3,960) | 29.4 (1,172/3,987) | 0.3 (6/2,019) |
| ES cells | |||
| WT p5 | 1.6 (66/4,022) | 42.9 (1,735/4,049) | 0 (0/2,014) |
| WT p85 | 1.0 (38/3,846) | 35.0 (1,361/3,888) | 0.9 (18/1,943) |
| mTert-/- p5 | 1.7 (63/3,638) | 33.1 (1,225/3,701) | 0.9 (17/1,855) |
| mTert-/- p85 | 8.7 (346/3,964)* | 44.0 (1,756/3,992) | 2.6 (52/2,001) |
Cells were incubated with BrdUrd/BrdC for one (first column) or two (second column and third column) rounds of population doublings. After removing influential factors (SCE and interspersed G + C tracts), the rate of T-SCEs was 4.8 times higher in mTert-/- ES cells p85 than mTert-/- ES cells p5 (P < 0.01).
P < 0.01
Previously, Erdmann et al. (12) described that splenocytes derived from G4 mTert-/- mice showed a statistically significant increase in SFEs, but only in certain sibling crosses was a significant increase in end-to-end fusions observed in mTert-/- splenocytes with critically shortened telomeres. However, end-to-end chromosome fusions were readily detected in mTert-/- ES cells that possessed SFEs (15). Consistent with previous observations in both murine splenocytes and ES cells (12, 15), no chromosome end-to-end fusions were detected in late generations of mTert-/- mice, whereas an increase in end-to-end fusions was readily detected in mTert-/- ES cells during prolonged growth in culture (Tables 1 and 2 and Fig. 4, which is published as supporting information on the PNAS web site). Furthermore, end-to-end fusions were found in mTert-/- ES cells even with few SFEs (Fig. 4 D and F-F″).
It is interesting that genomic rearrangements, i.e., SCE or nonhomologous end joining (including chromosome end-to-end fusion), are readily detected only in long-term culture ES cells, but not in mTert-/- splenocytes, possessing multiple SFEs. This observation suggests that these genomic rearrangements may be triggered by SFEs in a cell-type-dependent manner or, specifically, in cultured cells but not in mice. These rearrangements could protect or maintain critically short telomeres, thus conferring prolonged viability to germ cells (or long-term cultured cells).
In summary, we found that T-SCE and other genomic rearrangements (i.e., SCE or nonhomologous end joining) are more evident in long-term cultured ES cells than in splenocytes derived from mice, even though both had roughly similar frequencies of SFEs. Moreover, T-SCEs are not concomitant events with SFEs, because mTert-/- mice or mTert-/- ES cells that possess few SFE do not possess a significant increase in T-SCE events, and an increase in T-SCEs was only observed in a subset of cells and/or at chromosome ends with multiple SFEs. In addition, several other types of genomic rearrangements were observed in late passage of mTert-/- ES cells, suggesting that critically short telomeres can lead to, directly or indirectly, other forms of genomic instability. Thus, it is likely that a combination of different genomic rearrangements (i.e., T-SCE, SCE, and nonhomologous end joining) in late passage of mTert-/- ES cells may protect chromosome ends with eroded telomeres, and thus cell viability, during prolonged cellular growth in culture.
Supplementary Material
Acknowledgments
We sincerely thank Dr. Lea Harrington (Ontario Cancer Institute, Toronto), in whose laboratory the mouse breeding and subsequent isolation, activation, and BrdUrd labeling of murine splenocytes were carried out, for providing reagents and critical comments during this study; Dr. Liane Russell and Ms. Darla Miller for their critical reading of the manuscript; Ms. Cecilia Wang for assisting in the image analysis of FISH; and Dr. Susan Bailey (Colorado State University, Boulder) for generously supplying advice and protocols pertaining to CO-FISH and SCE. We acknowledge the support of the office of Biological and Environmental Research, U.S. Department of Energy under Contract DE-AC056-960R22464 with UT-Battelle LLC, and N.E. acknowledges the support of the National Institute of Health Grant AG16629-03.
Author contributions: Y.L. designed research; Y.W., N.E., R.J.G., J.W., M.G., and Y.L. performed research; Y.W. and Y.L. contributed new reagents/analytic tools; Y.W. and Y.L. analyzed data; and Y.L. wrote the paper.
Abbreviations: ALT, alternative lengthening of telomeres; SCE, sister chromatid exchange; T-SCE, telomere SCE; SFE, signal-free end; CO-FISH, chromosome orientation FISH; BC, back cross; pn, passage n; Gn, generation n.
References
- 1.Greider, C. W. (1996) Annu. Rev. Biochem. 65, 337-365. [DOI] [PubMed] [Google Scholar]
- 2.Klapper, W., Parwaresch, R. & Krupp, G. (2001) Mech. Ageing Dev. 122, 695-712. [DOI] [PubMed] [Google Scholar]
- 3.Djojosubroto, M. W., Choi, Y. S., Lee, H. W. & Rudolph, K. L. (2003) Mol. Cells 15, 164-175. [PubMed] [Google Scholar]
- 4.Karlseder, J. (2003) Cancer Lett. 194, 189-197. [DOI] [PubMed] [Google Scholar]
- 5.Blasco, M. A., Lee, H. W., Hande, M. P., Samper, E., Lansdorp, P. M., DePinho, R. A. & Greider, C. W. (1997) Cell 91, 25-34. [DOI] [PubMed] [Google Scholar]
- 6.Lee, H. W., Blasco, M. A., Gottlieb, G. J., Horner, J. W., Jr., Greider, C. W. & DePinho, R. A. (1998) Nature 392, 569-574. [DOI] [PubMed] [Google Scholar]
- 7.Herrera, E., Samper, E., Martin-Caballero, J., Flores, J. M., Lee, H. W. & Blasco, M. A. (1999) EMBO J. 18, 2950-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rudolph, K. L., Chang, S., Lee, H. W., Blasco, M., Gottlieb, G. J., Greider, C. & DePinho, R. A. (1999) Cell 96, 701-712. [DOI] [PubMed] [Google Scholar]
- 9.Yuan, X., Ishibashi, S., Hatakeyama, S., Saito, M., Nakayama, J., Nikaido, R., Haruyama, T., Watanabe, Y., Iwata, H., Iida, M., et al. (1999) Genes Cells 4, 563-572. [DOI] [PubMed] [Google Scholar]
- 10.Liu, Y., Snow, B. E., Hande, M. P., Yeung, D., Erdmann, N. J., Wakeham, A., Itie, A., Siderovski, D. P., Lansdorp, P. M., Robinson, M. O. & Harrington, L. (2000) Curr. Biol. 10, 1459-1462. [DOI] [PubMed] [Google Scholar]
- 11.Hemann, M. T., Strong, M. A., Hao, L. Y. & Greider, C. W. (2001) Cell 107, 67-77. [DOI] [PubMed] [Google Scholar]
- 12.Erdmann, N., Liu, Y. & Harrington, L. (2004) Proc. Natl. Acad. Sci. USA 101, 6080-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samper, E., Flores, J. M. & Blasco, M. A. (2001) EMBO Rep. 2, 800-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niida, H., Matsumoto, T., Satoh, H., Shiwa, M., Tokutake, Y., Furuichi, Y. & Shinkai, Y. (1998) Nat. Genet. 19, 203-206. [DOI] [PubMed] [Google Scholar]
- 15.Liu, Y., Kha, H., Ungrin, M., Robinson, M. O. & Harrington, L. (2002) Proc. Natl. Acad. Sci. USA 99, 3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niida, H., Shinkai, Y., Hande, M. P., Matsumoto, T., Takehara, S., Tachibana, M., Oshimura, M., Lansdorp, P. M. & Furuichi, Y. (2000) Mol. Cell. Biol. 20, 4115-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundblad, V. & Szostak, J. W. (1989) Cell 57, 633-643. [DOI] [PubMed] [Google Scholar]
- 18.Lundblad, V. & Blackburn, E. H. (1993) Cell 73, 347-360. [DOI] [PubMed] [Google Scholar]
- 19.Le, S., Moore, J. K., Haber, J. E. & Greider, C. W. (1999) Genetics 152, 143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, Q., Ijpma, A. & Greider, C. W. (2001) Mol. Cell. Biol. 21, 1819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackett, J. A., Feldser, D. M. & Greider, C. W. (2001) Cell 106, 275-286. [DOI] [PubMed] [Google Scholar]
- 22.Lundblad, V. (2002) Oncogene 21, 522-531. [DOI] [PubMed] [Google Scholar]
- 23.IJpma, A. S. & Greider, C. W. (2003) Mol. Biol. Cell 14, 987-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maringele, L. & Lydall, D. (2004) Genes Dev. 18, 2663-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maringele, L. & Lydall, D. (2004) Genetics 166, 1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riha, K., McKnight, T. D., Griffing, L. R. & Shippen, D. E. (2001) Science 291, 1797-1800. [DOI] [PubMed] [Google Scholar]
- 27.Riha, K. & Shippen, D. E. (2003) Chromosome Res. 11, 263-275. [DOI] [PubMed] [Google Scholar]
- 28.Espejel, S., Franco, S., Sgura, A., Gae, D., Bailey, S. M., Taccioli, G. E. & Blasco, M. A. (2002) EMBO J. 21, 6275-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Espejel, S., Franco, S., Rodriguez-Perales, S., Bouffler, S. D., Cigudosa, J. C. & Blasco, M. A. (2002) EMBO J. 21, 2207-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karlseder, J., Smogorzewska, A. & de Lange, T. (2002) Science 295, 2446-2449. [DOI] [PubMed] [Google Scholar]
- 31.d'Adda di Fagagna, F., Reaper, P. M., Clay-Farrace, L., Fiegler, H., Carr, P., Von Zglinicki, T., Saretzki, G., Carter, N. P. & Jackson, S. P. (2003) Nature 426, 194-198. [DOI] [PubMed] [Google Scholar]
- 32.Takai, H., Smogorzewska, A. & de Lange, T. (2003) Curr. Biol. 13, 1549-1556. [DOI] [PubMed] [Google Scholar]
- 33.d'Adda di Fagagna, F., Teo, S. H. & Jackson, S. P. (2004) Genes Dev. 18, 1781-1799. [DOI] [PubMed] [Google Scholar]
- 34.Espejel, S., Klatt, P., Menissier-de Murcia, J., Martin-Caballero, J., Flores, J. M., Taccioli, G., de Murcia, G. & Blasco, M. A. (2004) J. Cell Biol. 167, 627-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hao, L. Y., Strong, M. A. & Greider, C. W. (2004) J. Biol. Chem. 279, 45148-45154. [DOI] [PubMed] [Google Scholar]
- 36.Karlseder, J., Hoke, K., Mirzoeva, O. K., Bakkenist, C., Kastan, M. B., Petrini, J. H. & de Lange, T. (2004) PLoS Biol. 2, E240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarsounas, M., Munoz, P., Claas, A., Smiraldo, P. G., Pittman, D. L., Blasco, M. A. & West, S. C. (2004) Cell 117, 337-347. [DOI] [PubMed] [Google Scholar]
- 38.Wang, R. C., Smogorzewska, A. & de Lange, T. (2004) Cell 119, 355-368. [DOI] [PubMed] [Google Scholar]
- 39.Bradshaw, P. S., Stavropoulos, D. J. & Meyn, M. S. (2005) Nat. Genet. 37, 193-197. [DOI] [PubMed] [Google Scholar]
- 40.Wright, W. E. & Shay, J. W. (2005) Nat. Genet. 37, 116-118. [DOI] [PubMed] [Google Scholar]
- 41.Bailey, S. M., Meyne, J., Chen, D. J., Kurimasa, A., Li, G. C., Lehnert, B. E. & Goodwin, E. H. (1999) Proc. Natl. Acad. Sci. USA 96, 14899-14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilley, D., Tanaka, H., Hande, M. P., Kurimasa, A., Li, G. C., Oshimura, M. & Chen, D. J. (2001) Proc. Natl. Acad. Sci. USA 98, 15084-15088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu, H. L., Gilley, D., Galande, S. A., Hande, M. P., Allen, B., Kim, S. H., Li, G. C., Campisi, J., Kohwi-Shigematsu, T. & Chen, D. J. (2000) Genes Dev. 14, 2807-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smogorzewska, A. & de Lange, T. (2004) Annu. Rev. Biochem. 73, 177-208. [DOI] [PubMed] [Google Scholar]
- 45.Bryan, T. M., Englezou, A., Gupta, J., Bacchetti, S. & Reddel, R. R. (1995) EMBO J. 14, 4240-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bryan, T. M., Englezou, A., Dalla-Pozza, L., Dunham, M. A. & Reddel, R. R. (1997) Nat. Med. 3, 1271-1274. [DOI] [PubMed] [Google Scholar]
- 47.Reddel, R. R., Bryan, T. M. & Murnane, J. P. (1997) Biochemistry (Moscow) 62, 1254-1262. [PubMed] [Google Scholar]
- 48.Henson, J. D., Neumann, A. A., Yeager, T. R. & Reddel, R. R. (2002) Oncogene 21, 598-610. [DOI] [PubMed] [Google Scholar]
- 49.Yeager, T. R., Neumann, A. A., Englezou, A., Huschtscha, L. I., Noble, J. R. & Reddel, R. R. (1999) Cancer Res. 59, 4175-4179. [PubMed] [Google Scholar]
- 50.Wu, G., Jiang, X., Lee, W. H. & Chen, P. L. (2003) Cancer Res. 63, 2589-2595. [PubMed] [Google Scholar]
- 51.Nabetani, A., Yokoyama, O. & Ishikawa, F. (2004) J. Biol. Chem. 279, 25849-25857. [DOI] [PubMed] [Google Scholar]
- 52.Dunham, M. A., Neumann, A. A., Fasching, C. L. & Reddel, R. R. (2000) Nat. Genet. 26, 447-450. [DOI] [PubMed] [Google Scholar]
- 53.Varley, H., Pickett, H. A., Foxon, J. L., Reddel, R. R. & Royle, N. J. (2002) Nat. Genet. 30, 301-305. [DOI] [PubMed] [Google Scholar]
- 54.Bailey, S. M., Brenneman, M. A. & Goodwin, E. H. (2004) Nucleic Acids Res. 32, 3743-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bechter, O. E., Zou, Y., Walker, W., Wright, W. E. & Shay, J. W. (2004) Cancer Res. 64, 3444-3451. [DOI] [PubMed] [Google Scholar]
- 56.Londono-Vallejo, J. A., Der-Sarkissian, H., Cazes, L., Bacchetti, S. & Reddel, R. R. (2004) Cancer Res. 64, 2324-2327. [DOI] [PubMed] [Google Scholar]
- 57.Hakem, R., de la Pompa, J. L., Sirard, C., Mo, R., Woo, M., Hakem, A., Wakeham, A., Potter, J., Reitmair, A., Billia, F., et al. (1996) Cell 85, 1009-1023. [DOI] [PubMed] [Google Scholar]
- 58.Salmena, L., Lemmers, B., Hakem, A., Matysiak-Zablocki, E., Murakami, K., Au, P. Y., Berry, D. M., Tamblyn, L., Shehabeldin, A., Migon, E., et al. (2003) Genes Dev. 17, 883-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zijlmans, J. M., Martens, U. M., Poon, S. S., Raap, A. K., Tanke, H. J., Ward, R. K. & Lansdorp, P. M. (1997) Proc. Natl. Acad. Sci. USA 94, 7423-7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rizki, A. & Lundblad, V. (2001) Nature 411, 713-716. [DOI] [PubMed] [Google Scholar]
- 61.McEachern, M. J. & Iyer, S. (2001) Mol. Cell 7, 695-704. [DOI] [PubMed] [Google Scholar]
- 62.Shay, J. W. & Wright, W. E. (2005) Carcinogenesis 26, 867-874. [DOI] [PubMed] [Google Scholar]
- 63.Bailey, S. M., Cornforth, M. N., Ullrich, R. L. & Goodwin, E. H. (2004) DNA Repair 3, 349-357. [DOI] [PubMed] [Google Scholar]
- 64.Goytisolo, F. A., Samper, E., Martin-Caballero, J., Finnon, P., Herrera, E., Flores, J. M., Bouffler, S. D. & Blasco, M. A. (2000) J. Exp. Med. 192, 1625-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.