Abstract
Background
Campylobacter infections pose a significant challenge in low- and middle-income countries, contributing to child mortality. Campylobacter is linked to acute gastrointestinal illness and severe long-term consequences, including environmental enteric dysfunction (EED) and stunting. In 2018, our cross-sectional study in Ethiopia detected Campylobacter in 88% of stools from children aged 12–15 months, with an average of 11 species per stool using meta-total RNA sequencing. Building on these findings, we conducted a longitudinal study (December 2020–June 2022) to investigate Campylobacter colonization of infants and identify reservoirs and risk factors in rural eastern Ethiopia.
Results
After a preliminary screening of 15 Campylobacter species using species-specific quantitative PCR, we analyzed four target species in 2,045 samples from infants (first month to just one year of life) and biannual samples from mothers, siblings, and livestock (goats, cattle, sheep, and chickens). Candidatus C. infans (41%), C. jejuni (26%), and C. upsaliensis (13%) were identified as the predominant in the infant gut. Colonization of C. infans and C.jejuni increased (C. infans: 0.85%, C. jejuni-0.98% increase/ day in the odds of colonization) and abundance (P = 0.027, 0.024) with age. Enteric symptoms were strongly associated with C. infans (diarrhea: OR = 2.02 [95%CI: 35%,100%]; fever: OR = 1.62 [95%CI: 14%, 83%]) and C. jejuni (diarrhea: OR = 2.29 [95%CI: 46%,100%], fever: OR = 2.53 [95%CI: 56%,100%]). Based on linear mixed models, we found elevated cumulative loads of C. infans load in infants (especially females OR = 1.5 [95%CI: 10%, 67%]), consuming raw milk (OR = 2.3 [95%CI: 24%,100%]) or those exposed to areas contaminated with animal droppings (OR = 1.6 [95%CI: 7%,93%]), while C. jejuni cumulative loads were higher in infants ingesting soil or animal feces (OR = 2.2 [95%CI: 23%,100%]). C. infans was also prevalent in siblings (56%) and mothers (45%), whereas C. jejuni was common in chickens (38%) and small ruminants (goats 27%, sheep 21%).
Conclusions
Campylobacter was highly prevalent in rural Ethiopian infants. C. infans was primarily associated with human hosts, and C. jejuni was mainly linked to zoonotic sources. Our findings emphasize the need for targeted interventions addressing environmental, dietary, and behavioral factors to reduce Campylobacter transmission in resource-limited settings.
Keywords: Campylobacter, C. infans, C. jejuni, C. upsaliensis, qPCR, longitudinal study, eastern Ethiopia, infant stool, household determinants, environmental enteric dysfunction (EED)
Background
Campylobacter infections represent a substantial global health concern. They have been identified as a leading cause of bacterial gastroenteritis, resulting in 166 million cases of diarrhea globally in 2020, of which 96 million were foodborne (1). While most cases are self-limiting, the potential for severe, even life-threatening outcomes cannot be overlooked, particularly in infants, the elderly, and those with weakened immune systems (2).
Campylobacter infections in high- and middle-income countries are mainly foodborne (3), but other pathways are more important in settings with poor sanitation and hygiene infrastructure (4), contributing significantly to the disease spread. Beyond acute gastroenteritis, these infections have been linked to long-term gastrointestinal health issues, particularly in pediatric populations where the early-life gut microbiome is being established. This critical period is marked by rapid changes in the intestinal microbial community driven by diet and environment and generally stabilizes by three years of age (5). Research from the Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development (MAL-ED) project has associated Campylobacter infections with adverse health outcomes such as Environmental Enteric Dysfunction (EED) leading to compromised nutrient absorption, weakened immunity, and stunting (6–8). These early-life growth patterns are crucial indicators of nutritional status, with significant implications for future health outcomes, including mortality, chronic disease, neurodevelopment, and economic productivity in later life (7–10).
In 2018, a formative cross-sectional study in rural eastern Ethiopia conducted under the CAGED project’s umbrella (Campylobacter Genomics and Environmental Enteric Dysfunction) unveiled that the prevalence of Campylobacter at the genus level was 88% in stool samples from children under two years of age (11). Meta-total RNA sequencing revealed an average of 11 distinct Campylobacter species in positive stool samples from children, heightening concerns about their potential role in chronic outcomes such as EED and stunting (12).
Although by November 2024, 49 species have been validly published within the Campylobacter genus (https://lpsn.dsmz.de/genus/campylobacter), existing research disproportionately concentrates on thermophilic Campylobacter jejuni and Campylobacter coli (13). C. jejuni is the dominant diarrhea-associated species worldwide, with C. coli contributing 1–25% to Campylobacter-related gastroenteritis cases (14, 15). These species, while dominant, are not exclusive agents of Campylobacter-related gastroenteritis (16, 17). Moreover, the MAL-ED study revealed that when detecting all Campylobacter species using immunoassays, the impact on stunting was greater than when looking at just C. jejuni and C. coli alone (7–9).
Most data on Campylobacter epidemiology are from high-income countries and it remains understudied in low- and middle-income countries, particularly the non-thermophilic species abundant in vulnerable populations like Ethiopian children enrolled in the CAGED formative study (11). However, advancements in isolation and detection techniques have shed light on other emerging Campylobacter species, such as C. upsaliensis, C. lari, and C. hyointestinalis, due to their increased association with human illness (18). In addition, recent studies have described a new species Candidatus C. infans (C. infans from hereon), originally detected in infant stools from the Global Enteric Multicenter Study (GEMS) study(19). This species was detected by shotgun metagenomic sequencing in 59.1% of fecal samples from diarrheal and asymptomatic children under two years of age in Iquitos, Loreto, Peru. These samples were previously tested positive for the Campylobacter genus but negative for C. jejuni/coli by PCR. C. infans was identified as the dominant species in breastfed infants and is associated with diarrhea in humans and non-human primates (18). Furthermore, most studies have focused exclusively on human or animal hosts; however, the role of environmental factors, including soil and water, in Campylobacter transmission remains less understood.
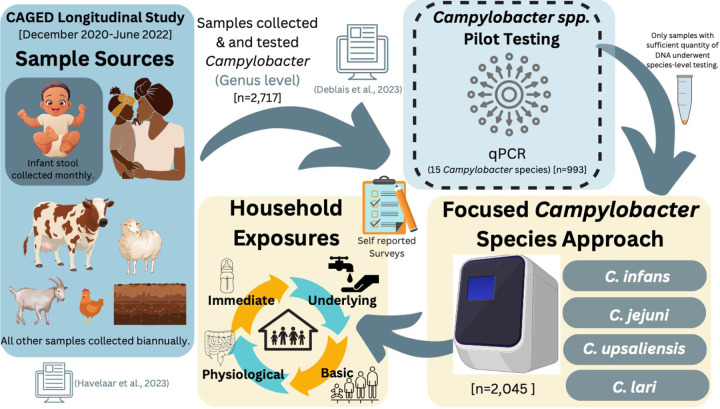
Building on these insights, a longitudinal study was conducted from December 2020 to June 2022 aimed at assessing the prevalence, species composition, and genomic diversity of thermotolerant and non-thermotolerant Campylobacter spp. in infants, adults, livestock, and environmental reservoirs in the Haramaya woreda in Eastern Ethiopia. The details of the study design have been previously described (20, 21). A high prevalence of Campylobacter at the genus level was observed, with 64% of stool samples from infants testing positive, with an age-dependent prevalence, as determined through TaqMan real-time PCR analysis (21). This study seeks to elucidate the temporal colonization patterns of Campylobacter at the species level in infants and the potential reservoirs, [humans (siblings, mothers), livestock (cattle, sheep, goats, chickens), the environment (soil samples collected at the front and inside the home)], and household determinants contributing to their infection. Using species-specific quantitative PCR, we assessed the prevalence and load of both thermotolerant and non-thermotolerant Campylobacter at the species level across these various sample types and conducted risk-factor analysis using metadata obtained through extensive face-to-face interviews (22).
Methods
Study design, sample size, sample collection, and interviews
A longitudinal study involving 106 infants was conducted. Participants were randomly selected from a birth registry in 10 kebeles (the smallest administrative unit in Ethiopia) in the Haramaya woreda, East Hararghe Zone, Oromia Region, Ethiopia (Fig. S1). Informed consent was obtained from parents in the local language (Afan Oromo). An average of 205 (minimum/maximum, 170/249) samples tested at a species level were collected per kebele. Each kebele included, on average, 11 households (minimum/maximum, 8/12), and an average of 20 (minimum/maximum, 4/28) samples per household were collected (Table S1). Infant stool samples (n = 907) were collected monthly, starting from enrollment (16–39 days after birth) until 353–375 days of age. Stools from the mother and closest-in-age sibling (n = 121 and 129) along with livestock (cattle, goat, sheep, chicken) feces (n = 640) and soil (n = 248) samples were collected twice in the first and second half year of life of the infants, (Fig. 1). Surveys were conducted during sampling to assess dietary intake and additional environmental factors. Further details on enrollment, sampling scheme, sample processing, and occurrence of Campylobacter at the genus level and determinants of infant colonization were described in previous publications (20–22).
Figure 1.
Flowchart Illustrating the Analysis Methods for the CAGED Longitudinal Cohort and Campylobacter Species Detection, Including the Investigation of Household Exposures Linked to Elevated Campylobacter Species Infections.
Detection of Campylobacter species using real-time quantitative PCR
Sample processing and DNA extraction were performed in Ethiopia using the QIAamp PowerFecal Pro kit (Qiagen, CA, USA) for human stool and livestock feces, and the DNase PowerSoil Pro DNA kit (Qiagen, Hilden, Germany) was used for soil samples as detailed in (21). The quality and quantity of DNA were analyzed using a UV5 Nano spectrophotometer, and poor-quality samples (260/230 ratio of < 1.0; 260/280 ratio of < 1.6) were cleaned using a Zymo genomic DNA clean and concentrator kit. DNA was shipped to the United States to detect Campylobacter species using SYBR Green quantitative PCR. Species-specific primers targeting hipO and cpn60 were used (Table 1). Each reaction mixture (25 µl final volume) contained 12.5 µl of Maxima SYBR Green/ROX qPCR Master Mix (2X), 0.5 nmol of forward and reverse primers, 50 ng of DNA, and nuclease-free water (Qiagen, Hilden, Germany). Real-time PCR runs were conducted using QuantStudio 5 (Applied Biosystems, Waltham, MA).
Table 1.
Summary of Campylobacter primers and annealing temperature.
| Species | Gene target | Primer sequence | Annealing (°C) | Product size (bp) | Product Tm (°C)* | Reference |
|---|---|---|---|---|---|---|
| C. jejuni* | hipO | GTGGTCATGGAAGTGCTCCAGAAA AGCTCCTATGCTTACAACTGCTGA |
58 | 131 | 78.4–79.8 | (42) |
| C. coli | cpn60 | TGACGGTAGAACTTTCAAATCC GCAAGTGCTTCACCTTCGATA |
64.6 | 148 | 78.5 | (43–45) |
| C. upsaliensis* | CGTTTTGGCACACGCTATTT CATCAACAATAGCCTCACAAGC |
64.6 | 110 | 79.5 | ||
| C. lari* | TCTGCAAATTCAGATGAGAAAA TTTTTCAGTATTTGTAATGAAATATGG |
54.3 | 180 | 78.5 | ||
| C. helveticus | TGAAGCGATTGTTGATGAGC AACGCCATCTTTTCCAACTC |
64.6 | 151 | 80 | ||
| C. fetus | TGAGGCTGTTACAAGCGAGTTA TGAGCTATCGCTATTTGCTGAA |
62.5 | 100 | 79 | ||
| C. hyointestinalis | GGGGCAAATCCTATTGAGGT TCGCTATTTGCAGAGATCGTAG |
62.5 | 137 | 79.5 | ||
| C. rectus | AGCTATCATCGCCGAGCTAA TGAATGGATTTTGCCTCCTC |
64.6 | 177 | 87 | ||
| C. showae | GCCACTATCTCGGCAAATTC ACGTTTAGCTCGTCGTGGAT |
60 | 125 | 85 | ||
| C. concisus | GGCTCAAAAGAGATCGCTCA CCCTCAACAACGCTTAGCTC |
64.6 | 158 | 83.5 | ||
| C. curvus | CTGATAGCTGATGCGATGGA CTCGACCTGCATTTTCTCG |
64.6 | 162 | 83 | ||
| C. gracilis | AAGAGATCGCACAGGTTGCT CAAACTGCATTCCCTCGACT |
64.6 | 162 | 85 | ||
| C. mucosalis | TGCTGATGCAATGGAGAAAG CCTGCATTTTCTCGGTGTTT |
64.6 | 152 | 81 | ||
| C.sputorum | CAAAGTTGCTGAGGCAATCA TGATCCAACAGCCTCATCTG |
64.6 | 118 | 78 | ||
| C. infans* | ACTCAGCCGATATCATCGCC TATCAAATCGCCAATGGCAC |
65 | 119 | 53–56.9 | This Study |
Note: species annotated with an asterisk represent the four tested species across all 2,045 samples.
A tiered approach to Campylobacter species testing
Our main objective was to evaluate the diversity and prevalence of Campylobacter species in infants during their first 12 months of life while identifying potential reservoirs within selected households, including family members, livestock, and environmental sources. To achieve this, we implemented a tiered testing strategy comprising the following sequential steps:
1. Initial Screening of 15 Species
We began with a preliminary screening of 15 distinct Campylobacter species, including both thermotolerant and non-thermotolerant types (C. infans, C. jejuni, C. upsaliensis, C. lari, C. concisus, C. mucosalis, C. showae, C. sputorum, C. rectus, C. gracilis, C. hyointestinalis, C. fetus, C. helveticus, C. curvus, and C. coli) (Fig. S2). These species were selected as they have been associated with humans and based on prevalence patterns identified during the CAGED formative research (11). The species screening was performed on a subset of samples; this approach aimed to identify species present in human stool and livestock feces.
2. Identification of Seven Predominant Species
The initial screening identified seven Campylobacter species (C. infans, C. jejuni, C. concisus, C. upsaliensis, C. mucosalis, C. showae, and C. lari) as predominant in human stool and livestock feces. Since our primary focus was on infants, a subset of samples (a minimum of 88 infant stool samples from infants aged 8 to 367 days, with a median age of 174 days) were tested for species diversity. C. infans, C. jejuni, C. concisus, C. upsaliensis, C. showae, and C. lari were the most frequently detected species in infant stool samples (Fig. S3A). Although C. mucosalis was detected in 3% of samples in the pilot screening, it was positive in only one infant stool sample and thus was excluded from further testing. The remaining species were rarely detected, accounting for less than 2% of infant stool samples.
3. Focused Testing of Four Dominant Species
Guided by the pilot qPCR results and shotgun metagenomic analysis performed on a subset of samples (n = 40 per source, further details on this approach and findings will be communicated in a separate manuscript (Mekuria et al., unpublished)), we narrowed our focus to the four most prevalent and abundant species found in infant stools—C. infans, C. jejuni, C. upsaliensis, and C. lari—for comprehensive testing across all available human, livestock, and environmental samples (n = 2,045).
Genomic DNAs extracted from C. infans (Christine Szymanski, University of Georgia, Athens, GA), C. jejuni (ATCC 81–176), C. upsaliensis (ATCC 49816), and C.lari (ATCC 43675) were used as positive controls. Nuclease-free water was used as a negative control. Species-specific primers were evaluated for primer sensitivity and cross-reactivity against multiple thermophilic and non-thermophilic Campylobacter spp. to assess their performance in targeted detection (Fig. S2). The following thermocycler conditions were used: 1 cycle at 50°C for 2 min, 1 cycle at 95°C for 10 min, and 40 cycles at 95°C for 15 s, annealing temperature (Table 1) 30s followed by 72°C for 30 s.
Standard curves were prepared for each species using a determined concentration of DNA. The DNA concentrations in these samples were converted into genome copy numbers using the following formula: genome copies = [concentration of DNA tested (ng) × 6.0221 × 1023]/[average mass of 1 bp of DNA (660 g/mol) × 109] (https://Campylobacter.idtdna.com/pages/education/decoded/article/calculations-converting-from-nanograms-to-copy-number). A linear regression analysis was used to determine the relationship between the cycle threshold (CT) values and the genome copy numbers obtained for the determined concentration of DNA tested. The following equations were obtained: log10 C. infans genome copies per 50ng DNA = [(41.04-CT)/3.74], log10 C. jejuni genome copies= [(37.62-CT)/3.75], log10 C. upsaliensis genome copies= [(39.39-CT)/3.69], log10 C. lari genome copies=[(38.32-CT)/3.94]. A CT value of < 35 was used to determine prevalence, as described in our previous publication (21).
Integrated approach to household determinant identification
We utilized an integrated conceptual framework that merges a modified version of the United Nations International Children’s Emergency Fund (UNICEF) model with insights from a systematic review of drivers of childhood undernutrition (12). This combined framework was used to identify potential determinants of key health outcomes, specifically Campylobacter infections at the species level and environmental enteric dysfunction (EED) and stunting. Details of this individual and household-level determinant analysis have been previously published (22).
The framework categorized variables into four levels: Basic, Underlying, Immediate, and Physiological causes. Basic causes included factors such as social/cultural conditions, economics/livelihoods, human capital, and basic demographics from the original UNICEF framework, along with variables reflecting benefits, risks, and control measures, as outlined in Chen, et al. (2021) (23). Underlying causes included household food insecurity, inadequate care and feeding practices, unhealthy household environmental conditions, and inadequate health services. Immediate causes encompass inadequate dietary intake and diseases. Physiological causes focused on gut health, assessed through diarrheal symptoms and biomarkers of EED (lactulose excretion and myeloperoxidase activity). Given that prolonged infections can affect gut health, we further explored the relationship between EED status (as indicated by biomarkers for gut inflammation and permeability) and the cumulative load of Campylobacter infections at the species level. EED was classified using a composite indicator of lactulose excretion (%L) and fecal myeloperoxidase (MPO) levels (22).Moderate gut permeability was defined as 0.2 < %L ≤ 0.45%, and severe permeability as %L > 0.45%. For gut inflammation, MPO thresholds were 2,000 ng/ml for moderate and 3,364 ng/ml for severe. EED was categorized as moderate if either %L or MPO was severe and as severe if both %L and MPO were in a severe category. Additional details on the variables included in each category are described in (Table S2).
Statistical analysis
All statistical analyses were executed in R v4.3.1, with data securely stored in REDCap and exported as CSV files for processing and analysis (24, 25). Differences in prevalence, co-occurrence, and abundance were assessed via Chi-square, Fisher’s exact, Wilcox, and Kruskal Wallis tests across potential exposures. Regression analyses were performed to explore the relationship between Campylobacter load, EED, and baseline determinants using the statistical approach described in (22). We regressed each infant’s average Campylobacter load and EED status over the follow-up period on baseline determinants, excluding birth-related feeding practices. To assess the short-term effects of colostrum feeding, early breastfeeding, and prelacteal feeding, we focused on Campylobacter load (C. infans, C. jejuni, C. upsaliensis) during early infancy (7–39 days). EED was assessed in infants 12–14 months of age and regressed on baseline and time-varying determinants over the entire follow-up period. For longitudinal analysis, the average Campylobacter load was regressed on the average time-varying determinants within age quartiles to capture immediate effects on abundance. The same approach was used to assess the impact of Campylobacter burden on current enteric disease symptoms (fever and diarrhea).
Linear mixed models with individual-level random intercepts were fitted using the R package lme4 to account for repeated measures (26). C. infans, C. jejuni, and C. upsaliensis load as a function of age were analyzed using generalized linear mixed models. To examine the prevalence of Campylobacter species in infant stool samples, logistic regression models were fitted using generalized estimating equations. Unconditional logistic regression models were used for symptomatic and asymptomatic infections, conditioned on the presence or absence of specific Campylobacter species, following methods we previously established (21). For all outcomes except enteric disease symptoms (diarrhea and fever), backward elimination was used to refine the multivariable models, starting with determinants that had adjusted p-values < 0.2 from initial screening, adjusting for sex and socio-economic status as fixed confounders.
Results
Based on a preliminary screening of 15 Campylobacter species (Table 1) using a subset of samples, we identified four species most detected in infant stool: C. infans (36%, n = 171/475), C. jejuni (14%, n = 66/475), C. upsaliensis (10%, n = 48/475), and C. lari (6%, n = 30/475) (Fig. S3). Subsequently, we targeted these four species in 2,045 samples across all sources.
Candidatus C. infans and C. jejuni predominated in infant stools.
C. infans was detected in 41% [95% CI: 38%, 44%] of infant stools, followed by C. jejuni at 26% [95% CI: 23%, 28%] and C. upsaliensis at 13% [95% CI: 11%,15%]. C. infans and C. jejuni were detected in early infancy, as early as in the first month of life (mean 23 days old), while C. upsaliensis was first detected at 3 months (mean 79 days). Overall, C. lari prevalence was the lowest across all source types (< 2%) and was excluded from further analysis.
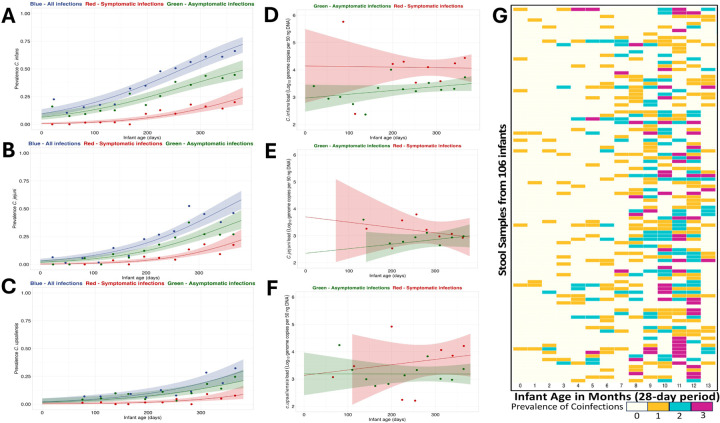
The prevalence of C. infans, C. jejuni, and C. upsaliensis increased as the infants grew older, with an 0.85% increase per day in the odds of C. infans (P < 0.0001), 0.98% per day for C. jejuni and 0.77% per day for C. upsaliensis colonization (P < 0.0001) (Fig. 2). This increase in prevalence was associated with an increased abundance of C. infans and C. jejuni with, on average, 0.0023 log10 genome copies per 50 ng DNA per day and 0.0029 log10 genome copies per 50 ng DNA per day. The load of C. infans increased linearly with age (P = 0.027), rising from 2.75 at 10 days to 3.59 log10 genome copies per 50 ng DNA at 370 days. A similar trend was observed for C. jejuni (P = 0.024), which increased from 2.04 log10 genome copies per 50ng DNA at 30 days to 3.05 log10 genome copies per 50ng DNA at 370 days.
Figure 2. Prevalence and load of predominant Campylobacter spp. in infant stool samples over time.
(A-C) Prevalence of C. infans, C. jejuni, and C. upsaliensis is shown by age, with points representing 4-week age groups. Lines indicate logistic regression models and shaded areas represent 95% confidence intervals. In blue are all species of infections, the red line shows symptomatic infections (presence of diarrhea) and asymptomatic infections in green. (D-F) C. infans, C. jejuni, and C. upsaliensis load in positive infant stool over time. Points are average loads by 4-week age group, lines are best-fitting linear regression models of load as a function of age as a continuous variable, and shaded areas are 95% confidence intervals. Confidence intervals are truncated between 2 to 6 log10 genome copies per 50 ng DNA. (G) Heatmap of Campylobacter spp. Coinfections (C. infans, C. jejuni, and C. upsaliensis) in infant stool samples, indicating prevalence by color intensity. X-axis: infant age; Y-axis: individual caged IDs. Data from 106 infants highlighting coinfections based on positive qPCR results at the genus and species level.
The co-occurrence of Campylobacter species in infant stool was analyzed in 28-day intervals (based on sampling scheme) to determine age-related patterns of infections. The first co-detection was observed at two months of age, and the proportion of samples with more than one species increased with age, peaking at 20% (39/95) by ten months. C. infans, C. jejuni, and C. upsaliensis were detected together between 4 and 13 months (Fig. 2G). The co-occurrence of Campylobacter species increased as infants aged, with significant associations observed at 114 to 207 days (β = 1.8123, P = 0.0009), 213 to 290 days (β = 2.9961, P < 0.0001), and 298 to 375 days (β = 3.4405, P < 0.0001).
Household Factors Associated with C. infans and C. jejuni Abundance in Infants.
Results of univariate analyses for individual- and household-level determinants (both with and without adjustment for confounders) from linear mixed modeling are summarized in (Table S2). Determinants with adjusted p values < 0.2 were included in multivariable analyses.
We observed significant associations between environmental and behavioral factors and the load of C. infans and C. jejuni in infants (Table 2). C. infans loads were significantly higher among infants consuming raw milk and those crawling in areas contaminated with animal droppings. Notably, female infants demonstrated consistently higher C. infans loads compared to males. Conversely, a lower average of C. infans load throughout the follow-up period was significantly associated with households with a higher sheep nighttime risk score (defined as households who confined sheep inside the home at night). Infants who put soil or animal feces in their mouths were at risk for higher C. jejuni loads (Table 2).
Table 2.
Associations between Campylobacter species load and household determinants.
| Outcome | Regression Coefficient | Odds Ratio (95%, Confidence Interval)^ | |
|---|---|---|---|
| Screening Analysisa | Multivariable Analysis | ||
| C. infans Load | Baseline Determinants & | ||
| Infant Sex (Female) | 0.608** | 1.471 (0.099, 0.674)** | |
| Sheep nighttime location risk score(≥ 1 vs. 0)# | −0.399** | 0.654 (−0.706, −0.142)** | |
| Disposal of infant’s stool, post-defecation | 0.211 | 1.278 (−0.039, 0.532)* | |
| Handwashing after field work | −0.304 | 0.769 (−0.628, 0.105) | |
| Longitudinal Determinants | |||
| HFIAS | 0.024* | 1.014 (−0.013, 0.042) | |
| Raw milk consumption | 0.962** | 2.335 (0.241, 1.455)* | |
| Crawling where animal droppings present | 0.597** | 1.647 (0.066, 0.932)* | |
| C. jejuni Load | Baseline Determinants & | ||
| Access to drinking water | −0.743* | 0.494 (−1.452, 0.039)* | |
| Access to any sanitation | −0.392* | 0.757 (−0.724, 0.165) | |
| Mother using soap to wash hands | −0.471 ** | 0.723 (−0.744, 0.100) | |
| Longitudinal Determinants | |||
| HFIAS | 0.040** | 1.027 (−0.009, 0.063) | |
| Putting soil/animal feces in mouth | 1.048** | 2.239 (0.232, 1.381)* | |
| Physical contact with livestock | 0.694** | 1.568 (−0.055, 0.955)· | |
| C. upsaliensis Load | Baseline Determinants & | ||
| Handwashing after handling raw food | −0.312 | 0.723 (−0.729, 0.082) | |
| Sheep daytime location risk score(≥ 1 vs. 0)# | −0.546** | 0.606 (−1.028, 0.026)* | |
| Collection of livestock waste | −0.544 | 0.653 (−1.067, 0.215) | |
| Longitudinal Determinants | |||
| Achieved MDD | 0.363 | 1.437 (−0.184, 0.910) |
Backward elimination suggested the final model was the single-exposure model. Further details on the screening results can be found in Table S2.
Adjusted for socio-economic status at baseline and sex.
≥1: Households kept the livestock species inside the house confined or unconfined; 0: Households did not have or keep the livestock species outside the house.
Abbreviations: HFIAS: household food insecurity access score.
The average Campylobacter load in an age quartile was regressed on each determinant’s concurrent proportion/mean, adjusting for age quartile, sex, and socioeconomic status at baseline. Linear-mixed models with individual-level random intercepts were used to estimate the coefficients.
Screening Analysis represents findings from univariate analysis in which determinants with adjusted p-values < 0.2 were included in the multivariable analysis.
Represents p ≤ 0.05.
Represents 0.05 ≤ p < 0.1.
Enteric Disease Symptoms
Most stool samples positive for Campylobacter were collected from asymptomatic infants (n = 682/907). However, C. infans colonization was significantly associated with diarrheal episodes (OR = 2.02, β = 0.70, P = 0.000105, [95%CI: 35%, 100%]) and fever (OR = 1.62, β = 0.48, P = 0.006, [95%CI: 14%, 83%]). Among the 346 stool samples positive for C. infans, 25% (85/346) were from infants experiencing diarrhea, and 24% (83/346) had a fever. Similarly, C. jejuni showed a significant association with diarrheal episodes (OR = 2.29, β = 0.83, P < 0.0001[95%CI: 46%, 100%]) and fever (OR = 2.53, β = 0.92, P < 0.0001, [95%CI: 56%,100%]), with 28% (61/214) of the 214 positive stool samples collected during episodes of diarrhea and 32% (68/214) with fever. Additionally, in symptomatic infants, we observed a daily increase of 0.54% in the prevalence of C. infans and 0.53% in the prevalence of C. jejuni (P = 0.0008 and P = 0.024, respectively) (Fig. 2A–F). When exploring the impact of Campylobacter abundance and enteric disease symptoms, we found higher C. infans loads were associated with diarrhea and fever (Table 3). However, fever was not statistically significant after adjustment for confounders. No statistical associations were found between enteric symptoms and the abundance of either C. jejuni or C. upsaliensis or the prevalence of C. upsaliensis.
Table 3.
Associations between Campylobacter species load and enteric disease symptoms. Odds Ratio (95%, Confidence Interval) ^
| Outcome | C. infans load | C. jejuni load | C. upsaliensis load | |||
|---|---|---|---|---|---|---|
| Unadjusted | Adjusted | Unadjusted | Adjusted | Unadjusted | Adjusted | |
| Diarrhea | 1.345 (0.056,0.539)* |
1.384 (0.061,0.589)* |
1.071 (−0.093,0.231) |
1.071 (−0.12,0.259) |
0.95 (−0.245,0.144) |
0.952 (−0.251,0.153) |
| Fever | 1.039 (0.014,0.063)* |
1.019 (−0.007,0.044) |
1.049 (0.033,0.063) |
1.039 (0.021,0.057) |
1.013 (−0.007,0.033) |
1.005 (−0.015,0.025) |
Using a linear mixed model with individual-level random intercept, the proportion of each symptom’s occurrence was regressed as the outcome on the concurrent average Campylobacter load using crude and adjusted models (adjusting for age quartile and infant sex at baseline).
p < 0.05.
EED
At 1 year of age, EED was identified in 54 out of 101 infants, with 56% [95%CI: 42%, 69%] classified as moderate and 44.4% [95%CI: 31%, 59%] as severe. Intestinal inflammation, based on MPO levels, was present in all EED cases, with severe inflammation observed in 46% [95%CI: 33%, 60%]. Elevated lactulose levels indicated intestinal permeability in 52 out of 99 infants, with severe permeability detected in 52% [95%CI: 38%, 66%]. Despite the high prevalence of EED, no significant associations were found between the bacterial load of C. infans, C. jejuni, or C. upsaliensis and EED. Similarly, there were no significant correlations between bacterial load and individual biomarkers of gut inflammation and permeability (Table 4).
Table 4.
Associations between Campylobacter species load and gut health indicators.
| Odds Ratio (95%, Confidence Interval) | ||||||
|---|---|---|---|---|---|---|
| Outcome | C. infans load | C. jejuni load | C. upsaliensis load | |||
| Unadjusted | Adjusted ^ | Unadjusted | Adjusted^ | Unadjusted | Adjusted^ | |
| Lactulose percentage | 1.038 (0.627,1.719) |
1.016 (0.603,1.713) |
1.024 (0.672,1.560) |
1.031 (0.674,1.579) |
0.992 (0.692,1.422) |
0.998 (0.695,1.433) |
| Fecal myeloperoxidase | 1.038 (0.626,1.721) |
0.984 (0.580,1.669) |
0.901 (0.591,1.373) |
0.910 (0.591,1.401) |
0.963 (0.671,1.381) |
0.980 (0.680,1.413) |
| Environmental enteric dysfunction | 1.386 (0.826,2.324) |
1.384 (0.808,2.372) |
1.025 (0.673,1.563) |
1.030 (0.669,1.585) |
1.020 (0.710,1.463) |
1.035 (0.718,1.492) |
Adjusted for infant sex and age at EED sampling.
C. infans and C. jejuni are Common in Ethiopian Household Environments
Family Members
C. infans was frequently detected in stools from both siblings (under 5 years of age) and mothers, with a prevalence of 56% [95% CI: 48%, 65%] in siblings and 45% [95% CI: 36%, 54%] in mothers (Fig. 3). The second most common species, C. jejuni, showed a higher prevalence in siblings at 24% [95% CI: 16%, 31%] compared to mothers at 9% [95% CI: 3%, 14%] (P = 0.07). C. upsaliensis was detected in 9% of all human stool samples (23/250), with siblings showing a higher prevalence at 12% [95% CI: 6%, 18%] versus 6% in mothers [95% CI: 1%, 9%].
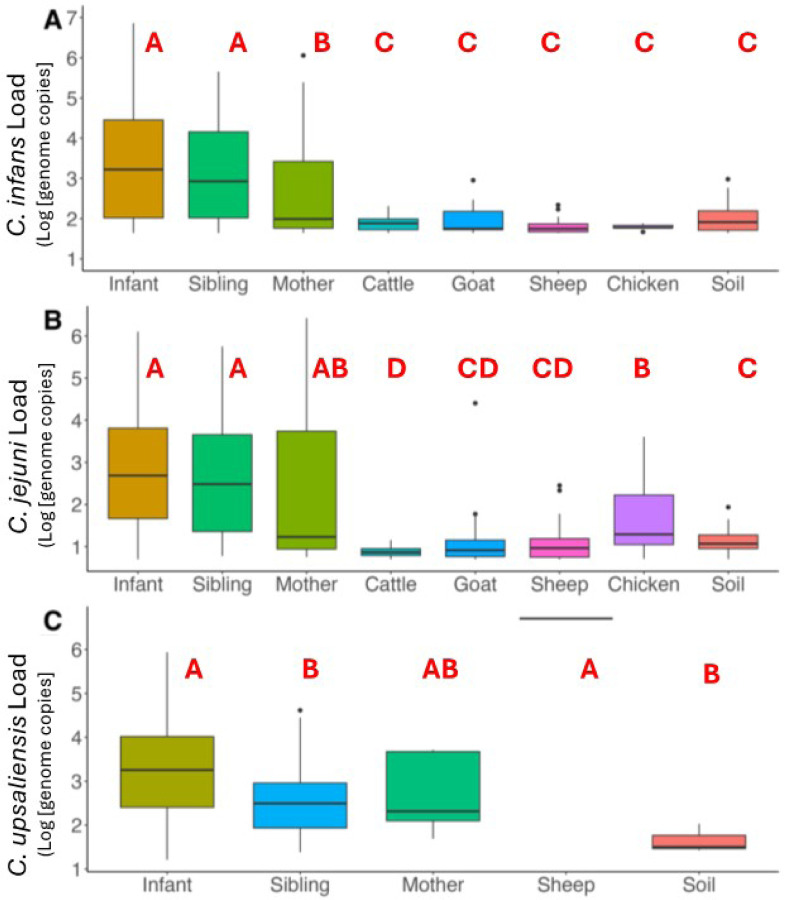
Figure 3. Abundance of C. infans, C. jejuni, and C. upsaliensis in 106 households.
The boxplot shows bacterial load in positive samples (log[genome copies per 50ng of DNA]). Letters (A to F) indicate statistical differences (P < 0.05) based on Kruskal Wallis and post hoc Dunn’s test.
While the abundance of C. infans was overall high in humans, siblings had the highest average load after infants, with an average of 3.15 log10 genome copies per 50ng DNA [95% CI: 2.86, 3.43], compared to mothers who had an average load of 2.67 [95% CI: 2.34, 2.99] (P = 0.0118) in positive C. infans samples. In contrast, no significant differences were observed in the loads of C. jejuni or C. upsaliensis between siblings and mothers. For C. jejuni, the average load was 2.67 [95% CI: 2.17, 3.15] in siblings and 2.39 [95% CI: 1.16, 3.62] in mothers. For C. upsaliensis, the average load was 2.59 [95% CI: 2.13, 3.06] in siblings and 2.75 [95% CI: 2.08, 2.75] in mothers.
Given the high prevalence of Campylobacter in humans, we examined the likelihood of multiple species co-occurring within the same sample. Like infants, siblings exhibited a high co-occurrence of multiple Campylobacter species that were significantly different from mothers (OR = 2.12, 95% CI: 1.07–4.35, β = 0.752, P = 0.033).
Zoonotic Sources
Among the livestock studied in the 106 households, C. jejuni prevalence was highest in chickens at 38% [ 95% CI: 28%, 47%] and small ruminants, including goats, at 27% [95% CI: 21%, 34%] and sheep at 21% [95%CI: 15%, 28%] (Fig. 3). Among livestock species, C. infans was most frequently detected in sheep at 13% [95% CI: 8%, 19%], followed by goats (10%, [95%CI: 6%, 15%] and cattle (9%, [95% CI: 6%, 15%] with chickens having the lowest occurrence at 4.8% [95%CI: 2%, 11%]. However, most of these samples had high CT values (around 35), suggesting low abundance of C. infans in these reservoirs (Fig.S4). The co-occurrence of multiple species was low across all zoonotic sources, at < 5% [95% CI: 5%, 13%].
In samples positive for C. jejuni, the load ranged from 0.7 to 4.4 log10 genome copies, with average of 1.17 [ 95% CI: 1.07, 1.26]. Chicken feces had higher C. jejuni loads (1.62 [95% CI: 1.37,186]) compared to cattle (0.87 [95% CI: 0.83,0.92] log10 genome copies per 50 ng of DNA (P = 0.0027, Fig. 3). In C. infans positive samples, the load ranged between 1.6 and 2.9 log10 genome copies. Goats had the highest average load of C. infans across all livestock, 1.95 [95% CI: 1.78, 2.12]; however, no statistical differences were observed in abundance between livestock species. C. upsaliensis was detected in one sheep sample with a load of 6.7 log10 genome copies per 50 ng of DNA.
Household Soil Samples
In soil samples, C. infans was the most prevalent at 33% [95% CI: 27%, 38%], followed by C. jejuni at 19% [95% CI: 14%, 23%], while C. upsaliensis was rare, constituting < 2% [95% CI: 0%, 3%]. Co-occurrence was observed in approximately 10% of soil samples [95%CI: 5%, 13%], in which samples collected from inside the home had a greater prevalence (17%, [95%CI: 5%, 29%]). The average load of C. infans and C. jejuni in positive soil samples was 2.04 [95% CI: 1.96, 2.11] and 1.18 [95% CI: 1.08, 1.29] log10 genome copies per 50 ng of DNA.
Comparative Analysis of Campylobacter Genus and Species Identification
Cross-tabulation was conducted to evaluate the concordance between genus-level Campylobacter detection (21) and the detection of specific Campylobacter species in this study. Among 2,045 samples from humans, livestock, and soil, 1,622 tested positive at the genus level. Of these, 865 were also positive for one or more of the tested species, while 340 samples tested negative for both genus and species-level tests. Interestingly, 83 samples tested positive for one or more species despite being negative at the genus level.
Among infant stool samples, 594 tested positive at the genus level, of which 444 were also positive for one or more of the tested Campylobacter species. While 264 samples tested negative for both genus and species-level PCRs, and 48 samples tested positive for one or more species but were negative at the genus level (Table 5). A significant association was observed between genus- and species-level positivity, with more likely to detect one or more species in genus-level positive samples compared to genus-negative samples (P < 0.0001). These findings suggest that the three species detected in our study account for a substantial proportion of the Campylobacter genus identified in these samples.
Table 5.
Concordance of Genus and Species-specific Campylobacter Detection in Infant Stool.
| Genus (−) | Genus (+) | |
|---|---|---|
| All sources (n = 2,045) | ||
| One species | 18% (75/423) | 37% (597/1622 |
| Multiple species | 2% (8/423) | 17% (268/1622) |
| No species | 80% (340/423) | 46% (757/1622) |
| Infant Stool (n = 907) | ||
| One species | 14% (43/313) | 43% (254/594) |
| Multiple species | 1% (5/313) | 32% (190/594) |
| No species | 85% (265/313) | 25% (150/594) |
This table presents a comparative analysis of Campylobacter species detection with genus-level test results across different sample sources. Of the 2,045 collected samples, 1,622 tested positive for Campylobacter at the genus level. Within the subset of infant stool samples (n = 907), 594 were confirmed positive for the Campylobacter genus.
Discussion
In low- and middle-income countries, diarrheal diseases remain a leading cause of mortality in children under 5 years of age, with Campylobacter species emerging as significant pathogens associated with both acute illness and chronic health consequences, including malnutrition, stunting, and cognitive deficits (8, 23, 27). The CAGED longitudinal study found Campylobacter is highly prevalent in rural households in eastern Ethiopia21. Here, we identified a high prevalence of C. infans and C. jejuni among infants residing in Haramaya, Ethiopia, using quantitative PCR (qPCR). Over 20% of infants were colonized with C. infans within the first month of life, and the prevalence plateaued at 61% by 11–12 months. In contrast, C. jejuni colonization was observed in 6% of infants during the first month, peaking at 53% by 10 months. C. upsaliensis colonization was observed later, emerging after the first two months of life, and remained relatively low (6–12%) throughout most of the first year. However, a notable increase to 28% at 11 months suggests that C. upsaliensis colonization may occur later in infancy. These findings align with research efforts to understand Campylobacter acquisition and clearance dynamics using the MAL-ED longitudinal data, which also observed increased Campylobacter acquisition rates in infants across multiple low- and middle-income countries during the first year of life (27). Both C. infans and C. jejuni abundances showed positive correlations with infant age, ranging between 2 to 3 log10 genome copies per 50 ng DNA, which is concerning as Campylobacter is highly infectious compared to other gastrointestinal pathogens, given that C. jejuni has a minimum infectious dose of 500 to 800 organisms (28, 29). In addition, the co-occurrence of Campylobacter species increased as infants grew older, with infants between 298–375 days old having 31 times higher odds of co-occurrence detection. Overall, the prevalence and loads of infants at approximately one year of age were higher than those in siblings, which in turn were higher than in mothers. These differences are likely due to the development of immune systems and differences in environmental exposure through behaviors such as crawling and play and different hygiene practices. The development of adaptive immunity may play a crucial role in age-related susceptibility to Campylobacter (27). Acute infections trigger the production of specific immunoglobulins (IgG, IgM, and IgA) in serum and intestinal secretions, with IgA and IgM antibodies particularly elevated during acute phases (30–33). This acquired immunity strengthens with repeated exposures throughout life(34), likely explaining the lower infection and co-occurrence rates observed in mothers compared to their children.
In our study, many Campylobacter-positive samples belong to asymptomatic infants; however, infants colonized with C. infans had 2.0 times higher odds of experiencing enteric symptoms, particularly diarrhea, which was linked to higher C. infans load. These findings contrast with recent observations from other LMICs, particularly from Peru, where researchers reported low C. infans prevalence in infants under two years of age and found no association between C. infans colonization and diarrheal symptoms (35, 36). While the Peru cohort was small, this discrepancy highlights the importance of conducting region-specific epidemiological studies. It suggests that the role C. infans in infant health may be more complex than initially understood, warranting further investigation into strain-specific characteristics and local determinants of pathogenicity. Additionally, we found that infants colonized with C. jejuni had 2.3 times higher odds of experiencing diarrhea, aligning with findings from other LMIC-oriented studies, including the GEMS and MAL-ED studies, which identified C. jejuni as a leading cause of bacterial diarrhea in LMICs during early childhood (7, 37, 38). Despite the high occurrence of Campylobacter infection and EED, with more than half of infants having EED by one year of life, no association was found between C. infans, C. jejuni, or C. upsaliensis load and EED in this population. These results align with previous work assessing the relationship between the overall Campylobacter load and gut function parameters (22) and is related to a small sample size in our study. Importantly, we observed similar loads of Campylobacter at the genus level and the three dominant species in samples from diarrheal and non-diarrheal infants, supporting the finding from the MAL-ED study that asymptomatic infants are also at risk of developing chronic gut inflammation and, consequently, EED and stunting (39).
Several household determinants were identified to impact Campylobacter species abundance. Household dietary practices significantly influenced Campylobacter abundance, with raw milk consumption strongly associated with higher C. infans loads. This association was also observed at the genus level, suggesting that introducing complementary foods, particularly animal-source foods, may be a critical source for microbial colonization (12, 23). Infant sex also influenced Campylobacter colonization, as females had a higher average load of C. infans, suggesting potential gender-based differences in susceptibility and/or exposure. Additionally, crawling in areas with animal droppings was linked to increased C. infans load. While C. infans was not prevalent among the four livestock species tested in this study, its detection in surface soil samples around the household underscores the potential for complex environmental exposure pathways contributing to elevated C. infans levels in infants. While sheep demonstrated the highest C. infans prevalence among tested livestock species, closer nighttime proximity to sheep was associated with lower infant C. infans loads, matching findings from genus-level testing. Similarly, keeping sheep confined inside the home was associated with a reduced abundance of C. upsaliensis. This pattern may suggest that though sheep are reservoirs of Campylobacter, the relationship between animal proximity and human colonization is complex and requires further investigation. Lastly, we found that infants who put soil or animal feces in their mouths were at risk for higher C. jejuni loads, a species predominant in zoonotic reservoirs. These findings emphasize the need for targeted interventions to address the complex interplay of environmental, dietary, and behavioral factors in Campylobacter transmission. Such interventions are urgently required to reduce infection rates and mitigate the burden of diarrheal diseases in resource-limited settings.
Building on our formative research, this longitudinal study provides deeper insights into the socio-demographic and exposure dynamics within the Haramaya woreda and may also be relevant for comparable smallholder environments. Our species-specific testing across human, livestock, and environmental samples revealed that three targeted Campylobacter species accounted for 54% of genus-level detections, suggesting the presence of additional, untested Campylobacter species and undetected species using the current method in these ecological niches. Indeed, shotgun metagenomic testing identified 21 dominant Campylobacter species across 280 samples (infants, mothers, siblings, cattle, goats, sheep, and chickens), revealing a diverse Campylobacter community in which many different species were found in ruminants that do not seem to be transmitted to humans. In infant stools, however, C. infans, C. jejuni, and C. upsaliensis collectively accounted for 75% of the Campylobacter genus-level signals, establishing these species as the predominant colonizers of the infant gut. This finding aligned with metagenomic data, where C. jejuni, C. infans, and C. upsaliensis were frequently detected alongside C. concisus (Mekuria et al., unpublished). In addition, 83 samples tested positive for one or more species, while the test at the genus level was negative. TaqMan was used for genus testing, whereas species testing was conducted with SYBR Green, and these observations may be partly explained by the different sensitivities of the two PCR methods. Further emphasizing these limitations, Parker et al. (2022) demonstrated that even validated primers for C. jejuni can yield false negatives. They found that a significant percentage of stool samples from children in Peru tested negative for C. jejuni using qPCR. Yet, the species was detected via whole-genome shotgun metagenomic sequencing performed on the same extracts. While we utilized validated primers for species testing, we acknowledge certain limitations in our approach. We observed some cross-reactivity in our primers, particularly for C. jejuni (with C. fetus, C. lari, and C. showae), C. upsaliensis (with C. helveticus), and C. lari (with C. helveticus and C. upsaliensis). C. infans primers showed minimal cross-reactivity with C. concisus at a high CT value, indicating low-level amplification. Future studies could benefit from a combined approach using both qPCR and metagenomic sequencing to address these limitations. While qPCR offers higher sensitivity and is more suitable for the absolute quantification of target genes, metagenomic sequencing provides broader coverage and an unbiased overview of microbial species within a sample (40). This complementary approach would leverage the strengths of both methods: the high sensitivity and quantification capabilities of qPCR and the comprehensive detection and novel species identification potential of metagenomics (41). By integrating these techniques, researchers can achieve a more robust surveillance strategy, gaining a deeper understanding of the microbial landscape while minimizing diagnostic gaps and potential biases introduced by primer-based methods.
This study contributes to the growing body of evidence elucidating the prevalence and diversity of Campylobacter species in human and animal populations. Our investigation revealed distinct distribution patterns: C. infans showed a high prevalence in human stool, which increased in the first year of life, and surface soil samples, while C. jejuni demonstrated substantial presence across environmental and zoonotic sources, particularly in livestock. Consistent with previous research, our findings reveal that human stools and livestock feces frequently harbor multiple Campylobacter species. Our species-specific quantitative PCR results, further validated through shotgun metagenomics analyses in a subset of samples (n = 280, 40 per source), not only confirmed the presence of targeted Campylobacter species but also suggested the existence of additional species, particularly in ruminants (Mekuria et al., unpublished). These complementary approaches strengthen our confidence in the observed prevalence patterns and underscore the value of refined detection methodologies.
Conclusions
In this study, we investigated the prevalence and dynamics of Campylobacter species colonization among 106 households residing in Haramaya, Ethiopia, using species-specific quantitative PCR. We identified C. infans, C. jejuni, and C. upsaliensis as the predominant colonizers of the infant gut, with notable age-dependent patterns and associations with enteric symptoms. Notably, C. infans and C. jejuni were linked to higher odds of diarrhea and fever, highlighting their clinical significance. Our findings support the recognition of Candidatus C. infans as a novel species with a distinct ecology. In this setting, C. infans was more prevalent in humans than C. jejuni, suggesting primarily human-to-human transmission. The low occurrence in livestock implies potential reverse zoonotic transmission or passage of this bacteria through the animal gut after exposure from a highly contaminated environment. The co-presence of anthroponotic C. infans and zoonotic C. jejuni and C. upsaliensis suggests that control methods in low-resource settings should address both transmission cycles. The identified risk factors and complex interplay of environmental, dietary, and behavioral factors offer potential targets for interventions to reduce Campylobacter colonization and associated health impacts.
Supplementary Material
Acknowledgments
This work is a result of the CAGED Research Team, whose members include: Amanda Evelyn Ojeda, Arie H. Havelaar, Abadir Jemal Seran, Abdulmuen Mohammed Ibrahim, Bahar Mummed Hassen, Belisa Usmael Ahmedo, Cyrus Saleem, Dehao Chen, Efrah Ali Yusuf, Gireesh Rajashekara, Getnet Yimer, Ibsa A. Ahmed, Ibsa Aliyi Usmane, Jafer Kedir Amin, Jemal Y. Hassen, Kedir A. Hassen, Kunuza Adem Umer, Karah Mechlowitz, Kedir Teji Roba, Loic Deblais, Mussie Bhrane, Mark J. Manary, Mawardi M. Dawid, Mahammad Mahammad Usmail, Nigel P. French, Nur Shaikh, Nitya Singh, Sarah L. McKune, Wondwossen A. Gebreyes,Xiaolong Li, Yenenesh Demisie Weldesenbet, Yang Yang, Zelalem Hailu Mekuria. This study would not have been possible without cooperation of study communities and local administration of the study kebeles. We want to express our appreciation to the study households, the Community Advisory Board and all who supported the study directly or otherwise.
We thank Jaap Wagenaar and Birgitta Duim (Utrecht University, Utrecht, the Netherlands) and Christine Szymanski (University of Georgia) for Candidatus C. infans strain 1900001.
Research reported in this publication was supported by the University of Florida Clinical and Translational Science Institute, which was supported in part by the NIH National Center for Advancing Translational Sciences under award number UL1TR001427. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Funding
The Bill and Melinda Gates Foundation funded the CAGED project to address food insecurity issues in Ethiopia and Burkina Faso through the project Equip—Strengthening Smallholder Livestock Systems for the Future (grant number OPP11755487). These funds are administered by the Feed the Future Innovation Lab for Livestock Systems, established with funding from the U.S. Agency for International Development (USAID) and co-led by the University of Florida’s Institute of Food and Agricultural Sciences and the International Livestock Research Institute. Support for the Feed the Future Innovation Lab for Livestock Systems is made possible by the generous support of the American people through USAID. The contents are the authors’ responsibility and do not necessarily reflect the views of USAID or the U.S. Government. REDCap is hosted at the University of Florida Clinical and Translational Science Institute (CTSI) and supported by NIH National Center for Advancing Translational Sciences grant UL1TR000064. This project is funded by the U.S. Agency for International Development Bureau for Food Security under agreement number AID-OAA-L-15-00003 as part of Feed the Future Innovation Lab for Livestock Systems. Any opinions, findings, conclusions, or recommendations expressed here are those of the authors alone. Research reported in this publication was supported by the University of Florida Clinical and Translational Science Institute, partly funded by the NIH National Center for Advancing Translational Sciences under award number UL1TR001427.
Funding Statement
The Bill and Melinda Gates Foundation funded the CAGED project to address food insecurity issues in Ethiopia and Burkina Faso through the project Equip—Strengthening Smallholder Livestock Systems for the Future (grant number OPP11755487). These funds are administered by the Feed the Future Innovation Lab for Livestock Systems, established with funding from the U.S. Agency for International Development (USAID) and co-led by the University of Florida’s Institute of Food and Agricultural Sciences and the International Livestock Research Institute. Support for the Feed the Future Innovation Lab for Livestock Systems is made possible by the generous support of the American people through USAID. The contents are the authors’ responsibility and do not necessarily reflect the views of USAID or the U.S. Government. REDCap is hosted at the University of Florida Clinical and Translational Science Institute (CTSI) and supported by NIH National Center for Advancing Translational Sciences grant UL1TR000064. This project is funded by the U.S. Agency for International Development Bureau for Food Security under agreement number AID-OAA-L-15-00003 as part of Feed the Future Innovation Lab for Livestock Systems. Any opinions, findings, conclusions, or recommendations expressed here are those of the authors alone. Research reported in this publication was supported by the University of Florida Clinical and Translational Science Institute, partly funded by the NIH National Center for Advancing Translational Sciences under award number UL1TR001427.
Footnotes
Additional Declarations: No competing interests reported.
Declarations
Ethics approval and consent to participate
Ethical approval was obtained from the University of Florida Internal Review Board (IRB201903141); the Haramaya University Institutional Health Research Ethics Committee (COHMS/1010/3796/20), and the Ethiopia National Research Ethics Review Committee (SM/14.1/1059/20). Written informed consent was obtained from all participating households (husband and wife) using a form in the local language (Afan Oromo).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Amanda Ojeda, University of Florida.
Loïc Deblais, Hypercell Technologies.
Bahar Mummed, Haramaya University.
Mussie Brhane, Haramaya University.
Kedir A. Hassen, Haramaya University.
Belisa Usmael Ahmedo, Haramaya University.
Yenenesh Demisie Weldesenbet, Haramaya University.
Dehao Chen, University of Florida.
Xiaolong Li, University of Florida.
Cyrus Saleem, University of Florida.
Mark J. Manary, Washington University.
Luiz F.W. Roesch, University of Florida.
Sarah L. McKune, University of Florida.
Arie H. Havelaar, University of Florida.
Gireesh Rajashekara, University of Illinois.
Availability of data and materials
Deidentified individual participant data will be available through Dataverse (https://dataverse.org/) after December 31, 2024.
References
- 1.Havelaar AH, Kirk MD, Torgerson PR, Gibb HJ, Hald T, Lake RJ et al. World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010. PLOS Med [Internet]. 2015. Dec 3 [cited 2024 Nov 19];12(12):e1001923. https://journals.plos.org/plosmedicine/article?id=10.1371/journal.pmed.1001923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chala G, Eguale T, Abunna F, Asrat D, Stringer A. Identification and Characterization of Campylobacter Species in Livestock, Humans, and Water in Livestock Owning Households of Peri-urban Addis Ababa, Ethiopia: A One Health Approach. Front Public Health [Internet]. 2021. [cited 2023 Aug 31];9. https://www.frontiersin.org/articles/10.3389/fpubh.2021.750551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beshearse E, Bruce BB, Nane GF, Cooke RM, Aspinall W, Hald T et al. Attribution of Illnesses Transmitted by Food and Water to Comprehensive Transmission Pathways Using Structured Expert Judgment, United States. Emerg Infect Dis [Internet]. 2021. Jan [cited 2024 Nov 19];27(1):182. https://pmc.ncbi.nlm.nih.gov/articles/PMC7774530/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hald T, Aspinall W, Devleesschauwer B, Cooke R, Corrigan T, Havelaar AH et al. World Health Organization Estimates of the Relative Contributions of Food to the Burden of Disease Due to Selected Foodborne Hazards: A Structured Expert Elicitation. PLOS ONE [Internet]. 2016. Jan 19 [cited 2024 Nov 19];11(1):e0145839. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0145839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pantazi AC, Balasa AL, Mihai CM, Chisnoiu T, Lupu VV, Kassim MAK et al. Development of Gut Microbiota in the First 1000 Days after Birth and Potential Interventions. Nutrients [Internet]. 2023. Aug 20 [cited 2024 Nov 19];15(16):3647. https://pmc.ncbi.nlm.nih.gov/articles/PMC10457741/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guerrant RL, DeBoer MD, Moore SR, Scharf RJ, Lima AAM. The impoverished gut—a triple burden of diarrhoea, stunting and chronic disease. Nat Rev Gastroenterol Hepatol [Internet]. 2013. Apr [cited 2024 Nov 19];10(4):220–9. https://www.nature.com/articles/nrgastro.2012.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amour C, Gratz J, Mduma E, Svensen E, Rogawski ET, McGrath M et al. Epidemiology and Impact of Campylobacter Infection in Children in 8 Low-Resource Settings: Results From the MAL-ED Study. Clin Infect Dis Off Publ Infect Dis Soc Am [Internet]. 2016. Nov 1 [cited 2023 Nov 7];63(9):1171–9. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5064165/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogawski ET, Liu J, Platts-Mills JA, Kabir F, Lertsethtakarn P, Siguas M et al. Use of quantitative molecular diagnostic methods to investigate the effect of enteropathogen infections on linear growth in children in low-resource settings: longitudinal analysis of results from the MAL-ED cohort study. Lancet Glob Health [Internet]. 2018. Dec 1 [cited 2024 Nov 19];6(12):e1319–28. https://www.sciencedirect.com/science/article/pii/S2214109X18303516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haque MA, Platts-Mills JA, Mduma E, Bodhidatta L, Bessong P, Shakoor S et al. Determinants of Campylobacter infection and association with growth and enteric inflammation in children under 2 years of age in low-resource settings. Sci Rep [Internet]. 2019. Nov 20 [cited 2023 Aug 27];9(1):17124. https://www.nature.com/articles/s41598-019-53533-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez JJ, Alam MA, Stride CB, Haque MA, Das S, Mahfuz M et al. Campylobacter infection and household factors are associated with childhood growth in urban Bangladesh: An analysis of the MAL-ED study. PLoS Negl Trop Dis [Internet]. 2020. May 14 [cited 2022 Oct 4];14(5):e0008328. https://journals.plos.org/plosntds/article?id=10.1371/journal.pntd.0008328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terefe Y, Deblais L, Ghanem M, Helmy YA, Mummed B, Chen D et al. Co-occurrence of Campylobacter Species in Children From Eastern Ethiopia, and Their Association With Environmental Enteric Dysfunction, Diarrhea, and Host Microbiome. Front Public Health [Internet]. 2020. Apr 15 [cited 2024 Nov 19];8. https://www.frontiersin.org/journals/public-health/articles/10.3389/fpubh.2020.00099/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen D, McKune SL, Singh N, Yousuf Hassen J, Gebreyes W, Manary MJ et al. Campylobacter Colonization, Environmental Enteric Dysfunction, Stunting, and Associated Risk Factors Among Young Children in Rural Ethiopia: A Cross-Sectional Study From the Campylobacter Genomics and Environmental Enteric Dysfunction (CAGED) Project. Front Public Health [Internet]. 2021. Jan 21 [cited 2024 Nov 19];8. https://www.frontiersin.org/journals/public-health/articles/10.3389/fpubh.2020.615793/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costa D, Iraola G. Pathogenomics of Emerging Campylobacter Species. Clin Microbiol Rev [Internet]. 2019. Jul 3 [cited 2024 Nov 19];32(4):10.1128/cmr.00072-18. https://journals.asm.org/doi/10.1128/cmr.00072-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Man SM. The clinical importance of emerging Campylobacter species. Nat Rev Gastroenterol Hepatol [Internet]. 2011. Dec [cited 2024 Nov 19];8(12):669–85. https://www.nature.com/articles/nrgastro.2011.191 [DOI] [PubMed] [Google Scholar]
- 15.Mansfield KG, Fox JG. Bacterial Diseases. In: The Common Marmoset in Captivity and Biomedical Research [Internet]. Elsevier; 2019. [cited 2024 Nov 19]. pp. 265–87. https://linkinghub.elsevier.com/retrieve/pii/B9780128118290000169 [Google Scholar]
- 16.Monte DFM, Ribeiro VB, Destro MT. Foodborne zoonosis. In: Dikeman M, editor. Encyclopedia of Meat Sciences (Third Edition) [Internet]. Oxford: Elsevier; 2024. [cited 2024 Nov 19]. pp. 229–36. https://www.sciencedirect.com/science/article/pii/B9780323851251001125 [Google Scholar]
- 17.François R, Yori PP, Rouhani S, Salas MS, Olortegui MP, Trigoso DR et al. The other Campylobacters: Not innocent bystanders in endemic diarrhea and dysentery in children in low-income settings. PLoS Negl Trop Dis [Internet]. 2018. Feb 7 [cited 2024 Nov 19];12(2):e0006200. https://journals.plos.org/plosntds/article?id=10.1371/journal.pntd.0006200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bian X, Garber JM, Cooper KK, Huynh S, Jones J, Mills MK et al. Campylobacter Abundance in Breastfed Infants and Identification of a New Species in the Global Enterics Multicenter Study. mSphere [Internet]. 2020. Jan 15 [cited 2024 Nov 19];5(1):10.1128/msphere.00735-19. https://journals.asm.org/doi/10.1128/msphere.00735-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duim B, van der Graaf-van Bloois L, Timmerman A, Wagenaar JA, Flipse J, Wallinga J et al. Complete Genome Sequence of a Clinical Campylobacter Isolate Identical to a Novel Campylobacter Species. Microbiol Resour Announc [Internet]. 2021. Feb 18 [cited 2024 Nov 26];10(7):e00721–20. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7892663/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Havelaar AH, Brhane M, Ahmed IA, Kedir J, Chen D, Deblais L et al. Unravelling the reservoirs for colonisation of infants with Campylobacter spp. in rural Ethiopia: protocol for a longitudinal study during a global pandemic and political tensions. BMJ Open [Internet]. 2022. Oct [cited 2024 Nov 19];12(10):e061311. https://bmjopen.bmj.com/lookup/doi/10.1136/bmjopen-2022-061311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deblais L, Ojeda A, Brhane M, Mummed B, Hassen KA, Ahmedo BU et al. Prevalence and Load of the Campylobacter Genus in Infants and Associated Household Contacts in Rural Eastern Ethiopia: a Longitudinal Study from the Campylobacter Genomics and Environmental Enteric Dysfunction (CAGED) Project. Appl Environ Microbiol [Internet]. 2023. Jun 13 [cited 2023 Jul 30];89(7):e00424–23. https://journals.asm.org/doi/10.1128/aem.00424-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen D, McKune SL, Yang Y, Usmane IA, Ahmed IA, Amin JK et al. Campylobacter colonization and undernutrition in infants in rural Eastern Ethiopia: a longitudinal community-based birth cohort study [Internet]. medRxiv; 2024. [cited 2024 Nov 19]. p. 2024.05.21.24307707. https://www.medrxiv.org/content/10.1101/2024.05.21.24307707v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen D, Mechlowitz K, Li X, Schaefer N, Havelaar AH, McKune SL. Benefits and Risks of Smallholder Livestock Production on Child Nutrition in Low- and Middle-Income Countries. Front Nutr [Internet]. 2021. Oct 27 [cited 2024 Nov 26];8. https://www.frontiersin.org/journals/nutrition/articles/10.3389/fnut.2021.751686/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R Core Team. R: A Language and Environment for Statistical Computing_. R Foundation for Statistical Computing, Vienna, Austria, <. https://www.R-project.org/. 2023. [Google Scholar]
- 25.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform [Internet]. 2009. Apr 1 [cited 2024 Dec 2];42(2):377–81. https://www.sciencedirect.com/science/article/pii/S1532046408001226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bates D, Mächler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using lme4. J Stat Softw [Internet]. 2015. Oct 7 [cited 2024 Nov 26];67:1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- 27.Chen D, Havelaar AH, Platts-Mills JA, Yang Y. Acquisition and clearance dynamics of Campylobacter spp. in children in low- and middle-income countries. Epidemics [Internet]. 2024. Mar 1 [cited 2024 Dec 15];46:100749. https://www.sciencedirect.com/science/article/pii/S1755436524000100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janssen R, Krogfelt KA, Cawthraw SA, van Pelt W, Wagenaar JA, Owen RJ. Host-Pathogen Interactions in Campylobacter Infections: the Host Perspective. Clin Microbiol Rev [Internet]. 2008. Jul [cited 2024 Dec 15];21(3):505–18. https://journals.asm.org/doi/10.1128/cmr.00055-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.dessouky YE, Elsayed SW, Abdelsalam NA, Saif NA, Álvarez-Ordóñez A, Elhadidy M. Genomic insights into zoonotic transmission and antimicrobial resistance in Campylobacter jejuni from farm to fork: a one health perspective. Gut Pathog [Internet]. 2022. Dec 5 [cited 2024 Dec 15];14(1):44. 10.1186/s13099-022-00517-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strid MA, Engberg J, Larsen LB, Begtrup K, Mølbak K, Krogfelt KA. Antibody Responses to Campylobacter Infections Determined by an Enzyme-Linked Immunosorbent Assay: 2-Year Follow-Up Study of 210 Patients. Clin Diagn Lab Immunol [Internet]. 2001. Mar [cited 2024 Nov 22];8(2):314–9. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC96055/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller G, Dunn GM, Reid TM, Ogden ID, Strachan NJ. Does age acquired immunity confer selective protection to common serotypes of Campylobacter jejuni? BMC Infect Dis [Internet]. 2005. Aug 23 [cited 2024 Nov 22];5:66. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1208888/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Callahan SM, Dolislager CG, Johnson JG. The Host Cellular Immune Response to Infection by Campylobacter Spp. and Its Role in Disease. Infect Immun [Internet]. 2021. Jul 15 [cited 2024 Nov 22];89(8). https://journals.asm.org/doi/10.1128/iai.00116-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Havelaar AH, van Pelt W, Ang CW, Wagenaar JA, van Putten JPM, Gross U et al. Immunity to Campylobacter: its role in risk assessment and epidemiology. Crit Rev Microbiol [Internet]. 2009. Feb 1 [cited 2024 Nov 27]; https://www.tandfonline.com/doi/abs/10.1080/10408410802636017 [DOI] [PubMed] [Google Scholar]
- 34.Simon AK, Hollander GA, McMichael A. Evolution of the immune system in humans from infancy to old age. Proc R Soc B Biol Sci [Internet]. 2015. Dec 22 [cited 2024 Nov 22];282(1821):20143085. https://royalsocietypublishing.org/doi/10.1098/rspb.2014.3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bardales PFG, Schiaffino F, Huynh S, Olortegui MP, Yori PP, Vasquez TP et al. Candidatus Campylobacter infans detection is not associated with diarrhea in children under the age of 2 in Peru. PLoS Negl Trop Dis [Internet]. 2022. Oct 17 [cited 2024 Nov 19];16(10):e0010869. https://journals.plos.org/plosntds/article?id=10.1371/journal.pntd.0010869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parker CT, Schiaffino F, Huynh S, Paredes Olortegui M, Peñataro Yori P, Garcia Bardales PF et al. Shotgun metagenomics of fecal samples from children in Peru reveals frequent complex co-infections with multiple Campylobacter species. PLoS Negl Trop Dis [Internet]. 2022. Oct 4 [cited 2024 Nov 22];16(10):e0010815. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9565744/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Platts-Mills JA, Juma J, Kabir F, Nkeze J, Okoi C et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet Lond Engl [Internet]. 2016. Sep 24 [cited 2024 Nov 20];388(10051):1291. https://pmc.ncbi.nlm.nih.gov/articles/PMC5471845/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donowitz JR, Drew J, Taniuchi M, Platts-Mills JA, Alam M, Ferdous T et al. Diarrheal Pathogens Associated With Growth and Neurodevelopment. Clin Infect Dis [Internet]. 2021. Aug 1 [cited 2024 Nov 22];73(3):e683–91. 10.1093/cid/ciaa1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Investigators MEN. Relationship between growth and illness, enteropathogens and dietary intakes in the first 2 years of life: findings from the MAL-ED birth cohort study. BMJ Glob Health [Internet]. 2017. Dec 1 [cited 2024 Nov 27];2(4):e000370. https://gh.bmj.com/content/2/4/e000370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferreira C, Otani S, Aarestrup FM, Manaia CM. Quantitative PCR versus metagenomics for monitoring antibiotic resistance genes: balancing high sensitivity and broad coverage. FEMS Microbes [Internet]. 2023. Jan 1 [cited 2024 Dec 16];4:xtad008. 10.1093/femsmc/xtad008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daw Elbait G, Daou M, Abuoudah M, Elmekawy A, Hasan SW, Everett DB et al. Comparison of qPCR and metagenomic sequencing methods for quantifying antibiotic resistance genes in wastewater. PLOS ONE [Internet]. 2024. Apr 5 [cited 2024 Dec 16];19(4):e0298325. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10997137/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivanova M, Singh R, Dharmasena M, Gong C, Krastanov A, Jiang X. Rapid identification of Campylobacter jejuni from poultry carcasses and slaughtering environment samples by real-time PCR. Poult Sci [Internet]. 2014. Jun 1 [cited 2024 Dec 2];93(6):1587–97. https://www.sciencedirect.com/science/article/pii/S0032579119323326 [DOI] [PubMed] [Google Scholar]
- 43.Chaban B, Musil KM, Himsworth CG, Hill JE. Development of cpn60-Based Real-Time Quantitative PCR Assays for the Detection of 14 Campylobacter Species and Application to Screening of Canine Fecal Samples. Appl Environ Microbiol [Internet]. 2009. May 15 [cited 2023 Sep 1];75(10):3055–61. https://journals.asm.org/doi/10.1128/aem.00101-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaban B, Ngeleka M, Hill JE. Detection and quantification of 14 Campylobacter species in pet dogs reveals an increase in species richness in feces of diarrheic animals. BMC Microbiol [Internet]. 2010. Mar 10 [cited 2023 Sep 1];10(1):73. 10.1186/1471-2180-10-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hill JE, Paccagnella A, Law K, Melito PL, Woodward DL, Price L, et al. Identification of Campylobacter spp. and discrimination from Helicobacter and Arcobacter spp. by direct sequencing of PCR-amplified cpn60 sequences and comparison to cpnDB, a chaperonin reference sequence database. J Med Microbiol. 2006;55(Pt 4):393–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deidentified individual participant data will be available through Dataverse (https://dataverse.org/) after December 31, 2024.