Abstract
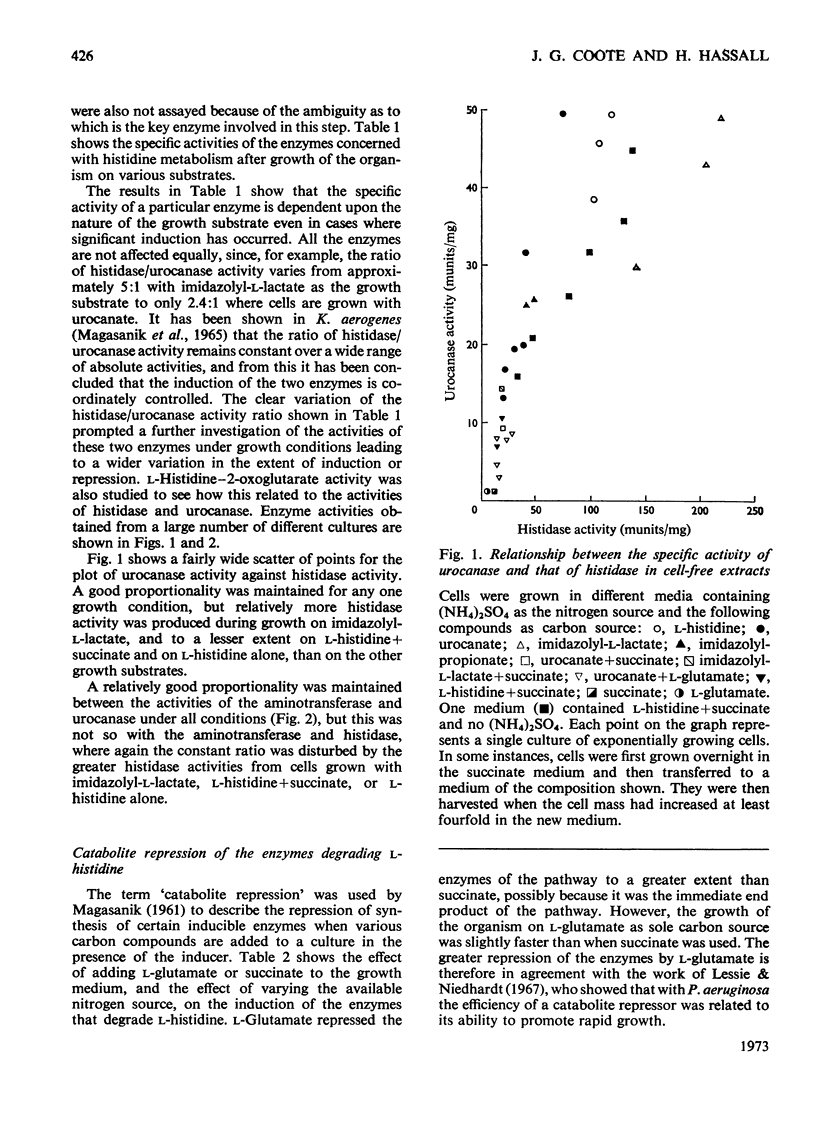
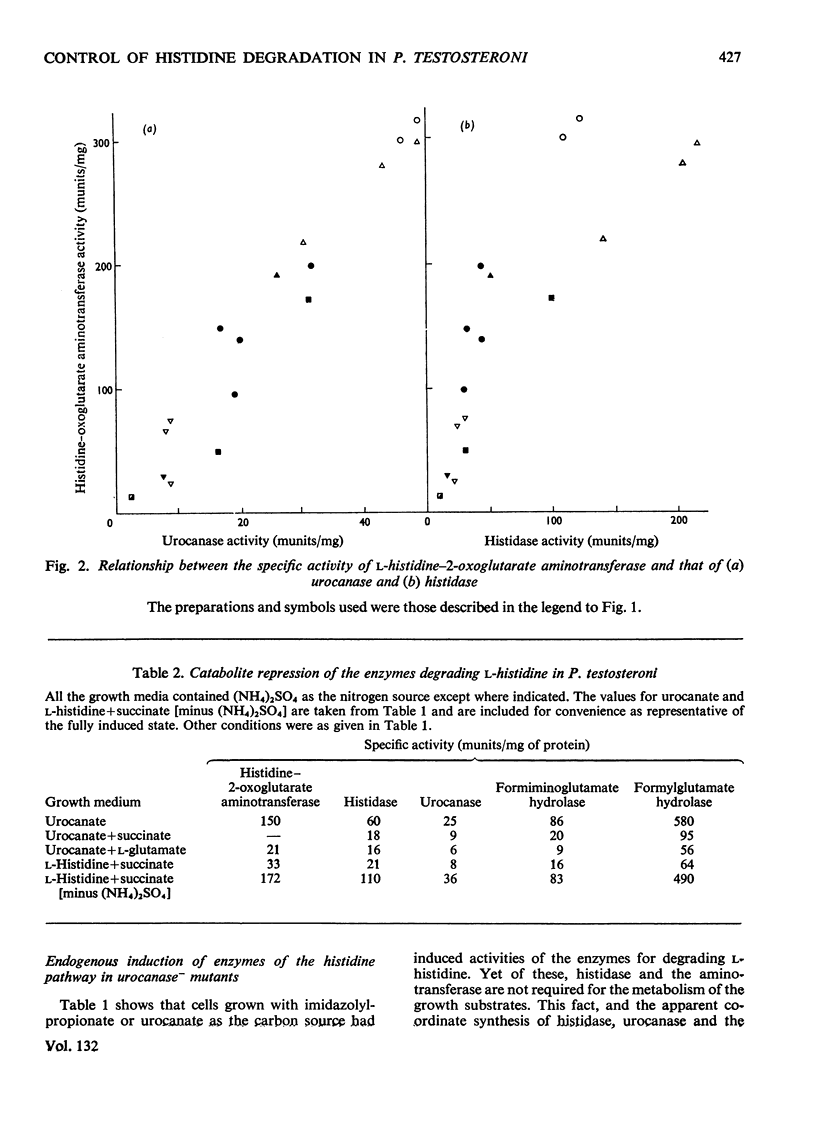
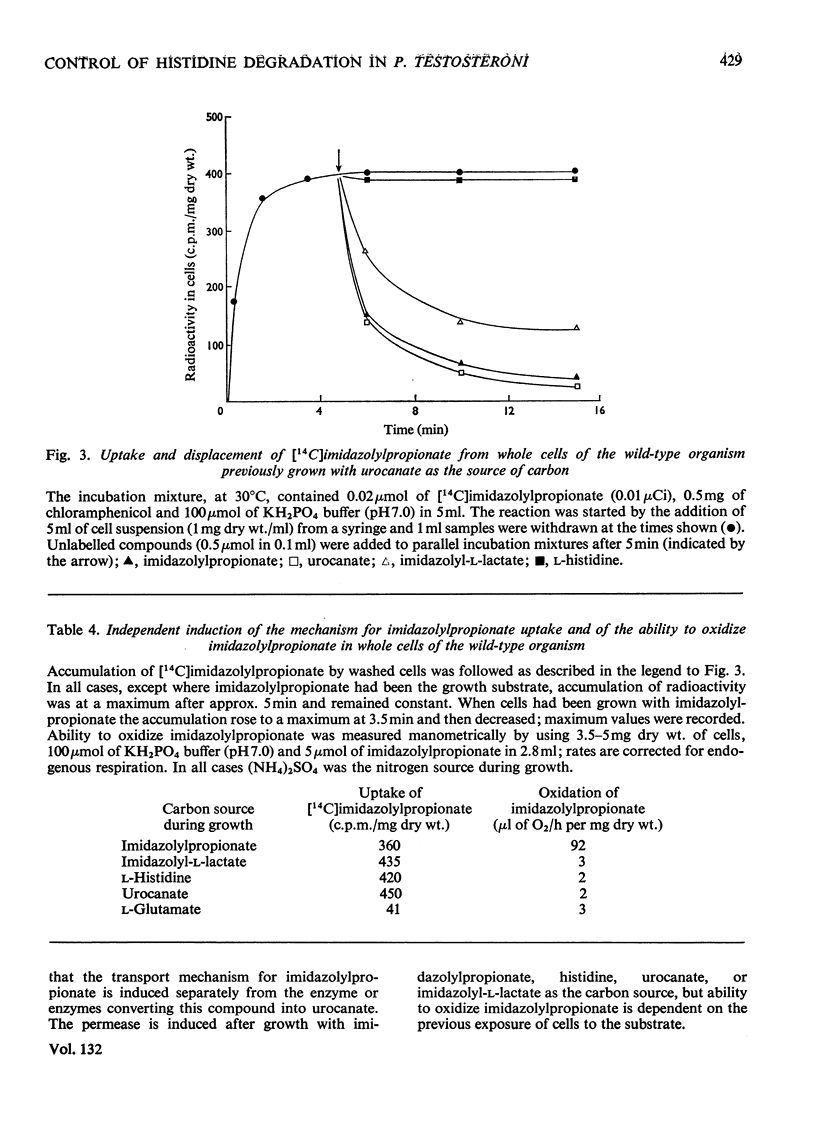
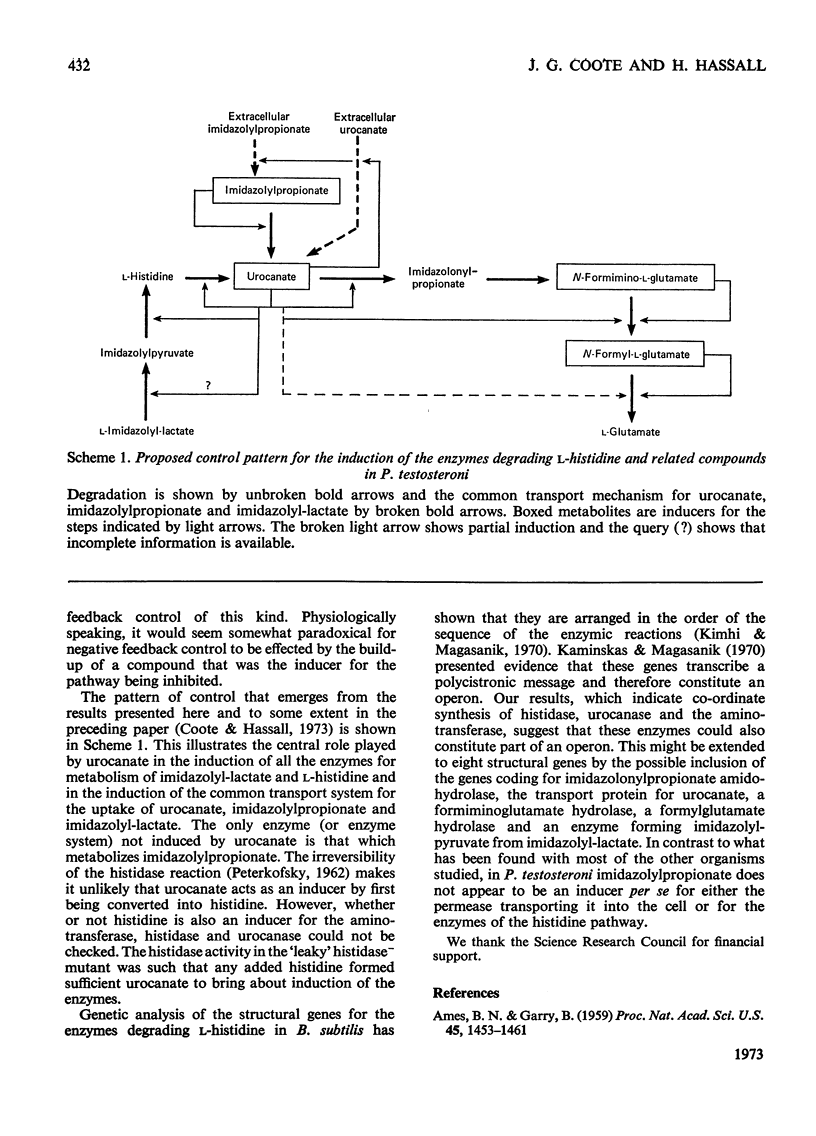
1. The induction of the enzymes for the degradation of l-histidine, imidazolylpropionate and imidazolyl-l-lactate in Pseudomonas testosteroni was investigated. 2. The activities of histidine ammonia-lyase, histidine–2-oxoglutarate aminotransferase and urocanase are consistent with these enzymes being subject to co-ordinate control under most growth conditions. However, a further regulatory mechanism may be superimposed for histidase alone under conditions where degradation of histidine must take place for growth to occur. 3. Experiments with a urocanase− mutant show that urocanate is an inducer for the enzymes given above and also for N-formiminoglutamate hydrolyase and N-formylglutamate hydrolase. 4. N-Formiminoglutamate hydrolase and N-formylglutamate hydrolase are also induced by their substrates, and it is suggested that these two enzymes may be different gene products from those expressed in the presence of urocanate. 5. Induction of the enzyme system for the oxidation of imidazolylpropionate is dependent on exposure of cells to this compound.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames B. N., Garry B. COORDINATE REPRESSION OF THE SYNTHESIS OF FOUR HISTIDINE BIOSYNTHETIC ENZYMES BY HISTIDINE. Proc Natl Acad Sci U S A. 1959 Oct;45(10):1453–1461. doi: 10.1073/pnas.45.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasin L. A., Magasanik B. Induction and repression of the histidine-degrading enzymes of Bacillus subtilis. J Biol Chem. 1968 Oct 10;243(19):5165–5178. [PubMed] [Google Scholar]
- Coote J. G., Hassall H. The degradation of L-histidine, imidazolyl-L-lactate and imidazolylpropionate by Pseudomonas testosteroni. Biochem J. 1973 Mar;132(3):409–422. doi: 10.1042/bj1320409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cánovas J. L., Ornston L. N., Stanier R. Y. Evolutionary significance of metabolic control systems. The beta-ketoadipate pathway provides a case history in bacteria. Science. 1967 Jun 30;156(3783):1695–1699. doi: 10.1126/science.156.3783.1695. [DOI] [PubMed] [Google Scholar]
- Hassall H., Rabie F. The bacterial metabolism of imidazolepropionate. Biochim Biophys Acta. 1966 Feb 28;115(2):521–523. doi: 10.1016/0304-4165(66)90460-0. [DOI] [PubMed] [Google Scholar]
- Hug D. H., Roth D., Hunter J. Regulation of histidine catabolism by succinate in Pseudomonas putida. J Bacteriol. 1968 Aug;96(2):396–402. doi: 10.1128/jb.96.2.396-402.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen D. E., Neidhardt F. C. Effect of growth rate on histidine catabolism and histidase synthesis in Aerobacter aerogenes. J Bacteriol. 1969 Apr;98(1):131–142. doi: 10.1128/jb.98.1.131-142.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminskas E., Magasanik B. Sequential synthesis of histidine-degrading enzymes in Bacillus subtilis. J Biol Chem. 1970 Jul 25;245(14):3549–3555. [PubMed] [Google Scholar]
- Kimhi Y., Magasanik B. Genetic basis of histidine degradation in Bacillus subtilis. J Biol Chem. 1970 Jul 25;245(14):3545–3548. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lessie T. G., Neidhardt F. C. Formation and operation of the histidine-degrading pathway in Pseudomonas aeruginosa. J Bacteriol. 1967 Jun;93(6):1800–1810. doi: 10.1128/jb.93.6.1800-1810.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund P., Magasanik B. N-formimino-L-glutamate formiminohydrolase of Aerobacter aerogenes. J Biol Chem. 1965 Nov;240(11):4316–4319. [PubMed] [Google Scholar]
- MAGASANIK B. Catabolite repression. Cold Spring Harb Symp Quant Biol. 1961;26:249–256. doi: 10.1101/sqb.1961.026.01.031. [DOI] [PubMed] [Google Scholar]
- Magasanik B., Lund P., Neidhardt F. C., Schwartz D. T. Induction and repression of the histidine-degrading enzymes in Aerobacter aerogenes. J Biol Chem. 1965 Nov;240(11):4320–4324. [PubMed] [Google Scholar]
- OHMURA E., HAYAISHI O. Enzymatic conversion of formylaspartic acid to aspartic acid. J Biol Chem. 1957 Jul;227(1):181–190. [PubMed] [Google Scholar]
- Ornston L. N. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. IV. Regulation. J Biol Chem. 1966 Aug 25;241(16):3800–3810. [PubMed] [Google Scholar]
- PETERKOFSKY A. The mechanism of action of histidase: amino-enzyme formation and partial reactions. J Biol Chem. 1962 Mar;237:787–795. [PubMed] [Google Scholar]
- PRICER W. E., Jr, RABINOWITZ J. C. Purine fermentation by Clostridium cylindrosporum. V. Formiminoglycine. J Biol Chem. 1956 Oct;222(2):537–554. [PubMed] [Google Scholar]
- ROSEN H. A modified ninhydrin colorimetric analysis for amino acids. Arch Biochem Biophys. 1957 Mar;67(1):10–15. doi: 10.1016/0003-9861(57)90241-2. [DOI] [PubMed] [Google Scholar]
- SPOLTER P. D., BALDRIDGE R. C. The metabolism of histidine. V. On the assay of enzymes in rat liver. J Biol Chem. 1963 Jun;238:2071–2074. [PubMed] [Google Scholar]
- Schlesinger S., Magasanik B. Imidazolepropionate, a nonmetabolizable inducer for the histidine-degrading enzymes in Aerobacter aerogenes. J Biol Chem. 1965 Nov;240(11):4325–4330. [PubMed] [Google Scholar]
- Schlesinger S., Scotto P., Magasanik B. Exogenous and endogenous induction of the histidine-degrading enzymes in Aerobacter aerogenes. J Biol Chem. 1965 Nov;240(11):4331–4337. [PubMed] [Google Scholar]
- TABOR H., MEHLER A. H. Isolation of N-formyl-L-glutamic acid as an intermediate in the enzymatic degradation of L-histidine. J Biol Chem. 1954 Oct;210(2):559–568. [PubMed] [Google Scholar]


