Abstract
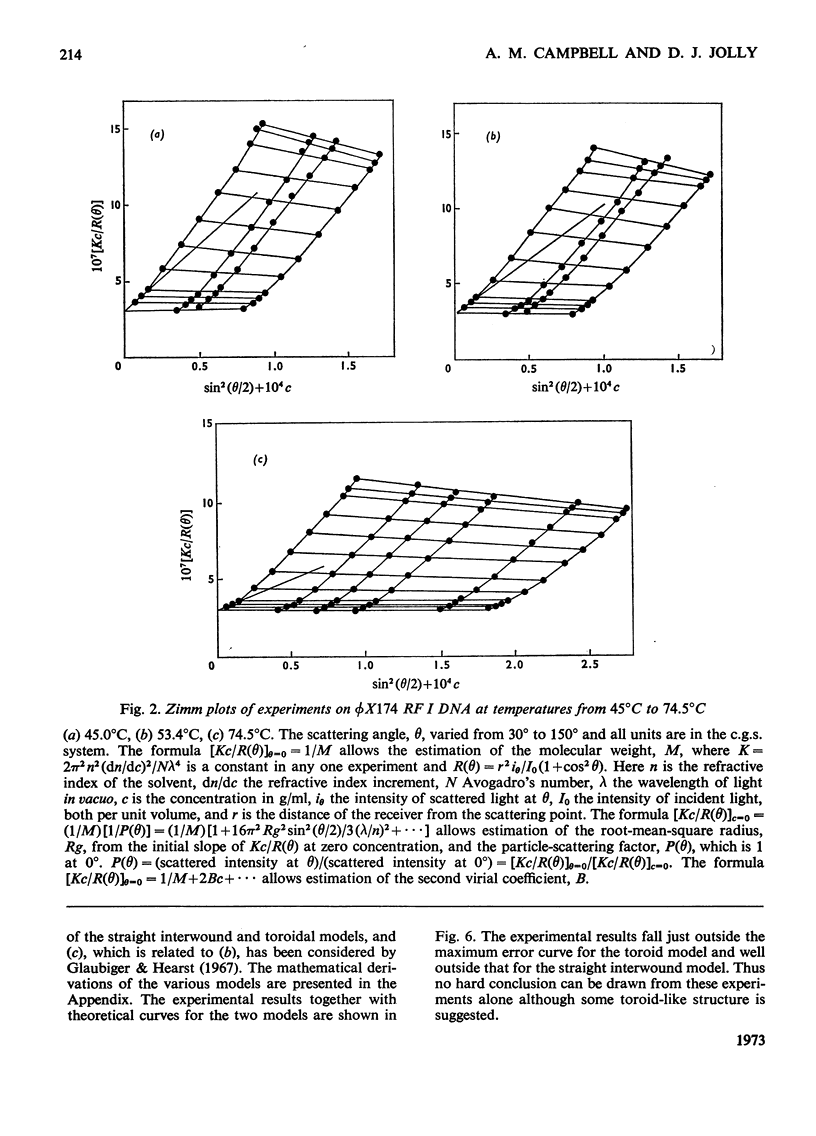
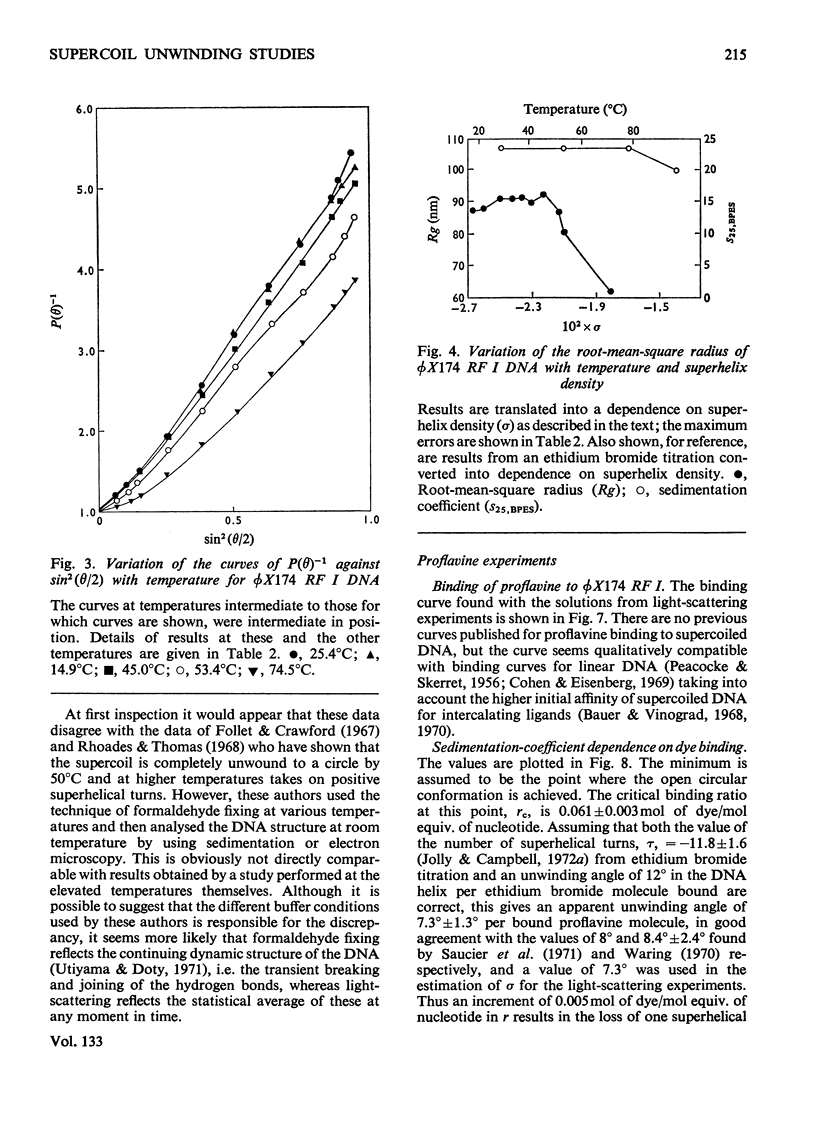
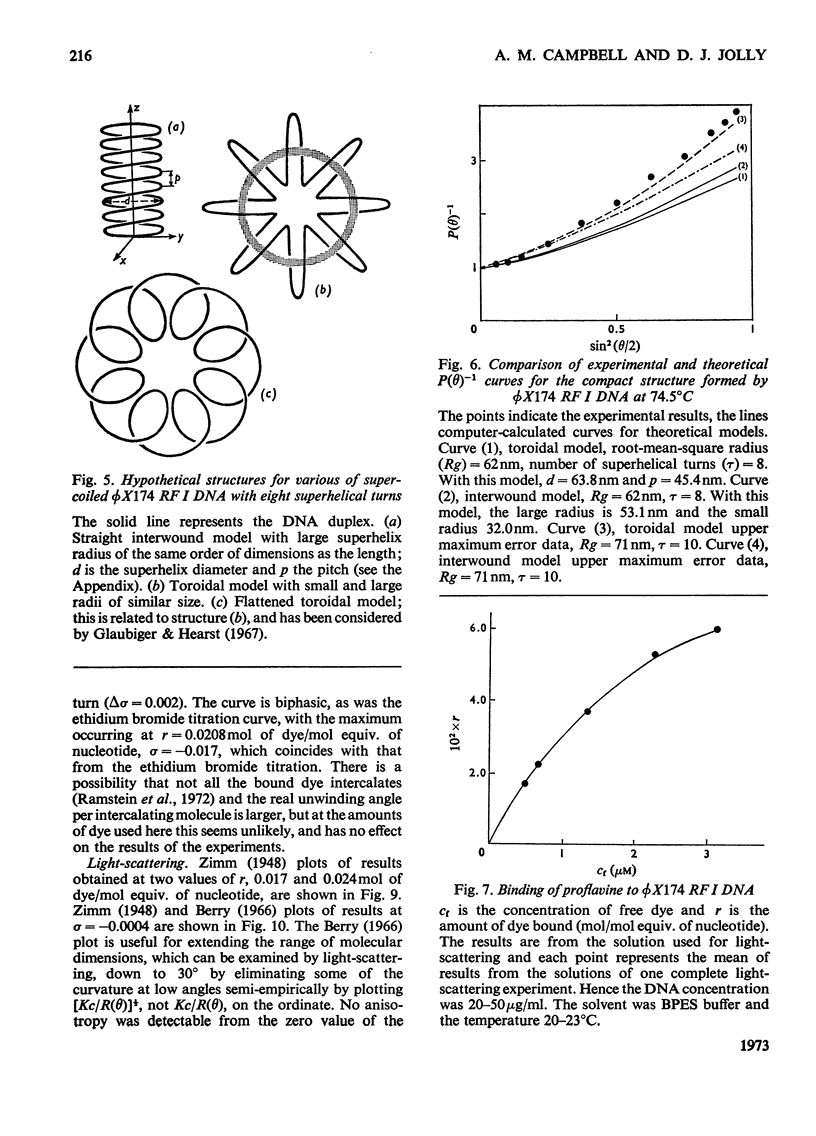
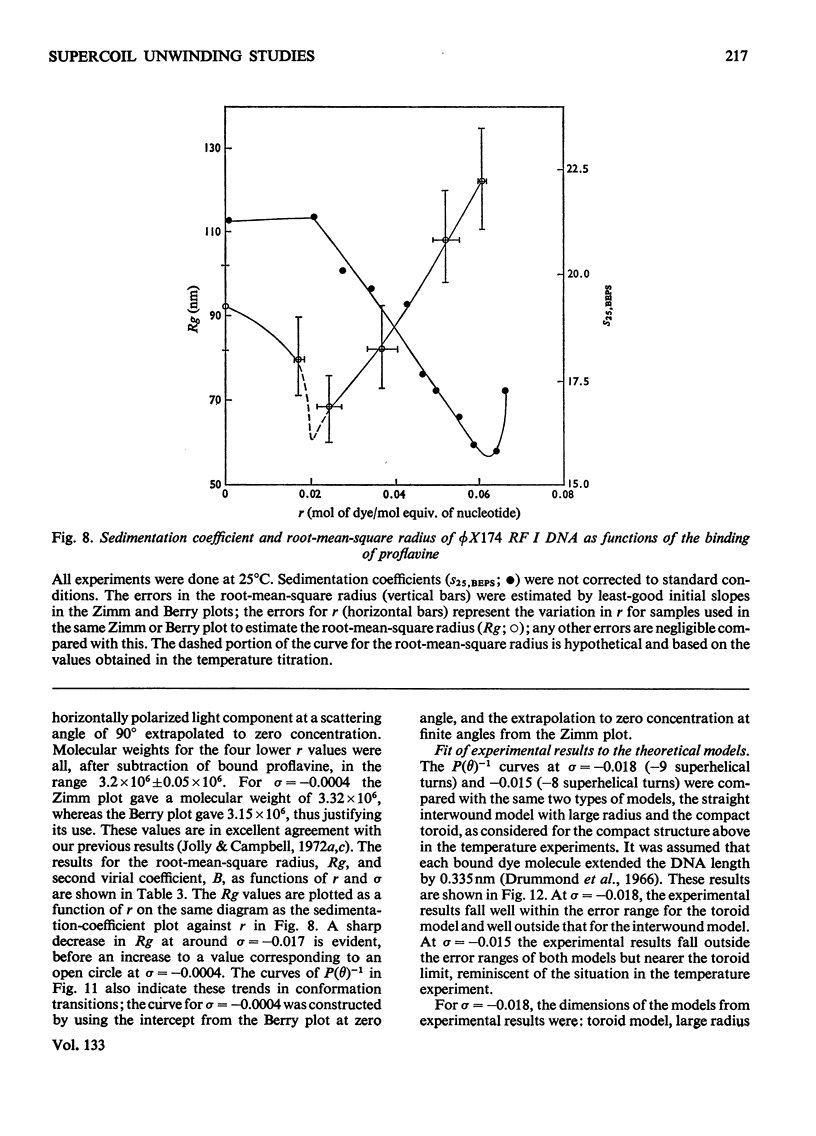
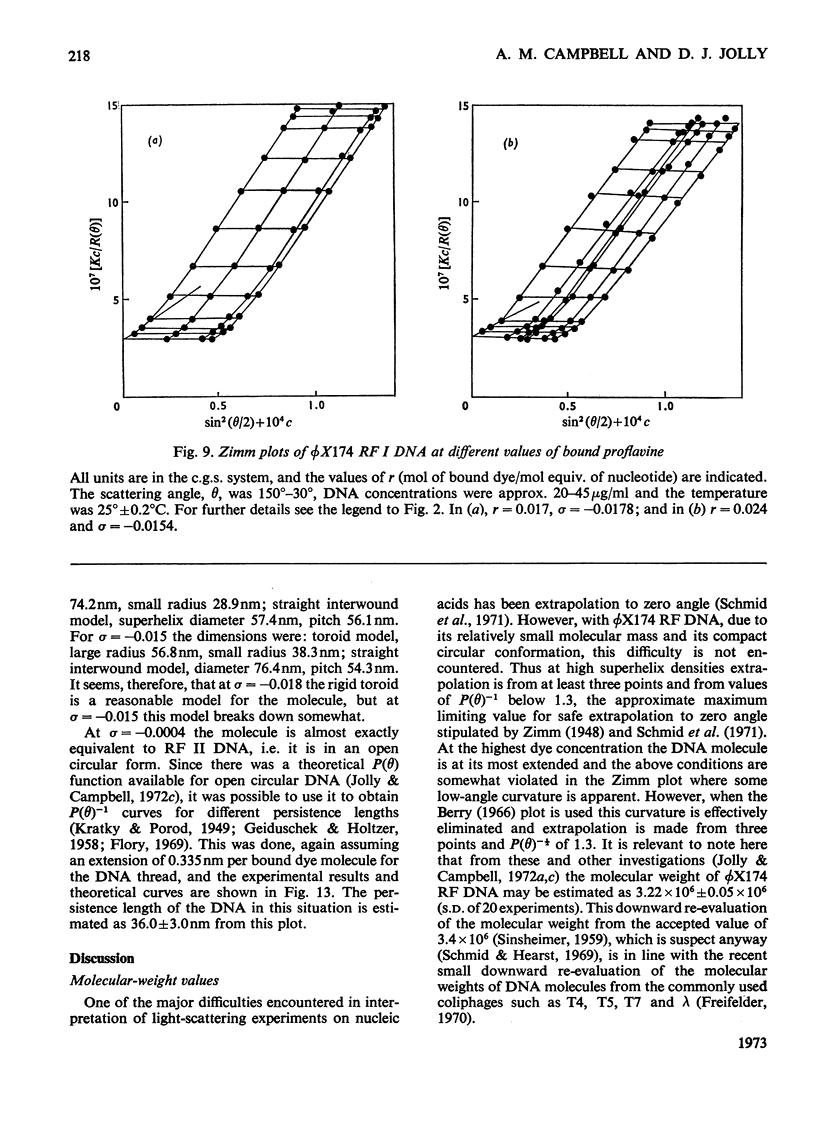
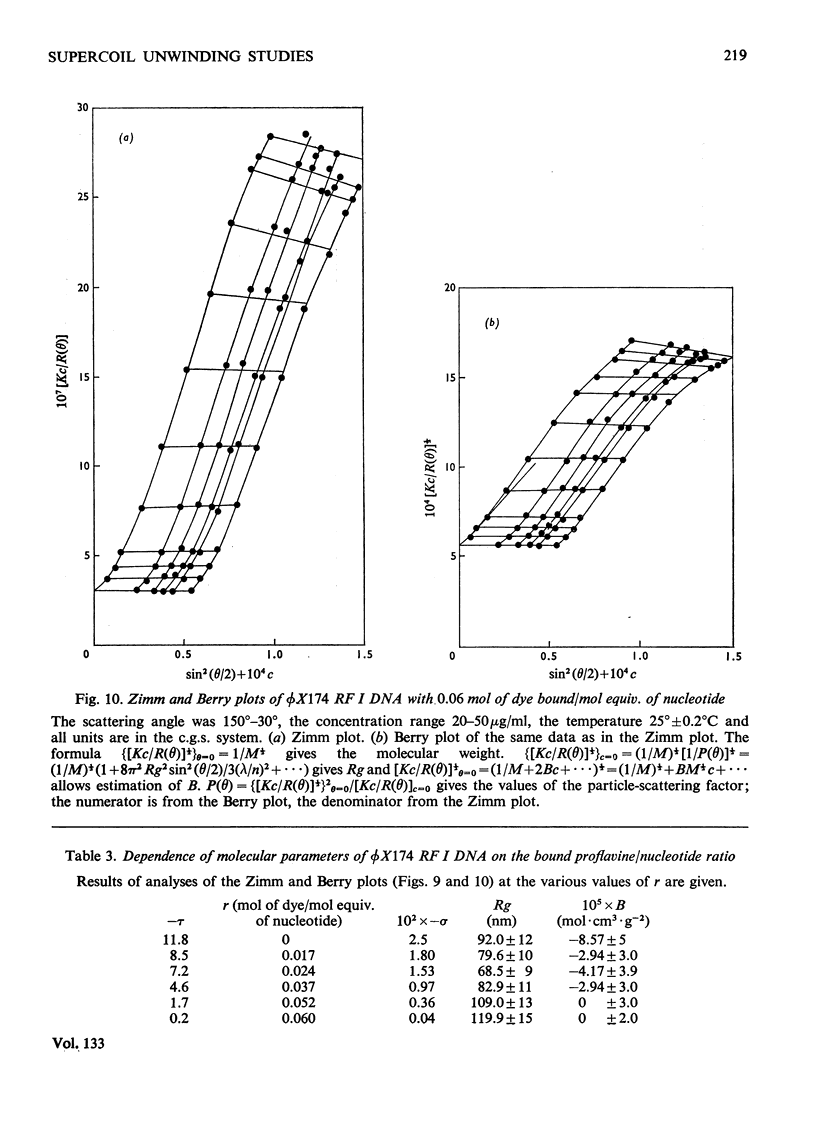
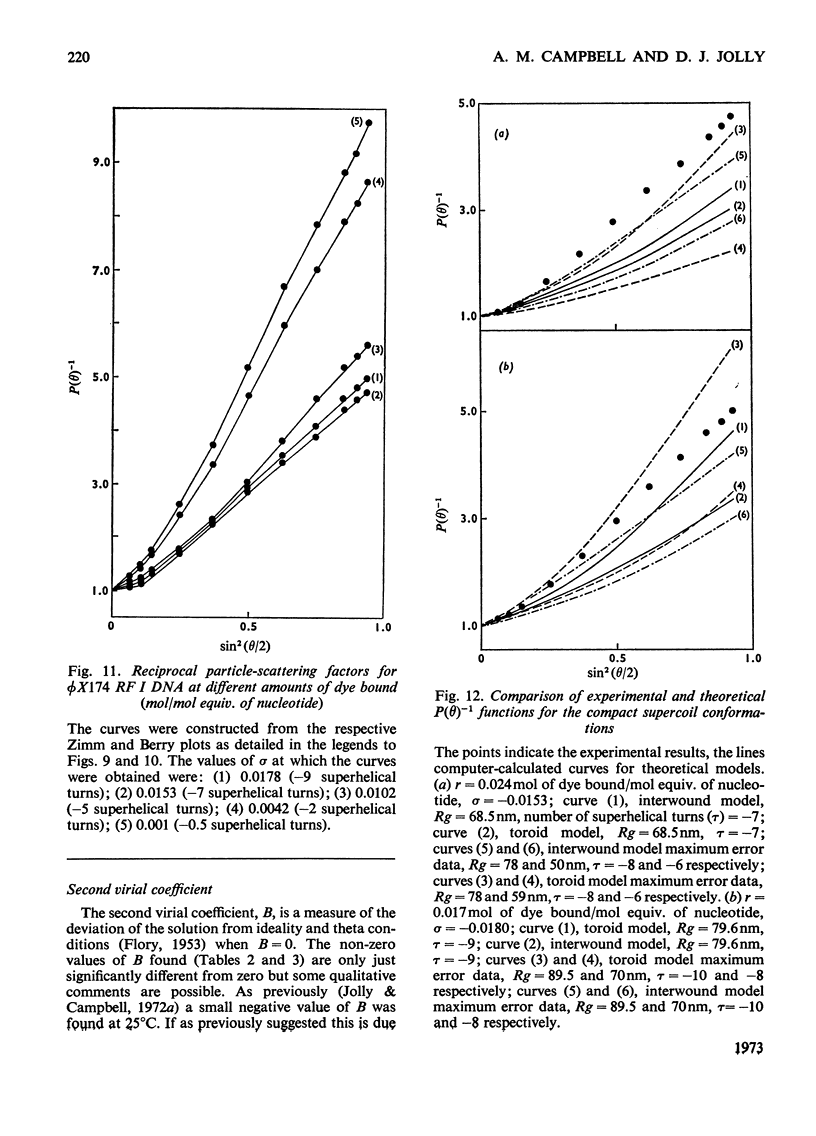
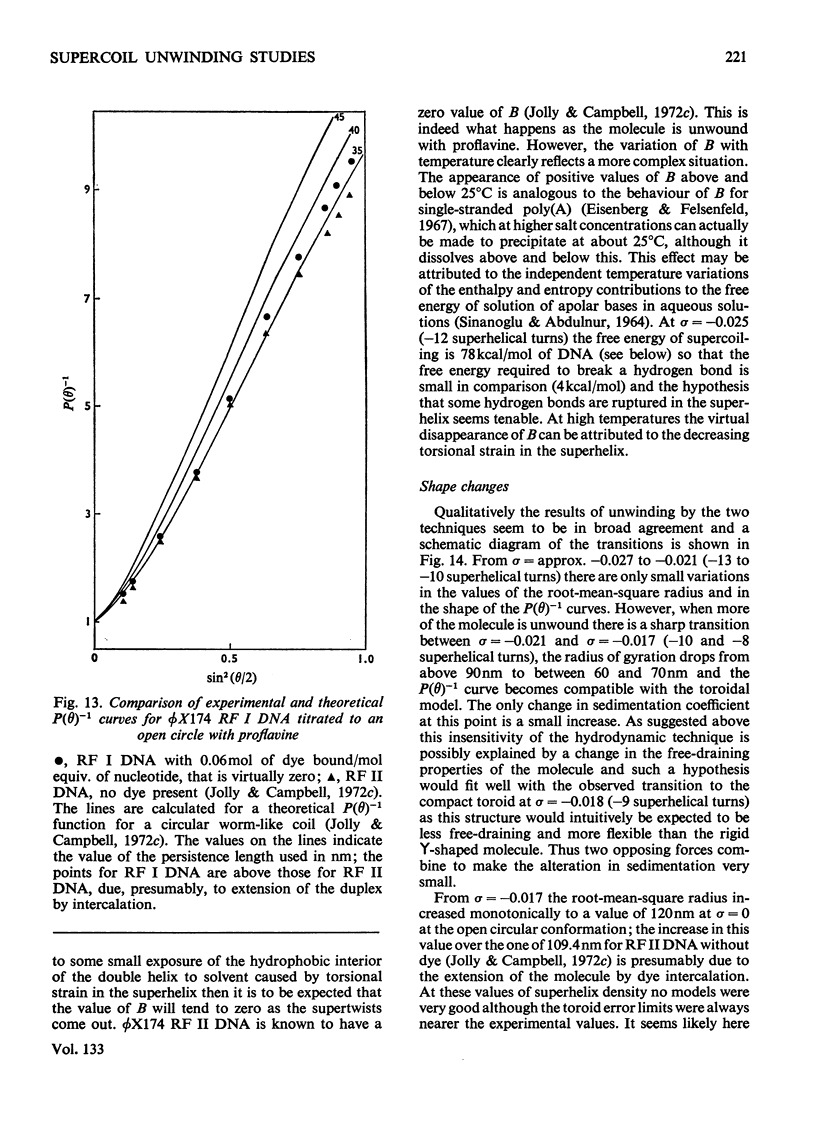
It has been shown previously that supercoiled [unk]X174 bacteriophage intracellular DNA (mol.wt. 3.2×106) with superhelix density, σ=−0.025 (−12 superhelical turns) at 25°C is best represented as a Y shape. In this work two techniques have been used to unwind the supercoil and study the changes in tertiary structure which result from changes in the secondary structure. The molecular weights from all experiments were in the range 3.2×106±0.12×106. In experiments involving temperature change little change in the Y shape was observed between σ=−0.027 (−13 superhelical turns, 14.9°C) and σ=−0.021 (−10 superhelical turns, 53.4°C) as evidenced by the root-mean-square radius and the particle-scattering factor P(θ). However, at σ=−0.0176 (−8 superhelical turns, 74.5°C) the root-mean-square radius fell to between 60 and 70nm from 90nm indicating a large structural change, as did alterations in the P(θ) function. In experiments with the intercalating dye proflavine from values of bound proflavine of 0–0.06mol of dye/mol equiv. of nucleotide which correspond to values of σ from −0.025 to −0.0004 (−12 to 0 superhelical turns) a similar transition was found when the superhelix density was changed by the same amount, and the molecule was shown to go through a further structural change as the unwinding of the duplex proceeded. At σ=−0.018 (−9 superhelical turns) the structure was compatible with a toroid, and at σ=−0.0004 it was compatible with a circle but at no point in the sequence of structure transitions was the structure compatible with the conventional straight interwound model normally visualized as the shape of supercoiled DNA.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauer W., Vinograd J. Interaction of closed circular DNA with intercalative dyes. II. The free energy of superhelix formation in SV40 DNA. J Mol Biol. 1970 Feb 14;47(3):419–435. doi: 10.1016/0022-2836(70)90312-8. [DOI] [PubMed] [Google Scholar]
- Bauer W., Vinograd J. The interaction of closed circular DNA with intercalative dyes. I. The superhelix density of SV40 DNA in the presence and absence of dye. J Mol Biol. 1968 Apr 14;33(1):141–171. doi: 10.1016/0022-2836(68)90286-6. [DOI] [PubMed] [Google Scholar]
- Blake A., Peacocke A. R. The interaction of aminocridines with nucleic acids. Biopolymers. 1968;6(9):1225–1253. doi: 10.1002/bip.1968.360060902. [DOI] [PubMed] [Google Scholar]
- Bloomfield V. A. Twisted circular DNA: sedimentation coefficients and the number of twists. Proc Natl Acad Sci U S A. 1966 Apr;55(4):717–720. doi: 10.1073/pnas.55.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode V. C., MacHattie L. A. Electron microscopy of superhelical circular lambda DNA. J Mol Biol. 1968 Mar 28;32(3):673–679. doi: 10.1016/0022-2836(68)90350-1. [DOI] [PubMed] [Google Scholar]
- Campbell A. M., Lochhead D. S. Optical rotatory dispersion and circular dichroism of superhelical deoxyribonucleic acid. Biochem J. 1971 Jul;123(4):661–663. doi: 10.1042/bj1230661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A. M., Lochhead D. S. Optical rotatory dispersion and circular dichroism of superhelical deoxyribonucleic acid. Biochem J. 1971 Jul;123(4):661–663. doi: 10.1042/bj1230661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford L. V., Waring M. J. Supercoiling of polyoma virus DNA measured by its interaction with ethidium bromide. J Mol Biol. 1967 Apr 14;25(1):23–30. doi: 10.1016/0022-2836(67)90276-8. [DOI] [PubMed] [Google Scholar]
- Crick F. General model for the chromosomes of higher organisms. Nature. 1971 Nov 5;234(5323):25–27. doi: 10.1038/234025a0. [DOI] [PubMed] [Google Scholar]
- Davidson N. Effect of DNA length on the free energy of binding of an unwinding ligand to a supercoiled DNA. J Mol Biol. 1972 May 14;66(2):307–309. doi: 10.1016/0022-2836(72)90482-2. [DOI] [PubMed] [Google Scholar]
- Dawson J. R., Harpst J. A. Light scattering and hydrodynamic properties of linear and circular bacteriophage lambda DNA. Biopolymers. 1971;10(12):2499–2508. doi: 10.1002/bip.360101211. [DOI] [PubMed] [Google Scholar]
- Dean W. W., Lebowitz J. Partial alteration of secondary structure in native superhelical DNA. Nat New Biol. 1971 May 5;231(18):5–8. [PubMed] [Google Scholar]
- Drummond D. S., Pritchard N. J., Simpson-Gildemeister V. F., Peacocke A. R. Interaction of aminoacridines with deoxyribonucleic acid: viscosity of the complexes. Biopolymers. 1966 Oct-Nov;4(9):971–987. doi: 10.1002/bip.1966.360040903. [DOI] [PubMed] [Google Scholar]
- Eisenberg H., Felsenfeld G. Studies of the temperature-dependent conformation and phase separation of polyriboadenylic acid solutions at neutral pH. J Mol Biol. 1967 Nov 28;30(1):17–37. doi: 10.1016/0022-2836(67)90240-9. [DOI] [PubMed] [Google Scholar]
- Follett E. A., Crawford L. V. Electron microscope study of the denaturation of human papilloma virus DNA. I. Loss and reversal of supercoiling turns. J Mol Biol. 1967 Sep 28;28(3):455–459. doi: 10.1016/s0022-2836(67)80095-0. [DOI] [PubMed] [Google Scholar]
- Freifelder D. Molecular weights of coliphages and coliphage DNA. IV. Molecular weights of DNA from bacteriophages T4, T5 and T7 and the general problem of determination of M. J Mol Biol. 1970 Dec 28;54(3):567–577. doi: 10.1016/0022-2836(70)90127-0. [DOI] [PubMed] [Google Scholar]
- Fuller F. B. The writhing number of a space curve. Proc Natl Acad Sci U S A. 1971 Apr;68(4):815–819. doi: 10.1073/pnas.68.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GEIDUSCHEK E. P., HOLTZER A. Application of light scattering to biological systems: deoxyribonucleic acid and the muscle proteins. Adv Biol Med Phys. 1958;6:431–551. doi: 10.1016/b978-1-4832-3112-9.50014-1. [DOI] [PubMed] [Google Scholar]
- GEIDUSCHEK E. P., HOLTZER A. Application of light scattering to biological systems: deoxyribonucleic acid and the muscle proteins. Adv Biol Med Phys. 1958;6:431–551. doi: 10.1016/b978-1-4832-3112-9.50014-1. [DOI] [PubMed] [Google Scholar]
- Gersch N. F., Jordan D. O. Interaction of DNA with aminoacridines. J Mol Biol. 1965 Aug;13(1):138–156. doi: 10.1016/s0022-2836(65)80085-7. [DOI] [PubMed] [Google Scholar]
- Glaubiger D., Hearst J. E. Effect of superhelical structure on the secondary structure of DNA rings. Biopolymers. 1967;5(8):691–696. doi: 10.1002/bip.1967.360050803. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr, Hearst J. E. Flexibility of native DNA from the sedimentation behavior as a function of molecular weight and temperature. J Mol Biol. 1968 Jul 14;35(1):111–129. doi: 10.1016/s0022-2836(68)80041-5. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr Sedimentation coefficient of polyoma virus DNA. Biopolymers. 1967;5(10):1009–1019. doi: 10.1002/bip.1967.360051012. [DOI] [PubMed] [Google Scholar]
- Hayashi M. A DNA-RNA complex as an intermediate of in vitro genetic transcription. Proc Natl Acad Sci U S A. 1965 Dec;54(6):1736–1743. doi: 10.1073/pnas.54.6.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helinski D. R., Clewell D. B. Circular DNA. Annu Rev Biochem. 1971;40:899–942. doi: 10.1146/annurev.bi.40.070171.004343. [DOI] [PubMed] [Google Scholar]
- Hirschman S. Z., Felsenfeld G. Determination of DNA composition and concentration by spectral analysis. J Mol Biol. 1966 Apr;16(2):347–358. doi: 10.1016/s0022-2836(66)80178-x. [DOI] [PubMed] [Google Scholar]
- Hussey H., Baddiley J. Lipid intermediates in the biosynthesis of the wall teichoic acid in Staphylococcus lactis 13. Biochem J. 1972 Mar;127(1):39–50. doi: 10.1042/bj1270039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly D. J., Campbell A. M. Light-scattering studies on deoxyribonucleic acid flexibility. The solution properties of a small circular deoxyribonucleic acid molecule. Biochem J. 1972 Dec;130(4):1019–1028. doi: 10.1042/bj1301019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly D. J., Campbell A. M. The three-dimensional structure of supercoiled deoxyribonucleic acid in solution. Evidence obtained from the angular distribution of scattered light. Biochem J. 1972 Jul;128(3):569–578. doi: 10.1042/bj1280569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly D. J., Campbell A. M. The three-dimensional structure of supercoiled deoxyribonucleic acid in solution. Evidence obtained from the angular distribution of scattered light. Biochem J. 1972 Jul;128(3):569–578. doi: 10.1042/bj1280569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KILKSON R., MAESTRE M. F. Structure of T-2 bacteriophage. Nature. 1962 Aug 4;195:494–495. doi: 10.1038/195494a0. [DOI] [PubMed] [Google Scholar]
- KLEINSCHMIDT A. K., BURTON A., SINSHEIMER R. L. ELECTRON MICROSCOPY OF THE REPLICATIVE FORM OF THE DNA OF THE BACTERIOPHAGE PHI-X174. Science. 1963 Nov 15;142(3594):961–961. doi: 10.1126/science.142.3594.961. [DOI] [PubMed] [Google Scholar]
- Khoury G., Byrne J. C., Martin M. A. Patterns of Simian Virus 40 DNA transcription after acute infection of permissive and nonpermissive cells. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1925–1928. doi: 10.1073/pnas.69.7.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasna A. I. Low-angle light-scattering studies on alkali- and heat-denatured DNA. Biopolymers. 1970;9(9):1029–1038. doi: 10.1002/bip.1970.360090906. [DOI] [PubMed] [Google Scholar]
- LUZZATI V., NICOLAUIEFF A. THE STRUCTURE OF NUCLEOHISTONES AND NUCLEOPROTAMINES. J Mol Biol. 1963 Aug;7:142–163. doi: 10.1016/s0022-2836(63)80043-1. [DOI] [PubMed] [Google Scholar]
- Lindstrom D. M., Dulbecco R. Strand orientation of simian virus 40 transcription in productively infected cells. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1517–1520. doi: 10.1073/pnas.69.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd P. H., Prutton R. N., Peacocke A. R. Sedimentation studies on the interaction of proflavine with deoxyribonucleic acid. Biochem J. 1968 Apr;107(3):353–359. doi: 10.1042/bj1070353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauss Y., Chambron Y. M., Bdaune M., Benoit H. Etude morphologique par diffusion de la lumiéredu complexe formé par le DNA et la proflavine. J Mol Biol. 1967 Aug 14;27(3):579–589. doi: 10.1016/0022-2836(67)90060-5. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Wilkins M. H., Richards B. M. Super-helical model for nucleohistone. Nature. 1967 Jul 29;215(5100):508–509. doi: 10.1038/215508a0. [DOI] [PubMed] [Google Scholar]
- Ramstein J., Dourlent M., Leng M. Interaction between proflavine and deoxyribonucleic acid influence of DNA base composition. Biochem Biophys Res Commun. 1972 May 26;47(4):874–882. doi: 10.1016/0006-291x(72)90574-8. [DOI] [PubMed] [Google Scholar]
- Rhoades M., Thomas C. A., Jr The P22 bacteriophage DNA molecule. II. Circular intracellular forms. J Mol Biol. 1968 Oct 14;37(1):41–61. doi: 10.1016/0022-2836(68)90072-7. [DOI] [PubMed] [Google Scholar]
- Révet B. M., Schmir M., Vinograd J. Direct determination of the superhelix density of closed circular DNA by viscometric titration. Nat New Biol. 1971 Jan 6;229(1):10–13. doi: 10.1038/newbio229010a0. [DOI] [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Saucier J. M., Festy B., Le Pecq J. B. The change of the torsion of the DNA helix caused by intercalation. II. Measurement of the relative change of torsion induced by various intercalating drugs. Biochimie. 1971;53(9):973–980. doi: 10.1016/s0300-9084(71)80065-2. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Hearst J. E. Molecular weights of homogeneous coliphage DNA's from density-gradient sedimentation equilibrium. J Mol Biol. 1969 Aug 28;44(1):143–160. doi: 10.1016/0022-2836(69)90410-0. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Rinehart F. P., Hearst J. E. Statistical length of DNA from light scattering. Biopolymers. 1971;10(5):883–893. doi: 10.1002/bip.360100511. [DOI] [PubMed] [Google Scholar]
- Shih T. Y., Fasman G. D. Circular dichroism studies of deoxyribonucleic acid complexes with arginine-rich histone IV (f2al). Biochemistry. 1971 Apr 27;10(9):1675–1683. doi: 10.1021/bi00785a027. [DOI] [PubMed] [Google Scholar]
- Triebel H., Reinert K. E., Strassburger J. Persistence length of DNA from hydrodynamic measurements. Biopolymers. 1971;10(12):2619–2621. doi: 10.1002/bip.360101222. [DOI] [PubMed] [Google Scholar]
- Upholt W. B., Gray H. B., Jr, Vinograd J. Sedimentation velocity behavior of closed circular SV40 DNA as a function of superhelix density, ionic strength, counterion and temperature. J Mol Biol. 1971 Nov 28;62(1):21–38. doi: 10.1016/0022-2836(71)90128-8. [DOI] [PubMed] [Google Scholar]
- Utiyama H., Doty P. Kinetic studies of denaturation and reaction with formaldehyde on polydeoxyribonucleotides. Biochemistry. 1971 Mar 30;10(7):1254–1264. doi: 10.1021/bi00783a024. [DOI] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J. Physical and topological properties of circular DNA. J Gen Physiol. 1966 Jul;49(6):103–125. doi: 10.1085/jgp.49.6.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Radloff R., Watson R., Laipis P. The twisted circular form of polyoma viral DNA. Proc Natl Acad Sci U S A. 1965 May;53(5):1104–1111. doi: 10.1073/pnas.53.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Watson R. Early and late helix-coil transitions in closed circular DNA. The number of superhelical turns in polyoma DNA. J Mol Biol. 1968 Apr 14;33(1):173–197. doi: 10.1016/0022-2836(68)90287-8. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Degree of superhelicity of covalently closed cyclic DNA's from Escherichia coli. J Mol Biol. 1969 Jul 28;43(2):263–272. doi: 10.1016/0022-2836(69)90266-6. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971 Feb 14;55(3):523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- Waring M. J. Complex formation between ethidium bromide and nucleic acids. J Mol Biol. 1965 Aug;13(1):269–282. doi: 10.1016/s0022-2836(65)80096-1. [DOI] [PubMed] [Google Scholar]
- Waring M. Variation of the supercoils in closed circular DNA by binding of antibiotics and drugs: evidence for molecular models involving intercalation. J Mol Biol. 1970 Dec 14;54(2):247–279. doi: 10.1016/0022-2836(70)90429-8. [DOI] [PubMed] [Google Scholar]
- Westphal H. SV40 DNA strand selection by Escherichia coli RNA polymerase. J Mol Biol. 1970 Jun 14;50(2):407–420. doi: 10.1016/0022-2836(70)90201-9. [DOI] [PubMed] [Google Scholar]


