Abstract
Background
The human genome contains at least 18 genes for Nudix hydrolase enzymes. Many have similar functions to one another. In order to understand their roles in cell physiology, these proteins must be characterised.
Results
We have characterised two novel human gene products, hAps1, encoded by the NUDT11 gene, and hAps2, encoded by the NUDT10 gene. These cytoplasmic proteins are members of the DIPP subfamily of Nudix hydrolases, and differ from each other by a single amino acid. Both metabolise diadenosine-polyphosphates and, weakly, diphosphoinositol polyphosphates. An apparent polymorphism of hAps1 has also been identified, which leads to the point mutation S39N. This has also been characterised. The favoured nucleotides were diadenosine 5',5"'-pentaphosphate (kcat/Km = 11, 8 and 16 × 103M-1s-1 respectively for hAps1, hAps1-39N and hAps2) and diadenosine 5',5"'-hexaphosphate (kcat/Km = 13, 14 and 11 × 103M-1s-1 respectively for hAps1, hAps1-39N and hAps2). Both hAps1 and hAps2 had pH optima of 8.5 and an absolute requirement for divalent cations, with manganese (II) being favoured. Magnesium was not able to activate the enzymes. Therefore, these enzymes could be acutely regulated by manganese fluxes within the cell.
Conclusions
Recent gene duplication has generated the two Nudix genes, NUDT11 and NUDT10. We have characterised their gene products as the closely related Nudix hydrolases, hAps1 and hAps2. These two gene products complement the activity of previously described members of the DIPP family, and reinforce the concept that Ap5A and Ap6A act as intracellular messengers.
Background
In addition to the canonical ribonucleoside and deoxyribonucleoside phosphates and cofactors, cells contain a large number of minor nucleotides. Among these are the diadenosine polyphosphates (ApnA, where n = 2–7 [1]). Ap3A and Ap4A are the most intensively studied of these and are generally present in the soluble fraction of eukaryotic and prokaryotic cells at concentrations between 10 nM and 5 μM [2]. Platelet dense granules, adrenal chromaffin granules and certain synaptic vesicles have been reported to contain high concentrations of Ap5A and Ap6A in addition to Ap3A and Ap4A, all of which can be exocytosed following appropriate stimuli and bind to target cell purinoceptors causing a variety of physiological responses in the cardiovascular and central and peripheral nervous systems [1,3-5]. However, although Ap6A has been detected in erythrocytes [6], there are no substantiated measurements of Ap5A and Ap6A in the soluble fraction of nucleated cells, and it is likely that they are typically present at concentrations much lower than those of Ap3A and Ap4A. The estimated 7 μM Ap5A reported in cardiac muscle [7] may also be confined to granules.
Whilst the extracellular diadenosine polyphosphates have partially characterised signalling properties, the possible functions of the soluble, intracellular compounds remain unclear. On the one hand, they may simply be by-products of several enzymic reactions (e.g. those catalysed by aminoacyl-tRNA synthetases and other ligases [8]). On the other, they may have one or more of a number of important regulatory roles, including involvements in DNA replication/repair, metabolic stress responses, the determination of cellular fate, and the regulation of enzyme activities and ion channels (see [1]). For example, Ap5A is a potent inhibitor of adenylate kinase [9].
It is clear that cells have a variety of relatively specific enzymes able to degrade these compounds. These include symmetrically-cleaving Ap4A hydrolases (in prokaryotes), members of the histidine triad protein family such as the FHIT tumour suppresser protein and (in lower eukaryotes) the related Ap4A phosphorylases, and most widespread of all, members of the Nudix hydrolase family (see [10,11] for reviews). Nudix hydrolases cleave predominantly the diphosphate linkage in compounds of general structure: nucleoside diphosphate linked to another moiety, X. Some family members have a relatively broad substrate specificity, while others have a much more restricted range. They all possess the Nudix sequence signature motif Gx5Ex5 [UA]xREx2EexGU (where U is an aliphatic hydrophobic amino acid), which represents the catalytic site of the enzyme. Animal and plant Ap4A hydrolases (EC 3.1.6.17) degrade Ap4A, Ap5A and Ap6A, always producing ATP as one of the products. They are most active towards Ap4A; however, the related enzymes from Escherichia coli [12] and Rickettsia prowazekii [13] show a marked preference for Ap5A.
More recently, distinct members of the Nudix family that prefer Ap6A have been purified and characterised. The three human enzymes termed diphosphoinositol polyphosphate phosphohydrolases 1 [14,15], 2α and 2β [16] (hDIPP-1, -2α and -2β) as well as Schizosaccharomyces pombe Aps1 [17] and Saccharomyces cerevisiae Ddp1p [18] all favour Ap6A over Ap5A. Activity with Ap4A is lower, and is in fact lacking in the case of Ddp1p. Remarkably these Ap6A hydrolases are also able to utilise the structurally unrelated non-nucleotide substrates, diphosphoinositol pentakisphosphate (PP-InsP5) and bis-diphosphoinositol tetrakisphosphate ([PP]2-InsP4), hence they are commonly referred to as the DIPP subfamily. These substrates are generally favoured in vitro, with kcat/Km ratios being 50–500-fold higher for PP-InsP5 compared to Ap6A, except for DIPP-2β, which has a significantly reduced relative activity with PP-InsP5 compared to the others. The intracellular levels of diphosphoinositol polyphosphates appear to be lower than those of the nucleotide substrates. PP-InsP5 levels are often between 1 [19] and 3% [20,21] of the levels of InsP6, which has been estimated as being between 15 and 100 μM (see [22]) and [PP]2-InsP4 levels are an order of magnitude lower [20,21], and as such are virtually undetectable in many cell types. PP-InsP5 turnover is regulated by the tumour promoter thapsigargin [19], while [PP]2-InsP4 turnover is regulated by cyclic AMP- and cyclic GMP-dependent processes via an undefined mechanism [23]. However, as for the ApnAs, no clear functions have yet been found for the diphosphoinositol polyphosphates, although they have been implicated in vesicle trafficking [24], apoptosis [25] and DNA repair [26].
We describe here two further members of the human DIPP subfamily with novel properties. These proteins, hAps1 and hAps2, products of the NUDT11 and NUDT10 genes respectively, closely resemble the other DIPPs in primary structure, but show a selectivity towards Ap5A and Ap6A and reduced activity towards PP-InsP5 and [PP]2-InsP4. They also display a novel pattern of tissue-specific expression.
Results
Sequence alignment
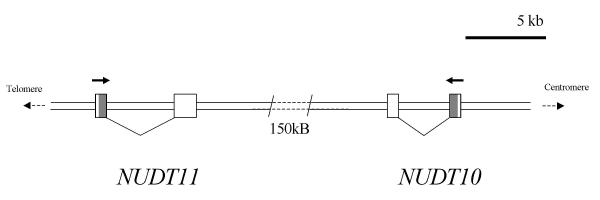
An initial BLAST search of the GenBank expressed sequence tag (EST) database with the hDIPP-1 sequence identified two closely related but previously uncharacterised predicted proteins, which we have called hAps1 and hAps2 (human ApsixA hydrolases 1 and 2). Alignment of the cDNA sequences with the human genome indicated that the genomic sequences encoding these proteins, FLJ10628 and LOC139770, lie about 150 kb apart on the X chromosome at Xp11.23 and are transcribed in opposite directions (Fig. 1). In accordance with the guidelines for the Nudix protein family, the genes for hAps1 and hAps2 have been designated NUDT11 and NUDT10 respectively by the HUGO Gene Nomenclature Committee (see http://www.gene.ucl.ac.uk/nomenclature/genefamily/npym.html). These proteins are each 164 amino acids long and are most similar to hDIPP-2β, hAps1 showing 76, 90 and 91% identity at the amino acid level to hDIPP-1, 2α and 2β, respectively. In particular, they both possess the additional Gln residue (Q86 in DIPP-2β) that distinguishes hDIPP-2β from hDIPP-2α and hDIPP-1 and which is responsible for the reduced activity of hDIPP-2β towards diphosphoinositol polyphosphates compared to the other DIPPs. hAps1 and hAps2 are also identical to the hDIPP-2 enzymes throughout the Nudix motif (Fig. 2). These observations led us to anticipate that hAps1 and hAps2 would be potent dinucleoside polyphosphate hydrolases with reduced activity towards diphosphoinositol polyphosphates. The existence of two such similar expressed gene products was surprising. Comparison of the open reading frames (ORFs) of hAps1 and hAps2 revealed 6 differences at the nucleotide level, leading to five silent changes (T8, E28, E69, D88 and stop), and an Arg-Pro substitution at position 89. A comparison of the cDNAs for the two gene products showed further divergence in the 3' untranslated region. It appears that the evolution of these closely related genes occurred recently, as hAps1 and hAps2 show greater similarity with each other than with the Aps-like sequences from mouse (of which two genes, XM_135784 and XM_135786 appear to yield the same protein product) or cow (Fig 3). Further examination of all available hAps1 ESTs revealed a sequence divergence, leading to a point mutation S39N, in the coding region. This is present in two ESTs (Table 1) and was detected in 10% (3 from 30) of polymerase chain reaction (PCR) products generated from a range of normal human cDNA samples from diverse sources. In all other respects investigated, including the 3' untranslated region, it appeared to be identical to hAps1. This would suggest that this minor variant, hAps1-39N, represents a polymorphism present in the human population. Its properties were also examined in this study.
Figure 1.
The NUDT11/NUDT10 region of chromosome Xp11.23. The two known exons of NUDT11, encoding hAps1, and NUDT10, encoding hAps2 are shown represented as boxes. Introns are represented by angled connecting lines. The coding region of each gene is shaded. Solid arrows indicate the direction of transcription of each gene.
Figure 2.
Sequence alignment of hAps1, hAps2, hDIPP-1 and hDIPP-2 (α and β). The figure shows an alignment of the amino acid sequences of hAps1, hAps2, hDIPP-1 and hDIPP-2 (α and β), generated by the MAP algorithm. Black shading denotes amino acids in at least two of the proteins were identical. Physicochemical similarity is denoted by grey shading.
Figure 3.
Comparison of hAps1 with translated EST sequences from mouse and cow. No full-length bovine EST for the hAps1 homologue is available, hence the composite of two separate sequences is shown. A predicted rat protein from genomic sequencing is also shown.
Table 1.
Tissue sources of known ESTs for hAps1 and hAps2
| Tissue | hAps1 | hAps2 |
| Brain | 15434059a, hippocampus, hAps1 | 15936725, medulla |
| 15495588, hypothalamus, hAps1 | 13980266, hypothalamus | |
| 15743679, pooled brain | ||
| Testis | 14001386, hAps1 | 14083118 |
| Breast | 15996913, hAps1 | |
| Ovary | 19353207, hAps1 | 18780184 |
| Uterus | 11970540, tumour cell line, hAps1 | |
| Eye | 19049709, hAps1 | |
| Teratocarcinoma | 7022778, 10989803, hAps1-39N |
aNumbers are GenBank gene identifiers
Purification and properties of hAps1 and hAps2
The ORFs for hAps1, hAps1-39N and hAps2 were cloned into the pGEX6P-1 expression vector and expressed in Escherichia coli (E. coli). After incubation with isopropyl β-D-thiogalactoside (IPTG) (100 μM) at 26°C for 8 hours, a major protein of the expected size was induced and expressed in a soluble form in each case. The glutathione S-transferase (GST)-tagged recombinant proteins were purified by chromatography on Glutathione Sepharose 4 Fast Flow resin (Fig. 4).
Figure 4.
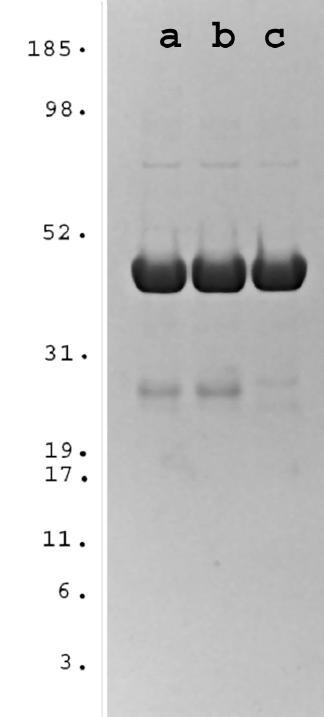
SDS-PAGE of purified hAps1, hAps1-39N and hAps2. GST-tagged hAps proteins were prepared as described under "Materials and methods". Approximately 10 μg of (a) hAps1, (b) hAps1-39N and (c) hAps2 were analysed by SDS-PAGE. Purity was determined by Coomassie-blue staining a 4–12% Bis-Tris NuPage gel (Novex). Molecular weight standards (MultiMark multi-colored standards) were from Novex.
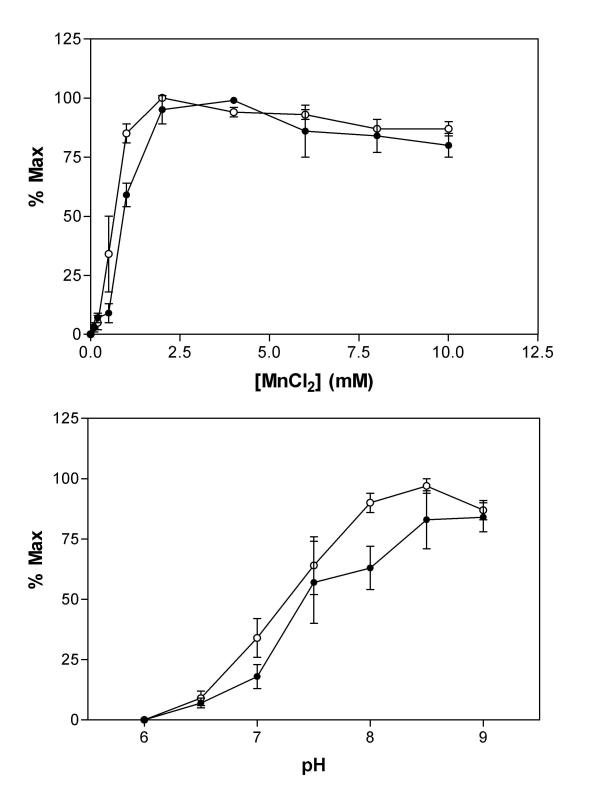
Initially, the ability of hAps1 and hAps2 to metabolise Ap5A was determined at 37°C in the presence of 50 mM HEPES, pH 7.6 and 100 μM substrate. A divalent cation was essential for activity, with Mn2+ by far the most effective between 2 and 6 mM (Fig. 5a). Cu2+ supported less than 30% and Zn2+ and Co2+ each less than 3% of the maximum activity. Ni2+, Ca2+ and, surprisingly, Mg2+ were unable to activate hAps1 or hAps2. When assayed in the presence of 1 mM MnCl2, hAps1 and hAps2 showed alkaline pH optima of pH 8.5 (Fig. 5b), a feature common among Nudix hydrolases. Activity increased from undetectable at pH 6 to over 70% maximum between 7.5 and 9. Previous characterisation of hDIPP-1, 2α and 2β, as well as S. pombe Aps1 was performed in a buffer containing 1 mM MnCl2 at pH 7.6. In order to compare results for hAps1 and hAps2 directly with hDIPP-1, hDIPP-2 and S. pombe Aps1, we have performed assays using identical conditions.
Figure 5.
Properties of hAps1 and hAps2. (a) Mn2+-dependency of hAps1 (filled circles) and hAps2 (open circles) activity towards Ap5A (100 μM). (b) pH dependency of hAps1 (filled circles) and hAps2 (open circles) activity towards Ap5A (100 μM).
When hAps1, hAps1-39N and hAps2 were assayed with a wide range of potential substrates all three proteins were found, as expected, to metabolise Ap5A and Ap6A. Of the other substrates tested, only p4A and p4G showed significant rates of hydrolysis by all three, while Ap4A and Gp4G were also effective substrates for hAps2. All three enzymes showed a marked preference for adenine over guanine nucleotides. Table 2 shows rates of metabolism and products formed where identified. Km and kcat values for each enzyme with Ap5A and Ap6A are shown in Table 3. The most rapidly metabolised substrate appeared to be Ap5A, although Ap6A was bound with higher affinity.
Table 2.
Nucleotide substrate utilisation by hAps1, hAps1-39N and hAps2
| Nucleotide | Rate (nmol/min/μg) | Major products | ||
| hAps1 | hAps1-39N | hAps2 | ||
| Ap6A | 0.25 ± 0.04 | 0.19 ± 0.01 | 0.28 ± 0.04 | AMP + p5A |
| Ap5A | 0.41 ± 0.05 | 0.37 ± 0.02 | 0.9 ± 0.1 | AMP + p4A |
| Ap4A | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.09 ± 0.03 | AMP + ATP |
| Ap3A | 0 | 0 | 0 | |
| Ap2A | 0 | 0 | 0 | |
| Gp5G | 0.04 ± 0.02 | 0.02 ± 0.01 | 0 | |
| Gp4G | 0.04 ± 0.02 | 0.04 ± 0.02 | 0.11 ± 0.04 | GMP, GDP, GTP |
| Gp3G | 0 | 0 | 0 | |
| Gp2G | 0 | 0 | 0 | |
| Ap5G | 0 | 0 | 0 | |
| Ap4G | 0 | 0 | 0 | |
| p4A | 0.07 ± 0.03 | 0.10 ± 0.03 | 0.13 ± 0.04 | ATP |
| ATP | 0 | 0.03 ± 0.02 | 0 | |
| ADP-ribose | 0 | 0 | 0 | |
| ADP-glucose | 0 | 0 | 0 | |
| dATP | 0 | 0.07 ± 0.04 | 0 | |
| p4G | 0.15 ± 0.05 | 0.21 ± 0.05 | 0.21 ± 0.03 | GTP |
| GTP | 0.02 ± 0.01 | 0.03 ± 0.01 | 0 | |
| GDP-glucose | 0.01 ± 0 | 0 | 0 | |
| GDP-mannose | 0 | 0 | 0 | |
| dGTP | 0.05 ± 0.03 | 0.05 ± 0.03 | 0 | |
| dUTP | 0.04 ± 0.03 | 0.05 ± 0.02 | 0 | |
| dTTP | 0.05 ± 0.04 | 0.05 ± 0.03 | 0 | |
| CTP | 0.03 ± 0.02 | 0.03 ± 0.02 | 0 | |
Table 3.
Kinetic parameters of hAps1, hAps1-39N and hAps2
| Enzyme | Substrate | Km (μM) | kcat (s-1) | kcat/Km (103M-1s-1) |
| hAps1 | Ap6A | 13 ± 5 | 0.17 ± 0.04 | 13.1 |
| Ap5A | 37 ± 9 | 0.4 ± 0.1 | 10.8 | |
| PP-InsP5 (n = 2) | 4 | 0.20 | 50 | |
| hAps1-39N | Ap6A | 11 ± 3 | 0.16 ± 0.02 | 14.5 |
| Ap5A | 50 ± 10 | 0.4 ± 0.1 | 8.0 | |
| PP-InsP5 (n = 2) | 0.3 | 0.09 | 290 | |
| hAps2 | Ap6A | 19 ± 1 | 0.20 ± 0.01 | 10.5 |
| Ap5A | 50 ± 10 | 0.8 ± 0.2 | 16.0 | |
| PP-InsP5 | 1.3 ± 0.6 | 0.14 ± 0.02 | 110 |
Prolonged treatment of Ap6A and Ap5A identified the major products as adenosine 5'-monophosphate (AMP) plus adenosine 5'-pentaphosphate (p5A) and AMP plus adenosine 5'-tetraphosphate (p4A) respectively. In neither case did p5A or p4A accumulate as these are metabolised almost as rapidly as they are formed, p5A being hydrolysed sequentially to p4A then ATP. In this respect, hAps1 and hAps2 show similar modes of action to hDIPP-1 [15] and hDIPP-2 (S.T. Safrany, data not shown). Nudix hydrolases have been found to be sensitive to inhibition by fluoride. Hydrolysis of Ap5A by hAps1 and hAps2 was found to be inhibited non-competitively with Ki values of 13.0 ± 0.5 and 30 ± 1 μM respectively (data not shown).
Despite the overall high degree of sequence similarity between hAps1, hAps2 and the DIPPs, hAps1 and hAps2 were found to have little activity towards PP-InsP5 or [PP]2-InsP4. hAps1 and hAps1-39N showed first order rate constants (k-1) with PP-InsP5 of 19 ± 4 and 7 ± 5 μg-1min-1 respectively. Activity of hAps2 was similar to that of hAps1, with a k-1 = 13 ± 6 μg-1min-1. These compare with k-1 values of 2200, 2000 and 220 μg-1min-1 for hDIPP-1, -2α and -2β respectively [16]. Km and kcat values were determined for each of the proteins (Table 3) and these suggested that the reduction in activity towards PP-InsP5 compared with hDIPP-1 and hDIPP-2α was primarily due to a reduced affinity for this substrate. Activity towards [PP]2-InsP4 was even weaker. hAps1 and hAps2 gave k-1 = 1.0 ± 0.2 and 0.9 ± 0.2 μg-1min-1 respectively. hAps1-39N showed no activity towards [PP]2-InsP4 under the conditions tested. In contrast, hDIPP-1, -2α and -2β gave k-1 values of 320, 90 and 32 μg-1min-1 respectively, whereas S. pombe Aps1 and S. cerevisiae Ddp1p showed similar activity with k-1 = 8.3 and 1.7 μg-1min-1 respectively [15].
Tissue distribution
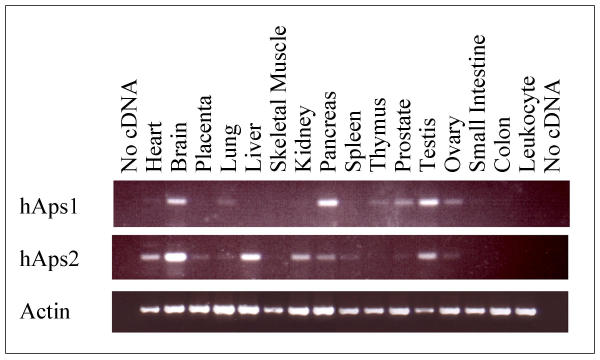
Analysis of the tissue distribution of hAps1 and hAps2 gene expression was performed using PCR and human tissue cDNA samples. Using primers selective for hAps1, PCR products were readily visible from brain, pancreas and testis and less so from lung, thymus, prostate, ovary, small intestine and heart (the least) (Fig. 6). Selective primers for hAps2 produced strong signals from brain and liver, but also from heart, placenta, lung, kidney, pancreas, spleen, prostate, testis and ovary (Fig. 6). No signal for hAps1 was detected in placenta, liver, skeletal muscle, kidney, spleen, colon or peripheral blood leukocytes. Likewise, no signal for hAps2 was found in skeletal muscle, thymus, small intestine, colon or peripheral blood leukocytes. A number of tumour cell line cDNA samples were also tested; hAps1 was present in prostate PC3, and ovary GI-102 cells. The prostate PC3 cell line was also found to express hAps2. Tumour cell lines from breast GI-101, lung LX-1, colon CX-1, lung GI-117, colon GI-112 and pancreas GI-103 were found to express neither hAps1 nor hAps2. These results are in broad agreement with the sources of ESTs for these two protein families (Table 1). This list is for comparison and is not exhaustive. In contrast, hDIPP-1 was found in all the tissues and cell types tested above (S.T. Safrany, unpublished data).
Figure 6.
Tissue distribution of hAps1, hAps2 and β-actin. PCR using human tissue first-strand cDNA as template was performed with primers specific for β-actin and for hAps1 or hAps2. 35 cycles of amplification were used for β-actin, 40 cycles for hAsp1 and hAps2.
Whilst an exhaustive analysis has not been performed for hDIPP-2, it was found to be highly expressed in heart, skeletal muscle, kidney and pancreas, and weakly expressed in brain, placenta lung and liver [16].
Subcellular localisation
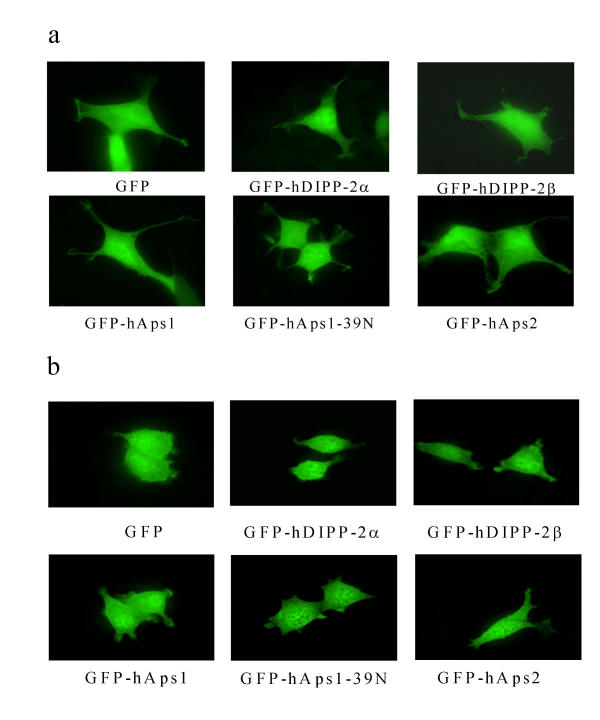
The subcellular localisation of hAps1, hAps1-39N and hAps2 was investigated by individual expression of the three proteins tagged at the N-terminus with EGFP in HEK293 and PC12 cells, and visualisation of the expressed fusion proteins by deconvolved fluorescence microscopy. These results show that in both cell types EGFP-tagged proteins showed a cytosolic location indistinguishable from EGFP-hDIPP-2α, EGFP-hDIPP-2β and EGFP alone (Fig. 7a and 7b). Similar data were obtained with proteins tagged at the C-terminus with EGFP (data not shown). This is in agreement with localisation predicted using PSORTII.
Figure 7.
Subcellular localisation of EGFP, EGFP-hDIPP-2 (α and β) and hAps1 and hAps2. Proteins fused to the C-terminus of EGFP were expressed in (a) HEK293 or (b) PC12 cells for 24 h and visualised by fluorescence microscopy.
Discussion
The human genome contains at least 18 Nudix genes, several of which encode multiple products, and 9 Nudix hydrolases have been characterised so far to varying degrees (see http://www.gene.ucl.ac.uk/nomenclature/genefamily/npym.html). These include hDIPP-1, hDIPP-2α and hDIPP-2β, encoded by the NUDT3 (hDIPP-1) and NUDT4 (hDIPP-2α and hDIPP-2β) genes, all of which hydrolyse both the higher order diadenosine polyphosphates and diphosphoinositol polyphosphates. In this report, we describe the cloning and functional characterisation of hAps1 and hAps2, two new members of the DIPP subfamily with a novel pattern of expression and substrate specificity. A polymorphism of hAps1 (hAps1-39N) has also been found, its activity and localisation appearing broadly similar to those of hAps1. Unlike the other subfamily members, hAps1 and hAps2 show little activity towards diphosphoinositol polyphosphates, both gene products favouring diadenosine polyphosphates, ApnA, where n = 5 or 6. In each case, Ap6A binds with higher affinity than Ap5A, whereas the kcat for the latter is greater. Given that no other PP-InsP5 hydrolase activity was observed during the purification of rat hepatic DIPP (most similar to hDIPP-1) and assuming that tissue expression and physiology are broadly conserved between human and rat, these results would suggest that Aps-like proteins do not contribute significantly to inositol phosphate metabolism.
Both hAps1 and hAps2 show typical Nudix hydrolase requirements, namely an alkaline pH optimum in vitro and a requirement for divalent cations [12]. An unusual feature of hAps1 and hAps2 is that they both require Mn2+ for activation, and that Mg2+ is without effect. Optimal [Mn2+] is approximately 4 mM, which is in contrast to that estimated for free intracellular [Mn2+] of below 0.5 μM [27]. Free [Mn2+] in our in vitro assays was not determined. It is well acknowledged that Mn2+ binds serum albumin very tightly, but despite this we would anticipate free [Mn2+] in the assays to be far in excess of intracellular levels. It is known that the activity of metal ion-requiring enzymes in different cellular compartments can be regulated through the controlled trafficking of these ions and therefore through the controlled access of these enzymes to their required ions. This principle has been most thoroughly studied in the case of Ca2+[28], although analogous observations have been made with Cu2+[29] and Mn2+[30]. Since the [Mn2+] throughout the cell is known to be non-uniform, the unusual requirement of the hAps enzymes suggests a possible mechanism of regulation through access to this ion.
Several biochemically distinct Ap5/6A hydrolases have been described previously. S. pombe Aps1, S. cerevisiae Ddp1p and hDIPP-1 and hDIPP-2 have all been shown to metabolise Ap5A and Ap6A. The activities of hAps1 and hAps2 towards Ap5A and Ap6A are comparable to hDIPP-1, but their activities towards the diphosphoinositol polyphosphates PP-InsP5 and [PP]2-InsP4 are greatly reduced, thus making them some 100–300-fold more selective towards Ap5A and Ap6A than hDIPP-1 or hDIPP-2. S. pombe Aps1 and S. cerevisiae Ddp1p, the sole representatives of the DIPP subfamily in the two yeast genomes, appear to favour the diphosphoinositol polyphosphates. Their modes of action have been shown to differ from their mammalian counterparts. The major products of Ap6A hydrolysis by S. pombe Aps1 and S. cerevisiae Ddp1p are ADP and p4A [17,18]. As with hDIPP-1 [15] and hDIPP-2 (Safrany, data not shown), hAps1 and hAps2 produced AMP and p5A as major products. With Ap5A as substrate, S. pombe Aps1 favoured the production of ADP plus ATP, whereas S. cerevisiae Ddp1p favoured production of AMP and p4A. In this respect hAps1 and hAps2 as well as hDIPP-1 and hDIPP-2 resemble Ddp1p.
Due to the chromosomal proximity of Aps1 and Aps2, and the greater degree of sequence identity with each other than with the Aps-like genes from mouse, rat or cow, it seems likely that this gene pair has arisen as the result of a recent gene duplication event on the short arm of the X chromosome [31]. A popular view of the evolution of catalytic motifs envisages the duplication of relatively non-specific progenitors followed by adaptation to perform special tasks, and the Nudix hydrolase motif appears to be a good illustration of this process. The existence of multiple forms of these enzymes would suggest that the diadenosine polyphosphates have significant biological functions, with the specialised task of hAps1 and/or hAps2 being the regulation of Ap5A and Ap6A levels.
Conclusions
Previously, the human DIPP family comprised hDIPP-1, a ubiquitous enzyme able to hydrolyse both inositol phosphates and diadenosine polyphosphates, and hDIPP-2α and -2β, which are predominantly diphosphoinositol polyphosphate phosphohydrolases highly expressed in heart, skeletal muscle, kidney and pancreas. We have now identified two further members, hAps1 and hAps2, which have high selectivity towards the diadenosine polyphosphates. The distinction between DIPP and ApnA hydrolase activities reinforces the importance of Q86 in DIPP-2β and the role of Q85 in hAps1 and hAps2. The addition of this amino acid in hDIPP-2 has been previously shown to have a substantial negative effect upon its activity towards diphosphoinositol polyphosphates, whereas Ap5A hydrolase activity was increased. The expression of hAps1 and hAps2 is also restricted. Of the tissues tested, only pancreas showed high levels of hDIPP-2 and hAps1, and only heart showed high levels of hDIPP-2 and hAps2, although ESTs were found suggesting that hDIPP-2 is expressed in testis, ovary and prostate. The presence of hAps1 and hAps2 in such a variety of tissues suggests that Ap5A and Ap6A may have roles other than as secreted molecules. We find that hAps1 is present in the pancreas, but there is no suggestion that it is secreted. For this reason, hAps1 and hAps2 are expected to control intracellular Ap5A and Ap6A levels.
In conclusion, we have identified two human Nudix hydrolases that share a novel substrate specificity, metabolising predominantly Ap5A and Ap6A. These two gene products complement the activity of previously described members of the DIPP family, and reinforce the concept of Ap5A and Ap6A acting as intracellular messengers.
Authors' contributions
NRL performed the cloning and subcloning, determined subcellular localisation in mammalian cells, participated in sequence analysis and obtained the genomic alignment. AGMcL assisted in the writing of the manuscript. STS conceived of this study, prepared recombinant enzymes, performed enzyme assays, determined the tissue distribution, participated in sequence analysis and drafted the manuscript. All authors read and approved the final manuscript.
Materials and Methods
Materials
Ap4G, Ap5G and p4A were purchased from Jena Bioscience, Jena, Germany. Other nucleotides were purchased from Sigma. Non-radioactive PP-InsP5 was a kind gift from J.R. Falck (University of Texas Southwestern Medical Center, Dallas, TX, USA). [3H]-PP-InsP5 and [3H]-[PP]2-InsP4 were prepared as previously described [15].
Cloning and plasmid construction
PCR products corresponding to the hAps1 sequence were amplified from a human testis library (Clontech), using the primers SSD019 (5'-ggcaggatccaagtgcaaacccaaccagacg-3') and SSD020 (5'-ggcaggatccttagggatcgctatctggcg-3') (BamHI sites are underlined). PCR was carried out using a HiFidelity Expand Kit (Roche), and the products cloned into the vector pCR2.1TOPO (Invitrogen) and sequenced. Surprisingly, three variant sequences were found (termed hAps1, hAps1-39N and hAps2). These were subcloned as BamHI fragments into the BamHI site of pGEX-6P1 (Amersham Pharmacia Biotech) (starting at K2, to compare with K3 of hDIPP-1 and hDIPP-2) and fully re-sequenced. The 3' nucleotide of SSD019 introduced a silent mutation into hAps2, and was allowed to remain. Likewise, SSD020 also introduced a silent mutation into the stop codon of hAps2. This was also allowed to remain. Vectors for the expression of Nudix hydrolase proteins fused to the C-terminus of Enhanced Green Fluorescent Protein (EGFP) were produced by ligating a cDNA encoding each protein as a BamHI digestion fragment from the GST-fusion vectors described above, into pEGFP-C1 (Clontech) previously digested with BglII and BamHI.
Bioinformatics
Multiple sequence alignments were performed using MAP and BoxShade, both run by EMBnet, Lausanne, Switzerland. Blast searches of EST and genomic databases were performed using the facilities provided by NCBI, Bethesda, MD, USA.
Expression and purification of hAps1, hAps1-39N and hAps2
Full-length hAps1, hAps1-39N and hAps2 were expressed and purified from E. coli as GST-fusion proteins. Expression plasmids based on pGEX-6P1 were transformed into E. coli strain BL21, and induced at 26°C overnight with 100 μM IPTG. Cells were harvested in buffer A (20 mM Tris, 150 mM NaCl, 2 mM DTT, 0.1 mM EGTA, pH 7.5), supplemented with 5 μg/ml leupeptin and 1 μg/ml aprotinin, and disrupted by sonication (3 × 15s). Particulate matter was removed by centrifugation. The supernatant was applied to (and subsequently eluted from) a 5 mm × 5 cm Glutathione Sepharose 4 Fast Flow column (Amersham Pharmacia Biotech) at a flow rate of 1 ml/min. The column was washed for 5 min with buffer A. Bound protein was eluted with a gradient generated by mixing buffer A with buffer B (buffer A plus 25 mM glutathione, pH 7.5) as follows: 0-10 min, 0-50% buffer B; 10-13 min, 100% buffer B. Fractions containing pure hAps1, hAps1-39N or hAps2 were supplemented with glycerol (10% v/v final), and stored at -80°C.
Tissue distribution
The tissue distribution of hAps1 and hAps2 expression was determined by PCR from human tissue cDNA samples (human cDNA panels, Clontech) and human cDNA libraries (testis from Clontech, brain from Dr Peter Cheung of the MRC Protein Phosphorylation Unit, Wellcome Trust Biocentre, Dundee, Scotland, UK). β-Actin-specific and hAps1 and hAps2-specific primers were used. The hAps1 primers were SSD049 (5'-cgtcttcgaacagaaccaggatcg-3'), and SSD045 (5'-caaaagccacacacatggtgcc-3') and the hAps2 primers were SSD050 (5'-cgtcttcgaacagaaccaggaccc-3') and SSD046 (5'-gtgcaacaacctggagaatagtcattgta-3'). Primers SSD045 and SSD046 were selective for the 3' untranslated regions of hAsp1 and hAsp2 respectively. β-Actin cDNA was amplified using β AS (5'-acactgtgcccatctacgaggg-3') and β AA (5'ccttctgcatcctgtcagcaatg-3'). PCR was carried out using a HiFidelity Expand Kit (Roche), and cDNA samples from heart, brain, placenta, lung, liver, skeletal muscle, kidney, pancreas (human MTC panel I #K1420-1, Clontech), spleen, thymus, prostate, testis, ovary, small intestine, colon, peripheral blood leukocytes (human MTC Panel II #K1421-1, Clontech). The following tumour cell lines were also tested: breast GI-101, lung LX-1, colon CX-1, lung GI-117, prostate PC3, colon GI-112, ovary GI-102 and pancreas GI-103 (human MTC Tumor Panel #K1422-1, Clontech).
Subcellular localisation
The subcellular localisation of hAps1, hAps1-39N and hAps2 proteins fused to EGFP was investigated using transfected HEK 293 epithelial cells and PC12 pheochromocytoma cells. PC12 cells were grown in DMEM/5% foetal bovine serum/5% horse serum. HEK293 were grown in DMEM/10% foetal bovine serum. Both cell types were grown on coverslips and transfected using Fugene-6 (Roche) with expression vectors for each Nudix hydrolase fused to the C-terminus of EGFP. 24 hours after transfection, cells were observed using a Leica inverted stage fluorescence microscope and a Hamamatsu Orca charge-coupled-device camera. Images were analysed using Improvision OpenLab deconvolution software.
Enzyme assays
For the analysis of nucleotide metabolism, assays were performed as previously described [15], unless otherwise stated. The UV-absorbing (260 nm) nucleotide reaction products were analysed using a 4.6 × 125 mm Partisphere 5 μm SAX HPLC column. Substrates and products were eluted at 1 ml/min by the following gradient generated by mixing water with buffer C (1.3 M (NH4)2HPO4, pH 4.8 with H3PO4): 0-5 min, 0% buffer C; 5-55 min, 0-50% buffer C; 56-70 min, 0% buffer C. pH optima of recombinant enzymes were determined in a series of buffers comprising 1 mM MnCl2, 100 μg/ml BSA and 15 mM MES, 15 mM HEPES or 15 mM Tris (pH 6-9 with HCl or NaOH). Divalent ion-dependency was determined in a buffer comprising 50 mM HEPES, 100 μg/ml BSA, pH 7.6, and 50 μM-10mM (where possible) divalent metal as the chloride salt. Inositol phosphate metabolism was determined exactly as described previously [14]. Km and kcat values were determined by hyperbolic regression analysis.
Authors' note
During the reviewing of this manuscript a paper was submitted and accepted in the Journal of Biological Chemistry (K Hidaka, JJ Caffrey, L Hua, T Zhang, JR Falck, GC Nickel, L Carrel, LD Barnes, SB Shears: An adjacent pair of human NudT genes on chromosome X are preferentially expressed in testis and encode two new isoforms of diphosphoinositol polyphosphate phosphohydrolase. J Biol Chem in press). DIPP-3β corresponds to hAps1, whereas DIPP-3α corresponds to hAps2.
Acknowledgments
Acknowledgements
We would like to thank Professor C.P. Downes and Dr. Alexander Gray for assistance, members of the Division of Cell Signalling, The University of Dundee for helpful discussion and the DNA sequencing service, The University of Dundee http://www.dnaseq.co.uk for sequencing of plasmids. This research was funded by grants from The Royal Society and Tenovus-Scotland to S.T.S. and the Wellcome Trust to A.G.McL.
Contributor Information
Nick R Leslie, Email: n.r.leslie@dundee.ac.uk.
Alexander G McLennan, Email: agmclen@liv.ac.uk.
Stephen T Safrany, Email: s.t.safrany@dundee.ac.uk.
References
- McLennan AG. Dinucleoside polyphosphates – friend or foe? Pharmacology & Therapeutics. 2000;87:73–89. doi: 10.1016/S0163-7258(00)00041-3. [DOI] [PubMed] [Google Scholar]
- Garrison PN, Barnes LD. Determination of dinucleoside polyphosphates. In: AG McLennan, editor. Ap4A and Other Dinucleoside Polyphosphates. Boca Raton, Fl.: CRC Press; 1992. pp. 29–61. [Google Scholar]
- Schluter H, Gross I, Bachmann J, Kaufmann R, van der Giet M, Tepel M, Nofer JR, Assmann G, Karas M, Jankowski J, et al. Adenosine(5') oligophospho-(5') guanosines and guanosine(5') oligophospho-(5') guanosines in human platelets. Journal of Clinical Investigation. 1998;101:682–688. doi: 10.1172/JCI119882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores NA, Stavrou BM, Sheridan DJ. The effects of diadenosine polyphosphates on the cardiovascular system. Cardiovasc Res. 1999;42:15–26. doi: 10.1016/S0008-6363(99)00004-8. [DOI] [PubMed] [Google Scholar]
- Miras-Portugal MT, Gualix J, Mateo J, Diaz-Hernandez M, Gomez-Villafuertes R, Castro E, Pintor J. Diadenosine polyphosphates, extracellular function and catabolism. Progress in Brain Research. 1999;120:397–409. doi: 10.1016/s0079-6123(08)63572-4. [DOI] [PubMed] [Google Scholar]
- Luo JK, Jankowski J, Tepel M, vonderGiet M, Zidek W, Schluter H. Identification of diadenosine hexaphosphate in human erythrocytes. Hypertension. 1999;34:872–875. doi: 10.1161/01.hyp.34.4.872. [DOI] [PubMed] [Google Scholar]
- Jovanovic A, Jovanovic S, Mays DC, Lipsky JJ, Terzic A. Diadenosine 5',5"-P1,P5-pentaphosphate harbors the properties of a signaling molecule in the heart. FEBS Lett. 1998;423:314–8. doi: 10.1016/S0014-5793(98)00114-8. [DOI] [PubMed] [Google Scholar]
- Sillero A, Sillero MAG. Synthesis of dinucleoside polyphosphates catalyzed by firefly luciferase and several ligases. Pharmacology & Therapeutics. 2000;87:91–102. doi: 10.1016/S0163-7258(00)00047-4. [DOI] [PubMed] [Google Scholar]
- Lienhard GE, Secemski II. P1,P5-Di(adenosine-5')pentaphosphate, a potent multisubstrate inhibitor of adenylate kinase. J Biol Chem. 1973;248:1121–1123. [PubMed] [Google Scholar]
- McLennan AG. The MutT motif family of nucleotide phosphohydrolases in man and human pathogens (review). Int J Mol Med. 1999;4:79–89. doi: 10.3892/ijmm.4.1.79. [DOI] [PubMed] [Google Scholar]
- Guranowski A. Specific and nonspecific enzymes involved in the catabolism of mononucleoside and dinucleoside polyphosphates. Pharmacol Ther. 2000;87:117–139. doi: 10.1016/S0163-7258(00)00046-2. [DOI] [PubMed] [Google Scholar]
- Bessman MJ, Walsh JD, Dunn CA, Swaminathan J, Weldon JE, Shen JY. The gene ygdP, associated with the invasiveness of Escherichia coli K1, designates a nudix hydrolase, Orf176, active on adenosine (5')-pentaphospho-(5')-adenosine (Ap(5)A). J Biol Chem. 2001;276:37834–37838. doi: 10.1074/jbc.M107032200. [DOI] [PubMed] [Google Scholar]
- Gaywee J, Xu W, Radulovic S, Bessman MJ, Azad AF. The Rickettsia prowazekii invasion gene homolog (invA) encodes a nudix hydrolase active on adenosine (5')-pentaphospho-(5')-adenosine. Mol Cell Proteomics. 2002;1:179–185. doi: 10.1074/mcp.M100030-MCP200. [DOI] [PubMed] [Google Scholar]
- Safrany ST, Caffrey JJ, Yang X, Bembenek ME, Moyer MB, Burkhart WA, Shears SB. A novel context for the 'MutT' module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. Embo J. 1998;17:6599–607. doi: 10.1093/emboj/17.22.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safrany ST, Ingram SW, Cartwright JL, Falck JR, McLennan AG, Barnes LD, Shears SB. The diadenosine hexaphosphate hydrolases from Schizosaccharomyces pombe and Saccharomyces cerevisiae are homologues of the human diphosphoinositol polyphosphate phosphohydrolase. Overlapping substrate specificities in a MutT-type protein. J Biol Chem. 1999;274:21735–40. doi: 10.1074/jbc.274.31.21735. [DOI] [PubMed] [Google Scholar]
- Caffrey JJ, Safrany ST, Yang X, Shears SB. Discovery of molecular and catalytic diversity among human diphosphoinositol-polyphosphate phosphohydrolases. An expanding Nudt family. J Biol Chem. 2000;275:12730–6. doi: 10.1074/jbc.275.17.12730. [DOI] [PubMed] [Google Scholar]
- Ingram SW, Stratemann SA, Barnes LD. Schizosaccharomyces pombe Aps1, a diadenosine 5',5' "-P1, P6-hexaphosphate hydrolase that is a member of the nudix (MutT) family of hydrolases: cloning of the gene and characterization of the purified enzyme. Biochemistry. 1999;38:3649–55. doi: 10.1021/bi982951j. [DOI] [PubMed] [Google Scholar]
- Cartwright JL, McLennan AG. The Saccharomyces cerevisiae YOR163w gene encodes a diadenosine 5', 5"'-P1,P6-hexaphosphate (Ap6A) hydrolase member of the MutT motif (Nudix hydrolase) family. J Biol Chem. 1999;274:8604–10. doi: 10.1074/jbc.274.13.8604. [DOI] [PubMed] [Google Scholar]
- Glennon MC, Shears SB. Turnover of inositol pentakisphosphates, inositol hexakisphosphate and diphosphoinositol polyphosphates in primary cultured hepatocytes. Biochem J. 1993;293:583–90. doi: 10.1042/bj2930583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright JL, Safrany ST, Dixon LK, Darzynkiewicz E, Stepinski J, Burke R, McLennan AG. The g5R (D250) gene of African swine fever virus encodes a nudix hydrolase that preferentially degrades diphosphoinositol polyphosphates. Journal of Virology. 2002;76:1415–1421. doi: 10.1128/JVI.76.3.1415-1421.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB, Ali N, Craxton A, Bembenek ME. Synthesis and metabolism of bis-diphosphoinositol tetrakisphosphate in vitro and in vivo. J Biol Chem. 1995;270:10489–97. doi: 10.1074/jbc.270.18.10489. [DOI] [PubMed] [Google Scholar]
- Shears SB. The versatility of inositol phosphates as cellular signals. Biochim Biophys Acta. 1998;1436:49–67. doi: 10.1016/S0005-2760(98)00131-3. [DOI] [PubMed] [Google Scholar]
- Safrany ST, Shears SB. Turnover of bis-diphosphoinositol tetrakisphosphate in a smooth muscle cell line is regulated by beta2-adrenergic receptors through a cAMP-mediated, A-kinase-independent mechanism. Embo J. 1998;17:1710–6. doi: 10.1093/emboj/17.6.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye W, Ali N, Bembenek ME, Shears SB, Lafer EM. Inhibition of clathrin assembly by high affinity binding of specific inositol polyphosphates to the synapse-specific clathrin assembly protein AP-3. J Biol Chem. 1995;270:1564–8. doi: 10.1074/jbc.270.4.1966. [DOI] [PubMed] [Google Scholar]
- Morrison BH, Bauer JA, Kalvakolanu DV, Lindner DJ. Inositol hexakisphosphate kinase 2 mediates growth suppressive and apoptotic effects of interferon-beta in ovarian carcinoma cells. J Biol Chem. 2001;276:24965–24970. doi: 10.1074/jbc.M101161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanakahi LA, Bartlet-Jones M, Chappell C, Pappin D, West SC. Binding of inositol phosphate to DNA-PK and stimulation of double-strand break repair. Cell. 2000;102:721–729. doi: 10.1016/S0092-8674(00)00061-1. [DOI] [PubMed] [Google Scholar]
- Wedler FC, Denman RB, Roby WG. Glutamine-Synthetase from Ovine Brain Is a Manganese(Ii) Enzyme. Biochemistry. 1982;21:6389–6396. doi: 10.1021/bi00268a011. [DOI] [PubMed] [Google Scholar]
- Ermak G, Davies KJA. Calcium and oxidative stress: from cell signaling to cell death. Molecular Immunology. 2002;38:713–721. doi: 10.1016/S0161-5890(01)00108-0. [DOI] [PubMed] [Google Scholar]
- Harrison MD, Jones CE, Solioz M, Dameron CT. Intracellular copper routing: the role of copper chaperones. Trends in Biochemical Sciences. 2000;25:29–32. doi: 10.1016/S0968-0004(99)01492-9. [DOI] [PubMed] [Google Scholar]
- Luk EEC, Culotta VC. Manganese superoxide dismutase in Saccharomyces cerevisiae acquires its metal co-factor through a pathway involving the Nramp metal transporter, Smf2p. J Biol Chem. 2001;276:47556–47562. doi: 10.1074/jbc.M108923200. [DOI] [PubMed] [Google Scholar]
- Eichler EE. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends in Genetics. 2001;17:661–669. doi: 10.1016/S0168-9525(01)02492-1. [DOI] [PubMed] [Google Scholar]