Abstract
1. The normalization of biochemical data to weight them appropriately for parameter estimation is considered, with reference particularly to data from tracer kinetics and enzyme kinetics. If the data are in replicate, it is recommended that the sum of squared deviations for each experimental variable at each time or concentration point is divided by the local variance at that point. 2. If there is only one observation for each variable at each sampling point, normalization may still be required if the observations cover more than one order of magnitude, but there is no absolute criterion for judging the effect of the weighting that is produced. The goodness of fit that is produced by minimizing the weighted sum of squares of deviations must be judged subjectively. It is suggested that the goodness of fit may be regarded as satisfactory if the data points are distributed uniformly on either side of the fitted curve. A chi-square test may be used to decide whether the distribution is abnormal. The proportion of the residual variance associated with points on one or other side of the fitted curve may also be taken into account, because this gives an indication of the sensitivity of the residual variance to movement of the curve away from particular data points. These criteria for judging the effect of weighting are only valid if the model equation may reasonably be expected to apply to all the data points. 3. On this basis, normalizing by dividing the deviation for each data point by the experimental observation or by the equivalent value calculated by the model equation may both be shown to produce a consistent bias for numerically small observations, the former biasing the curve towards the smallest observations, the latter tending to produce a curve that is above the numerically smaller data points. It was found that dividing each deviation by the mean of observed and calculated variable appropriate to it produces a weighting that is fairly free from bias as judged by the criteria mentioned above. This normalization factor was tested on published data from both tracer kinetics and enzyme kinetics.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Apps D. K., Marsh A. Kinetic studies of the interaction of pigeon-liver NAD kinase with adenine nucleotides and divalent cations. Eur J Biochem. 1972 Jun 23;28(1):12–19. doi: 10.1111/j.1432-1033.1972.tb01878.x. [DOI] [PubMed] [Google Scholar]
- Atkins G. L. A versatile digital computer program for non-linear regression analysis. Biochim Biophys Acta. 1971 Dec 21;252(3):405–420. doi: 10.1016/0304-4165(71)90142-5. [DOI] [PubMed] [Google Scholar]
- Atkins G. L. Some applications of a digital computer program to estimate biological parameters by non-linear regression analysis. Biochim Biophys Acta. 1971 Dec 21;252(3):421–426. doi: 10.1016/0304-4165(71)90143-7. [DOI] [PubMed] [Google Scholar]
- BERMAN M., WEISS M. F., SHAHN E. Some formal approaches to the analysis of kinetic data in terms of linear compartmental systems. Biophys J. 1962 May;2:289–316. doi: 10.1016/s0006-3495(62)86856-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland W. W. The statistical analysis of enzyme kinetic data. Adv Enzymol Relat Areas Mol Biol. 1967;29:1–32. doi: 10.1002/9780470122747.ch1. [DOI] [PubMed] [Google Scholar]
- DOWD J. E., RIGGS D. S. A COMPARISON OF ESTIMATES OF MICHAELIS-MENTEN KINETIC CONSTANTS FROM VARIOUS LINEAR TRANSFORMATIONS. J Biol Chem. 1965 Feb;240:863–869. [PubMed] [Google Scholar]
- Handa J., Handa H., Torizuka K., Hamamoto K., Kousaka T. Serial brain scanning with radioactive xenon and scintillation camera. Am J Roentgenol Radium Ther Nucl Med. 1970 Aug;109(4):701–706. doi: 10.2214/ajr.109.4.701. [DOI] [PubMed] [Google Scholar]
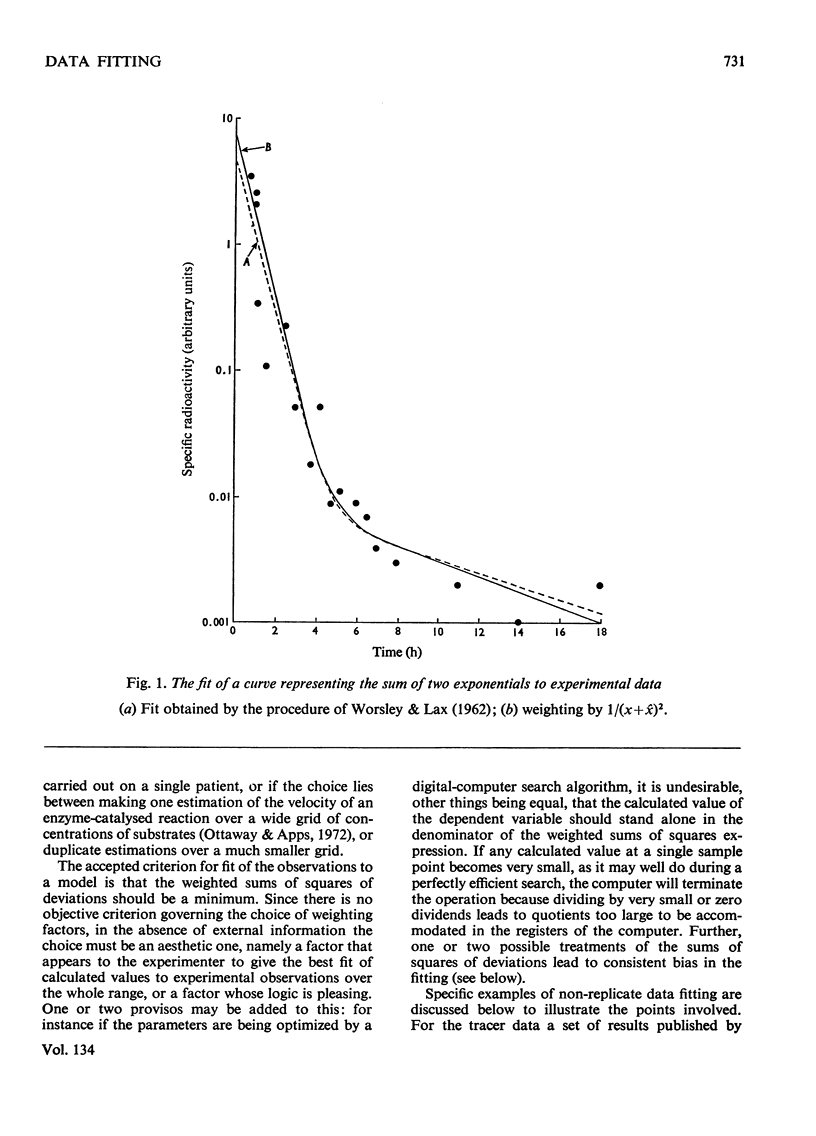
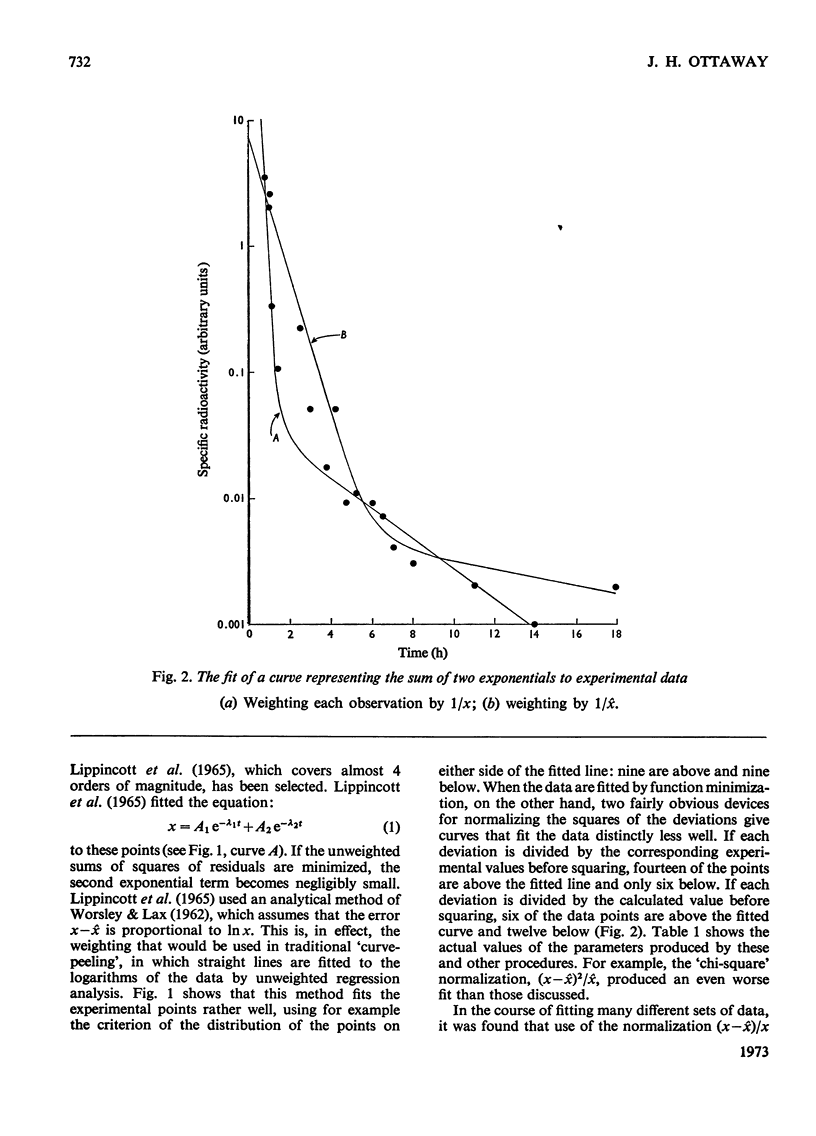
- Lippincott S. W., Korman S., Lax L. C., Corcoran C. Transfer rates of gamma globulin between cerebrospinal fluid and blood plasma (results obtained on a series of multiple sclerosis patients). J Nucl Med. 1965 Sep;6(9):632–644. [PubMed] [Google Scholar]
- Neer R., Berman M., Fisher L., Rosenberg L. E. Multicompartmental analysis of calcium kinetics in normal adult males. J Clin Invest. 1967 Aug;46(8):1364–1379. doi: 10.1172/JCI105629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaway J. H., Apps D. K. Some examples of the use of computer-produced contour plots in the fitting of enzyme rate equations to reaction-velocity measurements. Biochem J. 1972 Dec;130(3):861–870. doi: 10.1042/bj1300861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich J. G., Wangermann G., Falck M., Rohde K. A general strategy for parameter estimation from isosteric and allosteric-kinetic data and binding measurements. Eur J Biochem. 1972 Apr 11;26(3):368–379. doi: 10.1111/j.1432-1033.1972.tb01776.x. [DOI] [PubMed] [Google Scholar]
- Sauer F., Erfle J. D., Binns M. R. Turnover rates and intracellular pool size distribution of citrate cycle intermediates in normal, diabetic and fat-fed rats estimated by computer analysis from specific activity decay data of 14C-labeled citrate cycle acids. Eur J Biochem. 1970 Dec;17(2):350–363. doi: 10.1111/j.1432-1033.1970.tb01173.x. [DOI] [PubMed] [Google Scholar]
- WILKINSON G. N. Statistical estimations in enzyme kinetics. Biochem J. 1961 Aug;80:324–332. doi: 10.1042/bj0800324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WORSLEY B. H., LAX L. C. Selection of a numerical technique for analyzing experimental data of the decay type with special reference to the use of tracers in biological systems. Biochim Biophys Acta. 1962 May 7;59:1–24. doi: 10.1016/0006-3002(62)90694-7. [DOI] [PubMed] [Google Scholar]


