Abstract
The Kv4 A-type potassium currents contribute to controlling the frequency of slow repetitive firing and back-propagation of action potentials in neurons and shape the action potential in heart. Kv4 currents exhibit rapid activation and inactivation and are specifically modulated by K-channel interacting proteins (KChIPs). Here we report the discovery and functional characterization of a modular K-channel inactivation suppressor (KIS) domain located in the first 34 aa of an additional KChIP (KChIP4a). Coexpression of KChIP4a with Kv4 α-subunits abolishes fast inactivation of the Kv4 currents in various cell types, including cerebellar granule neurons. Kinetic analysis shows that the KIS domain delays Kv4.3 opening, but once the channel is open, it disrupts rapid inactivation and slows Kv4.3 closing. Accordingly, KChIP4a increases the open probability of single Kv4.3 channels. The net effects of KChIP4a and KChIP1–3 on Kv4 gating are quite different. When both KChIP4a and KChIP1 are present, the Kv4.3 current shows mixed inactivation profiles dependent on KChIP4a/KChIP1 ratios. The KIS domain effectively converts the A-type Kv4 current to a slowly inactivating delayed rectifier-type potassium current. This conversion is opposite to that mediated by the Kv1-specific “ball” domain of the Kvβ1 subunit. Together, these results demonstrate that specific auxiliary subunits with distinct functions actively modulate gating of potassium channels that govern membrane excitability.
The Kv4 subfamily of voltage-gated potassium channels underlie somatodendritic A-currents in several types of neurons (1–3) and Ito in cardiac myocytes (4–7). Operating at subthreshold membrane potentials, they contribute to controlling the frequency of slow repetitive firing in these excitable cells. The dendritic A-type K+ current in hippocampal neurons helps to integrate the back-propagating action potentials and excitatory postsynaptic potentials or inhibitory postsynaptic potentials, providing a rapid electric signal to initiate associative events such as long-term potentiation (LTP) and long-term depression (LDP) (8–12). In heart, Ito impacts on the early phase of repolarization of the action potential (13, 14).
We recently identified K-channel interacting protein 1–3 (KChIP1–3) that specifically modulate Kv4 currents (15). KChIP1–3 increase total Kv4 current, moderately slow channel inactivation, and considerably accelerate recovery from inactivation (15). They are EF-hand Ca2+-binding proteins that belong to the recoverin/neuronal calcium sensor-1 (NCS-1) family. KChIP1 and KChIP 3 are predominantly expressed in neuronal tissues, whereas KChIP2 is predominantly expressed in heart and brain (15).
Here we report an unexpected, distinct modulation of Kv4 currents by a K-channel inactivation suppressor (KIS) domain present in an additional KChIP, KChIP4a. We show that by eliminating fast inactivation in conjunction with changes in other kinetic parameters, the KIS domain effectively converts the fast inactivating A-type current to a slowly inactivating delayed rectifier type of currents. Also, we present evidence that KChIPs with and without the KIS domain modulate Kv4 currents in a combinatorial manner. The KIS domain acts oppositely to the Kvβ1 ball domain (16–19) and the ball-like domains of maxi-K β2, β3 subunits (20–22). These observations indicate that auxiliary subunits provide diverse mechanisms to control activity of potassium channels.
Materials and Methods
Electrophysiology.
Unitary potassium currents were recorded from cell-attached patches in the presence of 2 mM KCl in the recording pipette as described (23, 24). Macroscopic potassium currents were recorded by applying the two-electrode voltage clamp method in Xenopus oocytes and the tight-seal whole-cell method in Chinese hamster ovary cells and cerebellar granule neurons essentially as described (25), except noted as follows. To examine the kinetics of the macroscopic rising phase, the currents were evoked from a holding potential of −100 mV by 30-ms test pulses ranging from −20 to +30 mV in 10-mV intervals. In the presence of 33 mM external KCl, deactivation was measured from tail current relaxations evoked by a hyperpolarizing pulse to −100 mV after activating the outward current by a 30-ms pulse to +40 mV from a holding potential of −100 mV. Peak conductance-voltage (pG-V) relationship was derived from peak amplitudes of currents evoked by depolarizing steps from −90 mV to +75 mV at 15-mV increments. A best-fit fourth-order Boltzmann function was used to extract the observed activation parameters from the pG-V relationship. The prepulse inactivation curve was measured by determining the peak current at +40 mV after 10-s prepulses ranging from −90 to 0 mV, from a holding potential of −100 mV. The plot of the normalized peak conductance (G/Gmax) against the prepulse voltages was analyzed assuming a first-order Boltzmann function to determine the midpoint voltage of prepulse inactivation. The development of closed-state inactivation was determined by measuring the peak current evoked by a test pulse to +40 mV from a holding potential of −100 mV and from subsequent −70-mV prepulses of increasing length (−70 mV is approximately the midpoint of prepulse inactivation). Recovery from inactivation was measured by applying a two-pulse protocol essentially as described (15), except that the duration of the first pulse was 1 s. A best-fit exponential function was used to determine the time constants of macroscopic inactivation, the rising phase, tail current deactivation, and the kinetics of closed-state inactivation and recovery from inactivation. All recordings were performed at room temperature. All measurements are reported as mean ± SEM.
Real-Time Quantitative Reverse Transcription–PCR (TaqMan).
Total RNA was isolated from C57BL/6J male mice and treated with DNase. Reverse transcription was performed by using SuperScript preamplification system (GIBCO/BRL). TaqMan reactions were performed under conditions suggested by Applied Biosystems with 25 ng of cDNA, 750 nM primers, and 250 nM probe in each reaction. The sequences of KChIP4a splice variant-specific primers and probes are as follows: KChIP4a, 5′ primer CAGGGTGAGGAGCGCTTCT, 3′ primer GCCCCTCCAAGTTCATGGT, and probe TGGCTGTGGCTGGACCATGACCTA; KChIP4bl, 5′ primer GAGCTCCACAGGCGGTTTC, 3′ primer GCTTCATGAGCCGCTCTTTAA, and probe TCTACGCGCAGAACAACACCAAGCG. The primers and probes for the 18s rRNA internal control were obtained from Applied Biosystems; data were analyzed according to instructions. All reactions were run in duplicate. Values of the duplicates were averaged and shown as relative expression to that of the 18s.
Immunohistochemistry.
Mouse mAbs were raised by using recombinant KChIP4a as antigen. Selectivity of these Abs against KChIP1–3, and recognition for the core domain of KChIP4a, which is shared among N-terminal splice variants of KChIP4, were established by ELISA, Western blots, and immunocytochemistry assays. mAbs against Kv4.2 were as described (15). Details of the Ab characterization and specificity will be described elsewhere. Double-label, indirect immunofluorescence was performed on 20 μm-thick free floating sections by using a mouse mAb to Kv4.2 (IgG1 isotype) in combination with a second mouse mAb specific for KChIP4a core domain (IgG2a isotype). Localization of these two primary Abs was accomplished by using isotype-specific secondary Abs conjugated to Alexa-488 or Alexa-594 fluorophores, as described (15, 26). Images of double-labeled sections were captured by using a Zeiss laser scanning confocal microscope.
Immunoprecipitation/Immunoblotting.
Procedures by using adult rat brain membrane preparations and anti-KChIP4a Abs (see above) were performed as described (15, 27).
Herpes Simplex Virus (HSV) and Neuronal Transduction.
Helper-virus-free Herpes virus amplicons were generated by cotransfection of 2–2 cells (28) with HSV-1 δ-pac helper cosmid DNA and amplicon plasmids, and the resulting amplicon virus particles were concentrated and purified by centrifugation over sucrose, as described (29, 30). Concentrated amplicons were resuspended in Hanks' balanced salt solution, frozen in aliquots, and titered on 2–2 cells. Rat cerebellar granule neuron cultures were prepared as described (25). To transduce the cultured cerebellar neurons, 0.5–1 μl of purified amplicon (≈107–108 transducing units/ml) of HSV-green fluorescent protein (GFP) or HSV-KChIP4a-GFP was added to cerebellar cultures grown on poly-d-lysine-coated glass coverslips in 24-well tissue culture plates at 4 × 105 cells/well (multiplicity of infection <0.5). GFP-positive cells were examined electrophysiologically 24–48 h after transduction.
Results
KChIP4a Abolishes Fast Inactivation and Favors the Open State of Kv4 Channels.
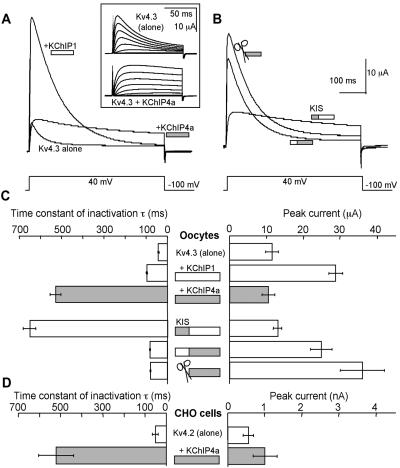
Mining of Millennium's internal databases led to the discovery of an additional member of the KChIP family, KChIP4a (Fig. 1). To our surprise, when coexpressed with Kv4.3 in Xenopus oocytes, KChIP4a abolished fast inactivation of the Kv4.3 current (Fig. 2A and Inset). The resulting current inactivated very slowly (inactivation time constant, τinact = 528 ± 25 ms) as opposed to the control Kv4.3 current (τinact = 41 ± 2 ms) (Fig. 2C). In contrast, KChIP1 increased the inactivation time constant only to 99 ± 2 ms (Fig. 2C). Similar results also were obtained with Kv4.2 in Chinese hamster ovary cells (Fig. 2D). This KChIP4a effect on inactivation is probably Kv4 subfamily-specific because KChIP4a similarly modulates Kv4.1 (the remaining member of Kv4 subfamily), but has no effect on Kv1.4 (data not shown).
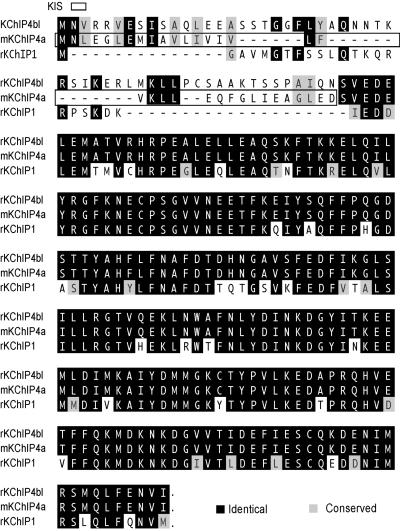
Figure 1.
Alignment of rodent KChIP4a, its splice variant KChIP4bl, and KChIP1 protein sequences. The KIS domain is boxed. Dashes between residues indicate space.
Figure 2.
The KIS domain of KChIP4a eliminates fast inactivation of Kv4. (A) Representative current traces of Kv4.3 alone, Kv4.3/KChIP4a, and Kv4.3/KChIP1 recorded from Xenopus oocytes. (Inset) Kv4.3 and Kv4.3/KChIP4a currents evoked by voltages from −90 mV to +45 mV at 15-mV increments. (B) Representative current traces of Kv4.3 with KChIP4a/KChIP1 chimeras and the KChIP4a deletion (Δ) mutant (✄; Δ2–44). (C) Inactivation time constants and peak current amplitudes for experiments in A and B. (D) Inactivation time constants and peak current amplitudes recorded from Chinese hamster ovary cells expressing Kv4.2 and Kv4.2/KChIP4a, respectively. n = 6–16.
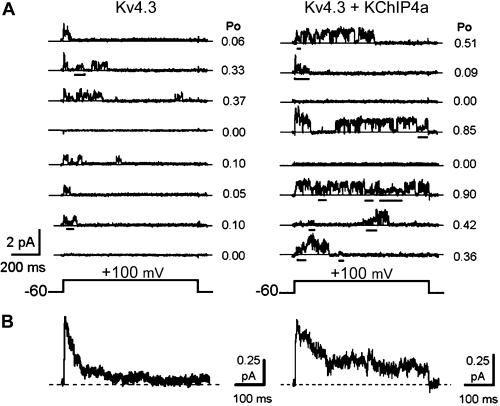
The novel properties of Kv4.3 macroscopic current conferred by KChIP4a were also apparent at the single-channel level (Fig. 3). Kv4.3 channels alone opened rapidly near the onset of the depolarizing pulse and reopened only a few times before undergoing complete inactivation (Fig. 3A Left). In contrast, KChIP4a dramatically prolonged the apparent open duration of Kv4.3 as the channel frequently reopened and fluctuated between multiple conductance levels (partial closures are indicated by the bars under the traces, Fig. 3A Right). The overall open probability per trace at +100 mV increased from 0.15 ± 0.01 to 0.25 ± 0.02 in the absence and presence of KChIP4a (n = 50 traces, each), respectively. Perhaps as a result of a more stable open level in the presence of KChIP4a, the main unitary conductance of Kv4.3 appeared to be slightly elevated in the presence of KChIP4a (from ≈4 to 5 pS). The ensemble average currents were qualitatively in agreement with the corresponding macroscopic currents (Fig. 3B).
Figure 3.
KChIP4a prolongs Kv4.3 opening at single-channel level. (A) Representative unitary current traces mediated by Kv4.3 and Kv4.3/KChIP4a. Partial closures are underlined. (B) Ensemble averages of 50 sweeps from A.
The N-Terminal KIS Domain Determines the Unique Effects of KChIP4a.
As KChIP4a and KChIP1 differ mainly at their N termini (Fig. 1), we reasoned that the KIS domain was located within the variable N terminus of KChIP4a. Two lines of evidence supported this hypothesis. First, the N-terminal deletion mutant of KChIP4a, Δ2−44KChIP4a, failed to produce the dramatic slowing of inactivation (Fig. 2 B and C). Instead, the modulation by Δ2−44KChIP4a resembled that by KChIP1–3 or other N-terminal deletion mutants of KChIP1–3 that carry only the conserved C-terminal core domains (Fig. 2 B and C; ref. 15). Second, the chimeric construct, 4N-1C, where the N-terminal domain of KChIP4a was fused to the C-terminal core domain of KChIP1, behaved almost identically to KChIP4a (Fig. 2 B and C), whereas the reciprocal chimera 1N-4C acted almost identically to Δ2−44KChIP4a or KChIP1 (Fig. 2 B and C). Therefore, we conclude that KIS domain is located at the N terminus of KChIP4a, is modular in nature, and can be dominant over the modulatory functions of the conserved core domains of KChIPs.
Kinetic Basis of KChIP4a Action on Kv4.3 Gating.
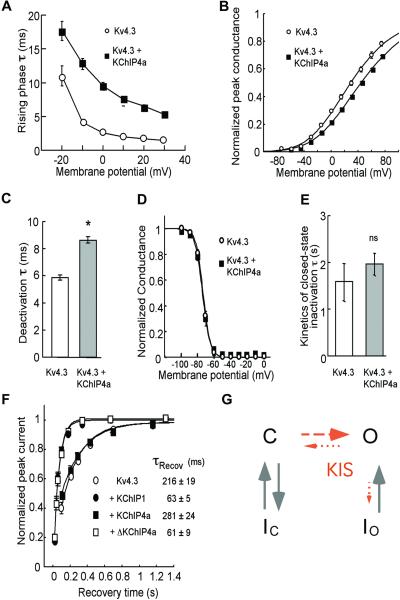
To gain further insights into the function of the KIS domain, we investigated additional kinetic and voltage-dependent properties of Kv4.3 and Kv4.3/KChIP4a currents. KChIP4a significantly slowed the rising phase of the macroscopic current at all voltages (Fig. 4A). Accordingly, single Kv4.3/KChIP4a channels sometimes failed to activate near the onset of the depolarization (Fig. 3A Right, seventh trace) and sometimes a single channel exhibited progressive ramp-like activation (Fig. 3A Right, first, seventh, and eighth traces). KChIP4a caused the pG-V relationship (pG-V curve) to appear slightly shallower (Fig. 4B). Note, however, that the voltage-dependent activation cannot be measured accurately because in Kv4 channels there is kinetic overlap between activation and inactivation. Interestingly, KChIP4a significantly slowed the tail current relaxation at −100 mV (which probably reflects slower channel closing) (Fig. 4C). On the other hand, KChIP4a had no effect on prepulse inactivation, development of closed-state inactivation, or recovery from inactivation (Fig. 4D–F). Note that, like KChIP1, the core domains of all KChIP1–4 also considerably speed up Kv4.3 recovery from inactivation (Fig. 4F; ref. 15), suggesting that the lack of effect on Kv4.3 recovery from inactivation is because of the KIS domain.
Figure 4.
Effects of the KIS domain on Kv4.3 channel's activation, closing, and inactivation. (A) Time constants (τ) of the rising phases of Kv4.3 and Kv4.3/KChIP4a currents at different voltages; n > 5. (B) pG-V relationship of Kv4.3 and Kv4.3/KChIP4a currents; n = 9. (C) Deactivation time constants of Kv4.3 and Kv4.3/KChIP4a at −100 mV. *, significant (P < 0.05); n > 20. (D) Prepulse inactivation as a function of voltages; n = 11. (E) Time constants of development of closed-state inactivation at −70 mV. ns, not significant; n = 4–6. (F) Recovery from inactivation of Kv4.3 and Kv4.3 coexpressed with KChIP1, KChIP4a, and Δ2–44KChIP4a at −80 mV; n = 5–8. (G) Working hypothesis of effects of KIS domain on Kv4.3 gating based on published Kv4 gating models (23, 24, 31, 32). Dashed and/or shortened arrows indicate weakened transition processes by KIS whereas solid arrows indicate lack of effect by KIS. The open state in this diagram probably represents an aggregate of open states with distinct conductances as suggested by the single channel recordings. C, closed; O, open; and I, inactivation.
Taken together, our data indicate two net effects of the KIS domain on Kv4.3 gating. First, the KIS domain delays channel activation, as suggested by the slower rising phase (Fig. 4A, 3). Second, once the channel is open, the KIS domain keeps it open longer by eliminating fast inactivation (Fig. 2) and hindering channel closing (Fig. 4C). Based on previously published Kv4-gating models (23, 24, 31, 32), we provide a conceptual scheme in Fig. 4G that summarizes how the KIS domain might influence Kv4 gating. Although it is harder for Kv4 channels to reach the open state in the presence of the KIS domain, the impaired (dashed arrows) open-state inactivation and channel closing effectively favor the open state of the channel once the channel opens.
Molecular Biology of KChIP4a.
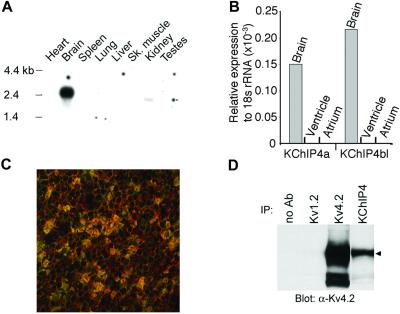
The presence of the KIS domain makes KChIP4a a unique member of the KChIP family as none of the other reported KChIP members possess similar function (ref. 15; Fig. 1). A group of splice variants of KChIP4a, named KChIP4b here, has been deposited in GenBank by others (as CALP/KChIP4/KChIP4.1–3) and by us. The GenBank accession nos. for the deposited KChIP4b entries are (h, human; m, mouse, and r, rat): AF453245(r), AF453246(h), AF302044(h), AF305071(m), AF305072(h), AF345445(r) (33), AF345444(r) (33), XM_017682(h), AY029176(h), AF367024(h), AF367023(h). All KChIP4b members carry alternatively spliced N-terminal domains that have no resemblance to that of KChIP4a and differ among themselves solely by length (one also differs at its C terminus). All KChIP4 variants are extremely conserved between rodent and human (e.g., 100% identity at amino acid level between mouse and human KChIP4a carrying the KIS domain). A probe derived from the 3′ untranslated region of a clone of rat KChIP4, presumably shared by all of the known splice variants, showed brain-predominant expression in Northern blot (Fig. 5A). Real-time reverse transcription–PCR (TaqMan) showed that both KChIP4a and the longest member of KChIP4b, KChIP4bl (Fig. 1), were present in mouse brain but absent in atrium or ventricle (Fig. 5B). We developed KChIP4-specific mAbs. These Abs selectively recognize the core domain of KChIP4 but not KChIP1–3. By using such Abs, it is clear that KChIP4a coassociated with Kv4.2 in COS cells (by coimmunoprecipitation and immunocytochemistry, data not shown). Further, immunoreactivity of these KChIP4-specific Abs was colocalized with that for Kv4.2 in multiple brain regions and coassociated with Kv4.2 in immunoprecipitates of rat hippocampal membranes (Fig. 5 C and D). The shortest member of KChIP4b is very similar to Δ2−44KChIP4a (the only difference being insertion of 2 amino acids, alanine and threonine, between positions 1 and 2 of Δ2−44KChIP4a). We found KChIP4bl to behave very similarly to KChIP1–3 electrophysiologically (not shown). This result further refines the location of KIS domain to be within the differentially spliced, N-terminal 34-aa module of KChIP4a.
Figure 5.
Association of KChIP4 and Kv4.2 in brain. (A) KChIP4 mRNA is predominantly expressed in rat brain by Northern analysis. The Northern mRNA blot was purchased from CLONTECH. (B) Both KChIP4a and KChIP4bl are expressed in mouse brain by real-time reverse transcription–PCR (TaqMan) analysis. (C) Confocal image shows colocalization (yellow) of Kv4.2 immunoreactivity (red) and KChIP4 core domain immunoreactivity (green) in the cell bodies and glomeruli formed by the dendrites of rat cerebellar granule neurons. (D) Coprecipitation of immunoreactivities against Kv4.2 and KChIP4 core domain from rat hippocampal lysate. IP, immunoprecipitation. The small triangle indicates position of full-length Kv4.2 protein.
Combinatorial Modulation of Kv4 Currents by KChIPs With and Without the KIS Domain.
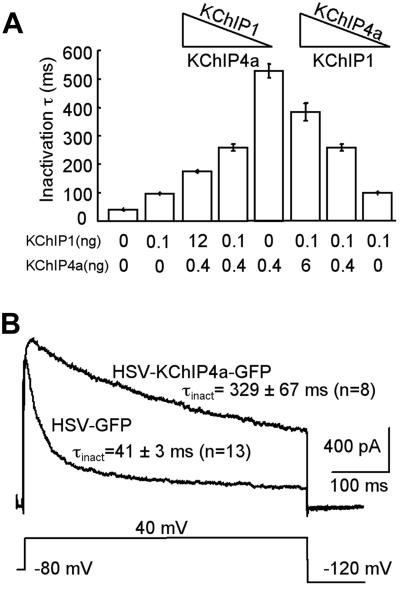
Because all KChIPs are highly expressed in neurons, their expression patterns may conceivably overlap in subsets of neurons under normal or pathological conditions. For example, immunoreactivities against the core domains of both KChIP1 and KChIP4a are detected in cerebellar granule neurons (Fig. 5C; ref. 15). We were thus intrigued in learning how Kv4 A-currents would be modified when both KChIP4a and KChIP1 are present in a single cell. We coinjected varying ratios of KChIP4a and KChIP1 cRNAs together with a fixed amount of Kv4.3 cRNA into oocytes. As shown in Fig. 6A, when the KChIP1 cRNA level was held constant, increasing KChIP4a cRNA caused the inactivation time constants of Kv4.3 current to gradually become more similar to that of Kv4.3/KChIP4a. The reciprocal was also true (Fig. 6A). These data suggest that, instead of an all-or-none effect, a spectrum of Kv4.3 current decay can be achieved solely by varying ratios of its auxiliary subunits. We then tested the same hypothesis in neurons. As shown in Fig. 6B, in cerebellar granule neurons transduced with HSV expressing KChIP4a-GFP, the inactivation of the A-type current was considerably slowed compared to the HSV-GFP control. We conclude that Kv4 currents show a mixed profile of KChIP1 and KChIP4a modulation when both KChIPs are present at the same time. This effect may be caused by mutually exclusive binding to Kv4, a mixed population of KChIP1- and KChIP4a-bound channels, a heteromeric channel complex with both KChIPs, or a combination of all of the above.
Figure 6.
KChIP4a and KChIP1 combinatorially modulate Kv4.3 inactivation. (A) KChIP4a and KChIP1 mixing experiments in oocytes. The amount of KChIP4a and KChIP1 cRNAs coinjected with 0.4 ng of Kv4.3 cRNA is indicated under each column. The triangles highlight constant levels and increasing/decreasing trends of KChIP1 or KChIP4a cRNAs in these experiments; n = 3–16. (B) KChIP4a expressed from a HSV vector slows inactivation of the transient A-current in cultured cerebellar granule neurons; n = 8–13.
Discussion
KChIPs with (e.g., KChIP4a) and without (e.g., KChIP1) the KIS domain produce distinct modulations of Kv4 currents. Although KChIP4a delays channel opening, when the channel opens, it favors the open state by disrupting fast inactivation and slowing channel closing (Fig. 4G). In contrast, KChIP1 speeds up inactivation from closed state and accelerates channel closing (34). As a result, Kv4 currents decay still reasonably fast in the presence of KChIP1–3 but very slowly in the presence of KChIP4a (Fig. 2). KChIP1 also differs from KChIP4a in that KChIP1 shifts the pG-V curve in the hyperpolarizing direction and speeds up recovery from inactivation (15), whereas KChIP4a modestly shifts the pG-V curve in the depolarizing direction and has no effect on recovery.
Our data clearly indicate that effects of the KIS domain on Kv4 currents are dominant over the effects of the core domain (Fig. 2 and ref. 15). The core domains alone of all KChIPs modulate Kv4 almost identically to full-length KChIPs without the KIS domain. Therefore, the core domains are sufficient to carry out two functions: channel binding and channel modulation. The KIS domain masks at least two aspects of the latter function. First, KIS domain keeps the channel open much longer as opposed to the core domain (Fig. 2). Second, the KIS domain completely reverses the acceleration of recovery from inactivation by the core domains (Fig. 4F). This dominant action highlights the importance of the KIS domain and provides an example of function mediated by the N-terminal domains of KChIPs.
The contribution of the KIS domain to native Kv4-mediated currents is not clear. We have not seen reports that attribute slowly inactivating potassium currents to the α-subunits of Kv4. We offer several possibilities for the apparent absence of such a connection. First, our mixing experiments (Fig. 6) suggest that the subthreshold A-current may be formed in native cells by Kv4 and a mixture of KChIPs with and without the KIS domain. Because it is the ratio of such KChIPs in a given cell that determines the electrophysiological properties of the underlying Kv4 currents, the effects of KChIP4a in most native neurons may not be as strong as those obtained in simplified expression systems in vitro. The best native system to reveal KChIP4a effect would be Kv4-positive neurons exclusively expressing KChIP4a. However, we do not know specifically where KChIP4a might be expressed because the Abs we used recognize the core domain of KChIP4. Second, although our TaqMan data obtained with a KIS-specific probe indicate that KChIP4a mRNA is indeed present significantly in brain (Fig. 5B), KChIP4a protein might not be made in all putative locations. Finally, the Kv4/KChIP4a current may be present in types of neurons whose native potassium currents have yet to be carefully characterized under normal or pathological conditions. It is encouraging to note, however, that the kinetic and voltage-dependent properties of several reported slowly inactivating K-currents bear many similarities with the Kv4/KChIP4a current studied here (35–38). Kv4/KChIP4a current is unlikely to be the D-type potassium current (ID) as they have distinct pharmacology. ID is sensitive to dendrotoxin (35, 36, 39), but Kv4/KChIP4a is not (unpublished data). Additional histochemical, pharmacological, and electrophysiological studies are necessary to establish a concrete connection between native currents and the Kv4/KChIP4a complex.
Auxiliary subunits of potassium channels play important roles in determining inactivation profiles of the pore-forming α subunits. The classic example is Kvβ1, the β subunit of Kv1 channels, which converts the noninactivating Kv1.1 current to a fast inactivating current using the ball domain (16–19, 40–43). Other examples include the β2 and β3 subunits of the Ca2+-activated large conductance potassium channel (maxi-K), which result in similar changes in inactivation (20–22). With the KIS domain, KChIP4a represents an auxiliary subunit that leads to the opposite conversion of inactivation of a potassium channel (i.e., from fast inactivating to very slow inactivating).
The interacting site of the KIS domain may involve residues around the inner vestibule of the pore because the kinetic effects of KChIP4a on Kv4.3 are remarkably similar to those of previously studied point mutations in the distal part of the S6 segment and the S4–5 loop of Kv4.1 (24). These residues, plus the intracellular N-terminal and C-terminal domains, determine the distinct non-N/non-C type of inactivation of Kv4 channels (24, 31). Despite the opposite roles in inactivation, the Kvβ family and the KChIP family have similar primary structural organizations, which include variable N-terminal domains and conserved C-terminal domains, and both families bind to the N-terminal domains of their cognate channels. Given recent evidence for the ball domain to reach its receptor site inside the pore through lateral internal “windows” (43), it also is conceivable that the KIS domain may similarly reach its receptor site at the pore, but achieve a distinct outcome with Kv4 channels that use different inactivation mechanisms (23, 24, 31, 32). In this sense, KIS also may differ from the N-type inactivation prevention domain present on the Kv1.6 α-subunit because it is thought that the N-type inactivation prevention domain antagonizes the inactivation ball instead of acting on residues around the inner mouth of the pore (44). Together with others (e.g., refs. 16, 17, 20–22, and 45), our data demonstrate that auxiliary subunits can dramatically drive potassium channel inactivation to opposite extremes. Such mechanisms actively tailor electrical signaling in excitable cells.
Acknowledgments
We thank Dr. Karl Clauser and Ms. Maria Knoppers for biochemical assistance, Drs. Gary Yellen, Morgan Sheng, Irwin Levitan, James Trimmer, Mark Bowlby, and Qiang Lu for critical reading of the manuscript, and Drs. Kevin Willis and James Barrett for support. M.C. and R.H.-P. were supported by the National Institutes of Health (NS-32337).
Abbreviations
- KChIP
K-channel interacting protein
- KIS
K-channel inactivation suppressor
- GFP
green fluorescent protein
- HSV
herpes simplex virus
- pG-V
peak conductance-voltage
Footnotes
References
- 1.Maletic-Savatic M, Lenn N J, Trimmer J S. J Neurosci. 1995;15:3840–3851. doi: 10.1523/JNEUROSCI.15-05-03840.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheng M, Tsaur M L, Jan Y N, Jan L Y. Neuron. 1992;9:271–284. doi: 10.1016/0896-6273(92)90166-b. [DOI] [PubMed] [Google Scholar]
- 3.Serodio P, Rudy B. J Neurophysiol. 1998;79:1081–1091. doi: 10.1152/jn.1998.79.2.1081. [DOI] [PubMed] [Google Scholar]
- 4.Dixon J E, Shi W, Wang H S, McDonald C, Yu H, Wymore R S, Cohen I S, McKinnon D. Circ Res. 1996;79:659–668. doi: 10.1161/01.res.79.4.659. [DOI] [PubMed] [Google Scholar]
- 5.Barry D M, Xu H, Schuessler R B, Nerbonne J M. Circ Res. 1998;83:560–567. doi: 10.1161/01.res.83.5.560. [DOI] [PubMed] [Google Scholar]
- 6.London B, Wang D W, Hill J A, Bennett P B. J Physiol (London) 1998;509:171–182. doi: 10.1111/j.1469-7793.1998.171bo.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu H, Dixon J E, Barry D M, Trimmer J S, Merlie J P, McKinnon D, Nerbonne J M. J Gen Physiol. 1996;108:405–419. doi: 10.1085/jgp.108.5.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffman D A, Magee J C, Colbert C M, Johnston D. Nature (London) 1997;387:869–875. doi: 10.1038/43119. [DOI] [PubMed] [Google Scholar]
- 9.Johnston D, Hoffman D A, Magee J C, Poolos N P, Watanabe S, Colbert C M, Migliore M. J Physiol (London) 2000;525:75–81. doi: 10.1111/j.1469-7793.2000.00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shibata R, Nakahira K, Shibasaki K, Wakazono Y, Imoto K, Ikenaka K. J Neurosci. 2000;20:4145–4155. doi: 10.1523/JNEUROSCI.20-11-04145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song W, Tkatch T, Baranauskas G, Ichinohe N, Kitai S T, Surmeier D J. J Neurosci. 1998;18:3124–3137. doi: 10.1523/JNEUROSCI.18-09-03124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serodio P, Kentros C, Rudy B. J Neurophysiol. 1994;72:1516–1529. doi: 10.1152/jn.1994.72.4.1516. [DOI] [PubMed] [Google Scholar]
- 13.Guo W, Li H, London B, Nerbonne J M. Circ Res. 2000;87:73–79. doi: 10.1161/01.res.87.1.73. [DOI] [PubMed] [Google Scholar]
- 14.Xu H, Li H, Nerbonne J M. J Physiol (London) 1999;519:11–21. doi: 10.1111/j.1469-7793.1999.0011o.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An W F, Bowlby M R, Betty M, Cao J, Ling H P, Mendoza G, Hinson J W, Mattsson K I, Strassle B W, Trimmer J S, et al. Nature (London) 2000;403:553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- 16.Rettig J, Heinemann S H, Wunder F, Lorra C, Parcej D N, Dolly J O, Pongs O. Nature (London) 1994;369:289–294. doi: 10.1038/369289a0. [DOI] [PubMed] [Google Scholar]
- 17.Scott V E, Rettig J, Parcej D N, Keen J N, Findlay J B, Pongs O, Dolly J O. Proc Natl Acad Sci USA. 1994;91:1637–1641. doi: 10.1073/pnas.91.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pongs O, Leicher T, Berger M, Roeper J, Bahring R, Wray D, Giese K P, Silva A J, Storm J F. Ann NY Acad Sci. 1999;868:344–355. doi: 10.1111/j.1749-6632.1999.tb11296.x. [DOI] [PubMed] [Google Scholar]
- 19.Zagotta W N, Hoshi T, Aldrich R W. Science. 1990;250:568–571. doi: 10.1126/science.2122520. [DOI] [PubMed] [Google Scholar]
- 20.Xia X M, Ding J P, Lingle C J. J Neurosci. 1999;19:5255–5264. doi: 10.1523/JNEUROSCI.19-13-05255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia X M, Ding J P, Zeng X H, Duan K L, Lingle C J. J Neurosci. 2000;20:4890–4903. doi: 10.1523/JNEUROSCI.20-13-04890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallner M, Meera P, Toro L. Proc Natl Acad Sci USA. 1999;96:4137–4142. doi: 10.1073/pnas.96.7.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beck E J, Covarrubias M. Biophys J. 2001;81:867–883. doi: 10.1016/S0006-3495(01)75747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jerng H H, Shahidullah M, Covarrubias M. J Gen Physiol. 1999;113:641–660. doi: 10.1085/jgp.113.5.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmqvist M H, Cao J, Knoppers M H, Jurman M E, Distefano P S, Rhodes K J, Xie Y, An W F. J Neurosci. 2001;21:4154–4161. doi: 10.1523/JNEUROSCI.21-12-04154.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhodes K J, Strassle B W, Monaghan M M, Bekele-Arcuri Z, Matos M F, Trimmer J S. J Neurosci. 1997;17:8246–8258. doi: 10.1523/JNEUROSCI.17-21-08246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhodes K J, Monaghan M M, Barrezueta N X, Nawoschik S, Bekele-Arcuri Z, Matos M F, Nakahira K, Schechter L E, Trimmer J S. J Neurosci. 1996;16:4846–4860. doi: 10.1523/JNEUROSCI.16-16-04846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith I L, Hardwicke M A, Sandri-Goldin R M. Virology. 1992;186:74–86. doi: 10.1016/0042-6822(92)90062-t. [DOI] [PubMed] [Google Scholar]
- 29.Fraefel C, Song S, Lim F, Lang P, Yu L, Wang Y, Wild P, Geller A I. J Virol. 1996;70:7190–7197. doi: 10.1128/jvi.70.10.7190-7197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraefel C. Current Protocols in Neuroscience. New York: Wiley; 1999. [Google Scholar]
- 31.Jerng H H, Covarrubias M. Biophys J. 1997;72:163–174. doi: 10.1016/S0006-3495(97)78655-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bahring R, Boland L M, Varghese A, Gebauer M, Pongs O. J Physiol (London) 2001;535:65–81. doi: 10.1111/j.1469-7793.2001.00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosati B, Pan Z, Lypen S, Wang H S, Cohen I, Dixon J E, McKinnon D. J Physiol (London) 2001;533:119–125. doi: 10.1111/j.1469-7793.2001.0119b.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beck, E. J., Bowlby, M. R., An, W. F., Rhodes, K. J. & Covarrubias, M. (2002) J. Physiol. (London), in press. [DOI] [PMC free article] [PubMed]
- 35.Storm J F. Nature (London) 1988;336:379–381. doi: 10.1038/336379a0. [DOI] [PubMed] [Google Scholar]
- 36.Wu R L, Barish M E. J Neurosci. 1992;12:2235–2246. doi: 10.1523/JNEUROSCI.12-06-02235.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greene R W, Haas H L, Reiner P B. J Physiol (London) 1990;420:149–163. doi: 10.1113/jphysiol.1990.sp017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spain W J, Schwindt P C, Crill W E. J Physiol (London) 1991;434:591–607. doi: 10.1113/jphysiol.1991.sp018488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stansfeld C E, Marsh S J, Halliwell J V, Brown D A. Neurosci Lett. 1986;64:299–304. doi: 10.1016/0304-3940(86)90345-9. [DOI] [PubMed] [Google Scholar]
- 40.Demo S D, Yellen G. Neuron. 1991;7:743–753. doi: 10.1016/0896-6273(91)90277-7. [DOI] [PubMed] [Google Scholar]
- 41.Isacoff E Y, Jan Y N, Jan L Y. Nature (London) 1991;353:86–90. doi: 10.1038/353086a0. [DOI] [PubMed] [Google Scholar]
- 42.Hoshi T, Zagotta W N, Aldrich R W. Science. 1990;250:533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- 43.Zhou M, Morais-Cabral J H, Mann S, MacKinnon R. Nature (London) 2001;411:657–661. doi: 10.1038/35079500. [DOI] [PubMed] [Google Scholar]
- 44.Roeper J, Sewing S, Zhang Y, Sommer T, Wanner S G, Pongs O. Nature (London) 1998;391:390–393. doi: 10.1038/34916. [DOI] [PubMed] [Google Scholar]
- 45.Xu J, Li M. J Biol Chem. 1997;272:11728–11735. doi: 10.1074/jbc.272.18.11728. [DOI] [PubMed] [Google Scholar]