Abstract
Nucleotide excision repair (NER) is the major DNA repair process that removes diverse DNA lesions including UV-induced photoproducts. There are more than 20 proteins involved in NER. Among them, XPC is thought to be one of the first proteins to recognize DNA damage during global genomic repair (GGR), a sub-pathway of NER. In order to study the mechanism through which XPC participates in GGR, we investigated the possible modifications of XPC protein upon UV irradiation in mammalian cells. Western blot analysis of cell lysates from UV-irradiated normal human fibroblast, prepared by direct boiling in an SDS lysis buffer, showed several anti-XPC antibody-reactive bands with molecular weight higher than the original XPC protein. The reciprocal immunoprecipitation and siRNA transfection analysis demonstrated that XPC protein is modified by SUMO-1 and ubiquitin. By using several NER-deficient cell lines, we found that DDB2 and XPA are required for UV-induced XPC modifications. Interestingly, both the inactivation of ubiquitylation and the treatment of proteasome inhibitors quantitatively inhibited the UV-induced XPC modifications. Furthermore, XPC protein is degraded significantly following UV irradiation in XP-A cells in which sumoylation of XPC does not occur. Taken together, we conclude that XPC protein is modified by SUMO-1 and ubiquitin following UV irradiation and these modifications require the functions of DDB2 and XPA, as well as the ubiquitin–proteasome system. Our results also suggest that at least one function of UV-induced XPC sumoylation is related to the stabilization of XPC protein.
INTRODUCTION
Nucleotide excision repair (NER) is a versatile DNA repair pathway to eliminate various structurally unrelated lesions that distort the double helix, including UV light-induced cyclobutane pyrimidine dimmers (CPDs) and pyrimidine (6-4) pyrimindone photoproducts (6-4PP), as well as intrastrand cross-links and bulky adducts induced by numerous chemical compounds (1). NER has two distinct subpathways, global genomic repair (GGR) and transcription-coupled repair (TCR). The former removes DNA lesions from the entire genome whereas the latter only removes DNA damage from the transcribed strands of transcriptionally active genes (2). Impaired NER activity has been associated with several human genetic disorders including Xeroderma Pigmentosum (XP), for which seven NER-deficient genetic complementation XP groups (XP-A to -G) have been identified. Unlike most XP complementation groups, XP-C patients show a defect only in GGR but TCR is normal. The gene defective in XP-C patients encodes the XPC protein, which exists in vivo as a heterotrimeric complex with hHR23B and centrin 2 (3–5). XPC-hHR23B appears to function as a damage recognition factor for GGR. Generally, XPC-hHR23B functions by recognizing and binding structural abnormalities introduced into double-stranded DNA by the lesions rather than recognizing any structural characteristics of the lesions themselves (6,7). Conformational changes in DNA induced by XPC-hHR23B could favor the subsequent binding of other NER factors such as TFIIH, XPA, RPA and two NER endonucleases XPG and ERCC1-XPF (6,8,9). Finally, the damage-containing oligonucleotide is removed by dual incisions and the gap is filled by DNA synthesis and ligation.
The changes of XPC protein levels during NER have been suggested in several studies using mouse and human cells. When XPC-GFP fusion protein was stably expressed in the mHR23A/B DKO MEFs (double knock out mouse embryo fibroblasts) together with hHR23B, UV irradiation resulted in dramatic accumulation of XPC-GFP (10). Compared to the exogenously expressed proteins, Okuda et al. (11) have shown that endogenous mouse XPC is relatively stable, and UV irradiation only induced ∼10% increase of XPC level at 9 h post-irradiation. However, experiments of Adimoolam and Ford (12) demonstrated that human endogenous XPC protein is significantly upregulated following UV irradiation in several human cells, and this response depends on the p53 status. Furthermore, studies in Saccharomyces cerevisiae indicated that the rapid degradation of ectopically expressed Rad4, the yeast homologue of XPC, appeared to be mediated by multi-ubiquitylation and DNA damage transiently stabilized the overexpressed Rad4 (13). In both yeast and mammalian systems, HR23B (in yeast, Rad23) has been shown to function in NER by governing XPC stability via partial protection against proteasomal degradation (10,13). However, the finding of UV-induced modest accumulation of mXPC in mHR23−/−, as well as DKO cells indicates the existence of additional mechanism for mXPC accumulation (e.g. the post-translational modification), for which the mHR23 proteins are not necessary (11).
Small ubiquitin-related modifier (SUMO) is the best-characterized member of a growing family of ubiquitin-like proteins involved in post-translational modifications (14–16). In mammals, there are three members of the SUMO protein family, SUMO-1, SUMO-2 and SUMO-3, which are implicated in partly overlapping, yet distinct functions (17,18). SUMO is covalently attached to other proteins through the activities of an enzyme cascade similar to that for ubiquitylation. There is only one known SUMO-activating enzyme, E1 and only one known SUMO-conjugating enzyme, E2 (Ubc9). The functional consequences of the SUMO attachment vary greatly from substrate to substrate, and in many cases, such consequences are not understood at the molecular level. Unlike ubiquitylation, sumoylation of proteins has not been linked to protein degradation. Proposed functions for sumoylation include regulation of protein–protein interaction and localization, inhibition of ubiquitin-mediated degradation, and regulation of transcription (16,19).
In the current study, we have shown that XPC protein is modified covalently following UV irradiation. The reciprocal immunoprecipitation (IP) and RNA interference studies demonstrated that UV-induced XPC modifications are both SUMO-1 and ubiquitin conjugated. We also present evidence suggesting that the UV-induced modifications of XPC are related to several NER factors including DDB2 and XPA, as well as ubiquitin–proteasome system.
MATERIALS AND METHODS
Cell culture and protein extraction
The normal human skin fibroblasts (OSU-2), expressing wild-type p53, were established and maintained in culture as previously described (20). Li-Fraumeni Syndrome (LFS) fibroblast 041 cell line (p53-Null) was provided by Dr Michael Tainsky (MD Anderson Cancer center, Austin, TX). Both cell lines were grown in DMEM supplemented with 10% fetal calf serum (FCS) and antibiotics at 37°C in a humidified atmosphere of 5% CO2. The primary NER-deficient fibroblast cell lines GM05509A (XP-A), GM15983(XP-C), GM01389A (XP-E), GM04313E (XP-F) and GM03021B (XP-G) were purchased from NIGMS Human Genetic Cell Repository (Coriell Institute for Medical Research, Camden, NJ) and were grown in MEM with 2× essential amino acid, 2× non-essential amino acid and 2× vitamin supplemented with 15% FCS and antibiotics. Human lung epithelial H460 cells and XPA siRNA-expressing H460 cells [Achieved by infection with a pRETRO-XPA retroviral vector encoding a hairpin-forming siRNA specific to XPA (21)] were obtained from Dr Anatoly Zhitkovich (Brown University, Providence, RI). Mouse embryo fibroblast ts20 (thermosensitive for E1 ubiquitin-activating enzyme) and its parental cell line A31N were kindly provided by Dr Harvey L. Ozer (UMDNJ-New Jersey Medical School). Both cell lines were cultured in 50% F-10 + 50% DMEM medium containing 10% FCS and antibiotics at 32°C in a humidified atmosphere of 5% CO2 (22). When needed, cells were transferred to the restrictive temperature (39°C) 16 h prior to the treatment and beginning of the experiments. UV irradiation was performed from a germicidal lamp at a dose rate of 1.0 J/m2/s as measured by a Kettering model 65 radiometer (Cole palmer Instrument Co., Vernon Hill, IL). At each time point, the cells were harvested and the cell lysates were prepared by direct boiling in SDS lysis buffer (62.5 mM Tris–HCl, pH 6.8, 10% glycerol, 2% SDS and protease inhibitors).
Antibodies and expression constructs
Anti-XPC antibody (XPC-2) was generated by immunizing rabbits with the synthetic peptide (KTKREKKAAASHLFPFEKL) corresponding to the C-terminus of human XPC protein and affinity-purified with the peptide (BioSource, Hopkinton, MA). This antibody was found to recognize not only the human but also the mouse XPC protein. Another polyclonal anti-XPC antibody that is specific to the full length of XPC protein was generously provided by Dr Fumio Hanaoka (Osaka University, Japan). Anti-DDB2 antibody (DDB2-A) was generated by immunizing rabbits with the synthetic peptide (KRPETQKTSEIVLRPRNKR) corresponding to the N-terminus of human DDB2 protein and affinity-purified with the peptide (BioSource). Polyclonal anti-XPA (FL-273), anti-XPB (S-19), anti-SUMO-1 (FL-101), anti-Ubiquitin (FL-76), anti-Lamin B (C-20), anti-Ubc9 (H-81) and monoclonal anti-p53 (DO-1) antibodies were from Santa Cruz Biotechnology, Inc (Santa Cruz, CA); rabbit anti-SUMO-2/3 was from Zymed (South San Francisco, CA); monoclonal anti-Actin was from Neomarkers (Fremont, CA); monoclonal anti-FLAG was from Stratagene (La Jolla, CA); monoclonal anti-HA was from BD Bioscience (San Jose, CA). The pcDNA3.1/XPC-V5-His and the pcDNA3.1/XPA-V5-His expression constructs were generated by respectively sub-cloning XPC and XPA cDNA into pcDNA3.1/V5-His vector (Invitrogen, Carlsbad, CA). The FLAG-tagged DDB2 construct (pBJ5-DDB2) was provided by Dr Gilbert Chu (Stanford University, Stanford, CA). SUMO-FLAG was cut out from a pRK-based expression vector for SUMO-FLAG (generously provided by Dr Xin-Hua Feng, Baylor College of Medicine) with ClaI and HindIII, and then sub-cloned into pcDNA3.1 vector (Invitrogen). Wild-type p53 expression vector was kindly provided by Dr Bert Vogelstein (Johns Hopkins University, Baltimore, MD). The expression vector for Ub-HA was obtained from Dr Dirk Bohmann (European Molecular Biology Laboratory, Heidelberg, Germany). Cell transfections were conducted with FuGene 6 (Roche, Indianapolis, IN) or Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction.
Immunoprecipitation and western blotting
IP was performed on the whole-cell lysates. The cell lysates prepared by direct boiling in SDS lysis buffer were diluted 10-folds with RIPA buffer (50 mM Tris–HCl, pH 7.4, 1% NP40, 0.25% Sodium Deoxycholate, 1 mM EDTA and 150 mM NaCl). Following preclearing with 30 μl of Protein G plus/A agarose beads (Oncogene Research Products, San Diego, CA), 2 mg of total protein was incubated with desired antibodies overnight at 4°C, and then incubated with 50 μl of Protein G plus/A agarose beads for another 1 h at 4°C. The beads were washed four times with RIPA buffer, and then boiled in 2× SDS sample buffer. The immunoprecipitates were then subjected to SDS–PAGE and the specific proteins were detected by immunoblotting with desired antibodies, as described earlier (23).
RNA interference
The target sequence of the Ubc9 siRNA was GCAGAGGCCTACACGATTTAC (Dharmacon Research, Lafayette, CO), which corresponds to nucleotides 391–411 of the C-terminus of human Ubc9 coding region. siRNA was transfected into OSU-2 cells using Lipofectamine 2000 (Invitrogen), as described by the manufacturer's instruction. Cells, after 48 h siRNA transfection, were UV irradiated at 20 J/m2 and incubated for another 1 h. The cell lysates were prepared by direct boiling in SDS lysis buffer and the same amount of lysates were loaded onto a 4–12% gradient gel for SDS–PAGE and western blotting.
RESULTS
UV treatment induces a series of novel covalent modifications of XPC
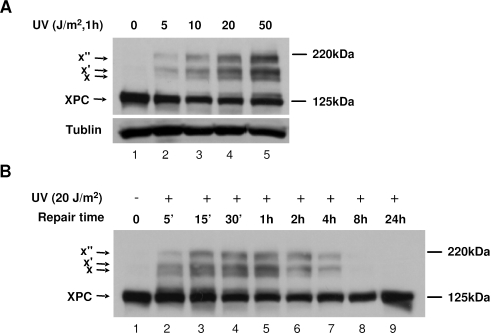
Western blotting of UV-irradiated normal human fibroblast OSU-2 cell lysates, prepared by direct boiling in SDS lysis buffer, revealed the existence of at least three protein species cross-reacting with an XPC-specific polyclonal antibody—one of the proteins migrating at 125 kDa and others at 145–220 kDa (Figure 1A). The predominant 125 kDa band is established to represent endogenous XPC protein. However, the identity of 145–220 kDa proteins (discrete bands denoted by X, X′ and X″), whose intensities are enhanced with the increased doses of UV treatment, was obscure. In addition, our results showed that the amount of 125 kDa XPC decreased following 1 h of UV irradiation at the dose ranges from 5 to 50 J/m2. In an extended time course experiment, the higher molecular mass forms of XPC appeared within 5 min following 20 J/m2 UV treatment, reached the maximal level by 1 h and were no longer detectable after 8 h (Figure 1B). The same UV radiation-induced pattern of XPC variants was also detected in extracts from several different human cell lines, e.g. HeLa, WI38, HCT116 (data not shown) as well as in extracts of mouse cells (see results below). Most importantly, these protein species as well as the endogenous XPC were not detected in XP-C cells (data not shown) but could be detected in XP-C cells ectopically expressing XPC-V5-His (see Results below), further confirming that 145–220 kDa proteins represented altered variants of endogenous XPC.
Figure 1.
Multiple high-molecular-weight bands of XPC resulted from UV irradiation of normal human cells. OSU-2 cells were UV irradiated at 0, 5, 10, 20 and 50 J/m2 and incubated for 1 h (A) or irradiated at 20 J/m2 and incubated for indicated times (B). Cell lysates were prepared by boiling in SDS lysis buffer as described in the Materials and Methods. Total protein (50 μg) was loaded for SDS–PAGE and detected by western blotting with rabbit anti-XPC antibody (XPC-2, 1:5000).
The higher molecular mass forms of XPC induced by UV radiation are SUMO-1 and ubiquitin conjugated
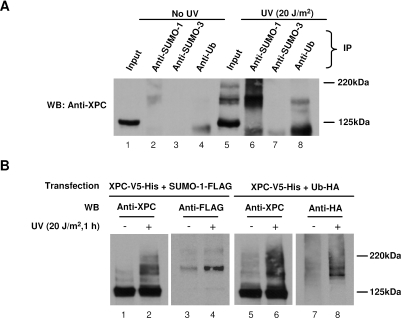
Since the UV-induced high-molecular-weight forms of XPC were detected under the denaturing condition (boiled in SDS lysis buffer), we reason that XPC is modified covalently. The common covalent modifications of proteins include phosphorylation, acetylation, ubiquitylation and sumoylation, etc. Because the modified XPC has a high molecular-weight which is at least 20 kDa more than that of native XPC, we speculated that these modifications might be ubiquitylation or sumoylation. In order to characterize such modifications, we immunoprecipitated the lysates from both UV-treated and mock-treated OSU-2 cells with anti-SUMO-1, anti-SUMO-2/3 or anti-ubiquitin antibodies. The immunoprecipitates were subjected to immunoblot analysis with rabbit anti-XPC antibody. As shown in Figure 2A, no high-molecular-weight form of XPC could be detected in the lysate from mock-treated sample (lane 1), whereas the distinct high-molecular-weight forms of XPC could be detected in the lysate from UV-treated sample (lane 5). Anti-SUMO-1, anti-SUMO-2/3 and anti-ubiquitin antibodies did not pull down any detectable XPC variant in mock-treated sample (lanes 2–4). In contrast, these XPC variants were clearly detected in the immunoprecipitates from anti-SUMO-1, but not anti-SUMO-2/3 antibody in the UV-treated sample (lanes 6 and 7), indicating that UV-induced covalent conjugates of XPC might be SUMO-1. It seems that XPC is also modified by ubiquitin following UV irradiation as we also detected a faint anti-XPC reactive band (compared to lane 6) in the position of modified XPC within the immunoprecipitates derived with anti-ubiquitin antibody. To confirm these results, we performed a reciprocal IP experiment in which we immunorecovered the XPC proteins from the lysates and detected the SUMO-1 or ubiquitin conjugated proteins. Because our anti-XPC antibody was deemed unsuitable for IP, we expressed XPC protein tagged with V5 and His along with SUMO-1-FLAG or ubiquitin-HA in XP-C cells and followed by UV irradiation of transfected cells. The XPC protein was purified with Ni-NTA agarose beads and detected by western blotting with anti-XPC for XPC protein, anti-FLAG for sumoylated XPC and anti-HA for ubiquitylated XPC. As shown in Figure 2B, the modified XPC protein could be detected in XP-C cells ectopically expressing XPC-V5-His following UV irradiation (Figure 2B, lanes 1, 2, 5 and 6). In XP-C cells transfected with both XPC and SUMO-1-FLAG, the high-molecular weight band in the position of the modified XPC could be detected by anti-FLAG antibody in UV-treated but not mock-treated samples (Figure 2B, lanes 3 and 4). Similarly, in XP-C cells transfected with both XPC and ubiquitin-HA, the high-molecular weight bands in the position of the modified XPC could also be detected by anti-HA antibody in UV-treated but not mock-treated samples (Figure 2B, lanes 7 and 8). These results further confirmed that XPC protein can be modified by both SUMO-1 and ubiquitin following UV irradiation.
Figure 2.
UV-induced high-molecular-weight bands of XPC are SUMO-1 conjugated (A) OSU-2 cells were UV irradiated at 20 J/m2 and incubated for 1 h. The cell lysates were prepared by direct lysis in SDS lysis buffer and diluted in RIPA buffer as described in Materials and Methods. Total protein (2 mg) was subjected to IP with anti-SUMO-1, anti-SUMO-2/3 or anti-Ubiquitin antibody. The precipitated proteins were detected by western blotting with anti-XPC antibody. Whole cell lysates (50 μg) were loaded on SDS–PAGE as input. (B) XP-C cells were transfected with either XPC-V5-His plus SUMO-1-FLAG or XPC-V5-His plus Ub-HA, then UV irradiated at 20 J/m2 and allowed to repair for 1 h. Total protein (2 mg) was used for pull-down with Ni-NTA agarose beads. The precipitated proteins were detected with anti-XPC, anti-FLAG or anti-HA antibody.
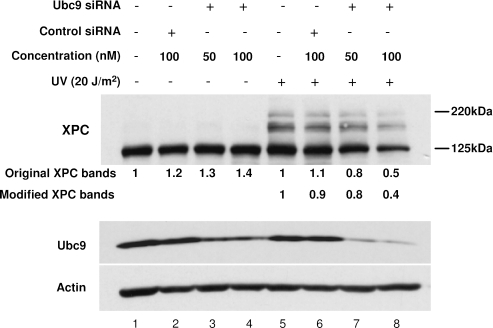
To further test whether one of the UV-induced XPC modifications is a result of sumoylation, we sought to assess the impact of disrupting the endogenous Ubc9 expression on the UV-induced XPC modification. Since Ubc9 is the only E2 enzyme for modification by members of SUMO family, inhibition of its expression would disrupt sumoylation of its substrates. We used a 21 nt siRNA that specifically targets to the human Ubc9 coding region and examined its effectiveness in silencing Ubc9 expression in OSU-2 cells as well as its influence on UV-induced XPC modification. After 48 h of transfection with the siRNA, OSU-2 cells expressed a significantly lower level of Ubc9 protein (Figure 3, lanes 3, 4, 7 and 8). In contrast, treatment of OSU-2 cells with unrelated control siRNA did not reduce the level of Ubc9 protein (Figure 3, lanes 2 and 6). As expected, no higher molecular-weight form of XPC was detected in mock-treated OSU-2 cells, irrespective of the siRNA transfection. On the contrary, UV irradiation indeed induced the higher molecular-weight forms of XPC and the amount of modified forms of XPC decreased significantly in cells transfected with Ubc9 siRNA, but not control siRNA (Figure 3, lanes 5–8). Relative inhibition measurement of modified forms, compared with those from cells without transfection, indicated ∼20 and 60% decrease in cells transfected with 50 and 100 nM Ubc9 siRNA, respectively (Figure 3, lanes 7 and 8). These results further confirmed that at least part of UV-induced modifications of XPC is sumoylation. Interestingly, transfection of Ubc9 siRNA also induced a decrease of the XPC level following UV irradiation, with a 50% decrease in cells transfected with 100 nM Ubc9 siRNA (Figure 3, lane 8 versus lane 5), indicating that Ubc9 acts as a stabilizer of XPC protein following UV irradiation possibly through conjugating SUMO to XPC protein.
Figure 3.
Ubc9 siRNA knocks down the expression of Ubc9 and compromises the UV-induced high-molecular-weight bands of XPC. OSU-2 cells were transiently transfected with either control siRNA or siRNA targeted to Ubc9 for 48 h. The cells were UV irradiated at 20 J/m2 and then incubated for another 1 h. The cell lysates were subjected to SDS–PAGE and proteins detected by western blotting with anti-XPC, anti-Ubc9 or anti-Actin antibody. The intensity of each band was determined by densitometry and the relative intensity of each band calculated in reference to control without transfection.
DDB2 is required in UV-induced XPC modifications
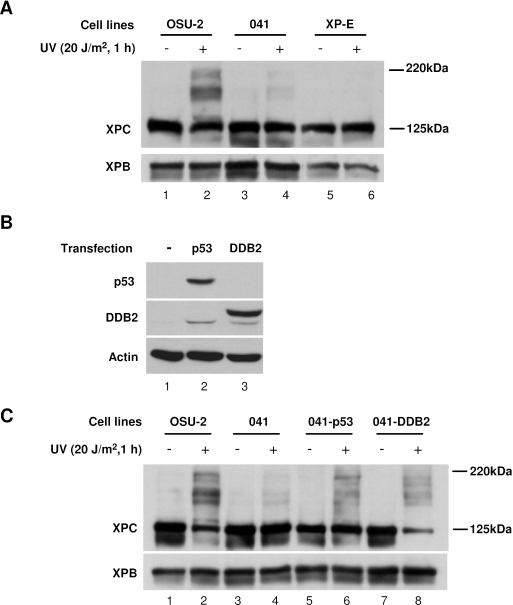
Our studies, and others, have shown that DDB2 is a key factor in regulating GGR of CPD, presumably through the recruitment of XPC to the DNA damage sites (24–26). Moreover, p53 could be participating in GGR via transactivating the expression of DDB2 protein (27). Thus, we tested whether DDB2 or p53 is involved in UV-induced XPC modifications. Normal human fibroblast OSU-2 cells, p53-deficient 041 cells and DDB2-deficient XP-E cells were treated with 20 J/m2 UV and allowed to repair for 1 h. Western blot analysis of cell lysates showed very weak modifications of XPC in 041 cells, but no modification in XP-E cells following UV irradiation (Figure 4A, lanes 3–6), whereas such modifications were very obvious in OSU-2 cells upon treatment with UV (Figure 4A, lanes 1 and 2). It seems that both p53 and DDB2 are required for UV-induced XPC modification. To further attest this result, we transfected p53 or DDB2 cDNA into 041 cells to determine whether the UV-induced XPC modifications could be visualized upon restoration of these proteins. It is known that DDB2 deficiency in 041 cells is due to the lack of its upstream transactivator p53 (27). Therefore, 041 cells with the ectopically expressed DDB2 alone can be used to study the function of DDB2 without possible interference of p53. At 48 h following transfection, the expression of p53 or DDB2 protein was confirmed in the transfected 041 cells. As shown in Figure 4B, 041 cells have undetectable level of p53 and DDB2 (lane 1). However, transfection of p53 not only restored the expression of p53 but also increased the endogenous DDB2 level (lane 2). In contrast, transfection of DDB2 only restored the expression of DDB2 while the p53 is still undetectable (lane 3). Thus, these cell lines can be used to study the role of p53 or DDB2 in UV-induced XPC modification. As shown in Figure 4C, the 041 cells transiently transfected with p53 construct (p53+, DDB2+) showed the restoration of UV-induced XPC modifications (Figure 4C, lane 6 versus lane 4). Furthermore, the 041 cells transfected with DDB2 construct alone (p53−, DDB2+) also demonstrated the restoration of UV-induced XPC modifications (Figure 4C, lane 8 versus lane 4). Thus, UV-induced sumoylation and ubiquitylation of XPC appear to require normal DDB2 gene product.
Figure 4.
p53 and DDB2 are involved in UV-induced XPC modification by SUMO-1 and ubiquitin (A) OSU-2, p53-deficient 041 and DDB2-deficient XP-E cells were UV irradiated at 20 J/m2 and incubated for 1 h. The cell lysates were subjected to western blotting with anti-XPC antibody. (B) and (C) 041 cell lines were transiently transfected with either p53 or DDB2-FLAG construct for 48 h. The cells were irradiated with UV at 20 J/m2 and repaired for 1 h. The segregated proteins were detected by western blotting with anti-p53 or anti-DDB2 to confirm the ectopic expression in transfectants (B). The XPC and its modifications were detected with rabbit anti-XPC antibody (C).
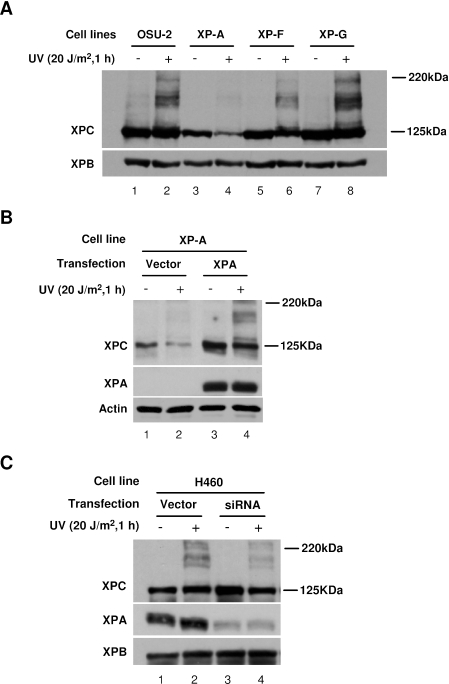
XPA is required for UV-induced modifications of XPC as well as preventing UV-induced XPC degradation
It is well known that there are six core repair factors in GGR, e.g. XPC-hHR23B, TFIIH, XPA, RPA, ERCC1-XPF and XPG (28–31) and these factors are assembled sequentially at the damage sites during the excision repair process (9). Amongst them, XPA is considered as another damage recognition factor to verify the damage and its recruitment to damage sites requires XPC (6,9). ERCC1-XPF and XPG, two endonucleases needed to incise the oligonucleotide containing damage site, are recruited to the damage sites during the late steps. In order to determine whether any of these relatively late congregating factors are required for UV-induced XPC modifications, we analyzed the XPC modifications in several cell lines including XPA-deficient XP-A cell line, XPF-deficient XP-F cell line and XPG-deficient XP-G cell line. Western blotting clearly showed that XPC modification is not induced upon UV irradiation in XP-A cells (Figure 5A, lane 4), whereas a substantial amount of XPC modification could be detected in XP-F cells following UV treatment (Figure 5A, lane 6). In contrast, a very strong UV-induced signal of XPC modification, comparable to that seen in normal OSU-2 cells, was detected in XP-G cells (Figure 5A, lane 8). Interestingly, the steady-state level of XPC in XP-A cells decreased significantly upon UV irradiation (Figure 5A, lane 3 versus 4). These results indicate that XPA is required for UV-induced modifications of XPC, and it seems that more XPC protein is degraded following UV irradiation in the absence of XPA protein. To verify the involvement of XPA in UV-induced XPC modification, we transfected XPA cDNA into this XP-A cells. As expected, the transfection restored not only the expression of XPA but also UV-induced XPC modifications in the XP-A cells (Figure 5B, lanes 3 and 4 versus lanes 1 and 2). Furthermore, the expression of XPA also reduced the UV-induced XPC degradation. We further tested another pair of isogenic H460 cell lines that are different only in the expression level of XPA. H460 cells stably transfected with either control vector or siRNA targeting to XPA were UV irradiated at 20 J/m2 and allowed to repair for 1 h. Western blotting with anti-XPA antibody showed that XPA protein level is decreased significantly in H460 cells with stably transfected XPA siRNA (Figure 5C). Immunoblot analysis of XPC showed that the amount of UV-induced XPC modifications was also reduced in correspondence to the decreased XPA in this cell line (Figure 5C). Meanwhile, the steady-state level of XPC was seen to decrease slightly following UV irradiation in H460 cells expressing XPA siRNA, whereas no obvious change of XPC level was observed in H460 cells with transfection of control vector. These data further indicate that XPA is a key determining factor in UV-induced modifications of XPC, and XPA might be a stabilizer of XPC protein during NER.
Figure 5.
UV-induced XPC modifications are compromised in XP-A cells (A) OSU-2, XP-A, XP-F and XP-G cells were UV irradiated at 20 J/m2 and incubated for another 1 h. The proteins were subjected to SDS–PAGE and detected with anti-XPC and anti-XPB (loading control) antibodies. (B) XP-A cells were transiently transfected with either empty vector or pcDNA3.1/XPA for 48 h, UV irradiated at 20 J/m2 and incubated for another 1 h. The proteins were subjected to SDS–PAGE and detected with anti-XPC, anti-XPA and anti-Actin (loading control) antibodies. (C) H460 cells, expressing either control vector or siRNA targeted to XPA, were UV irradiated at 20 J/m2 and incubated for another 1 h. The proteins were subjected to SDS–PAGE and detected with anti-XPC, anti-XPA and anti-XPB antibodies.
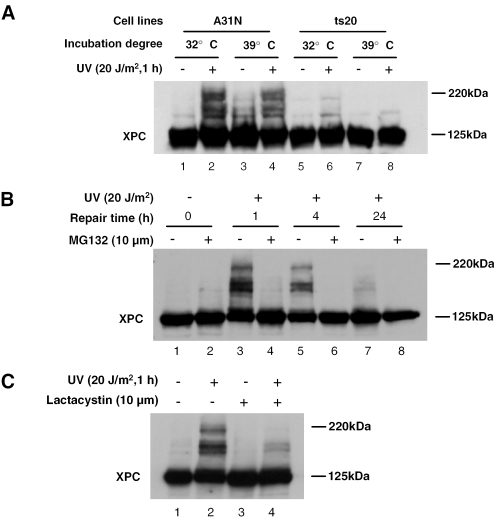
Ubiquitin–proteasome system is required in UV-induced XPC modifications
We have shown that ubiquitin–proteasome system, affecting XPC recruitment to the damage sites, is involved in NER in mammalian cells (32). Here, we assessed whether the ubiquitin–proteasome system is also involved in UV-induced XPC modifications. We first tested the role of ubiquitylation in UV-induced XPC modifications by using a mouse ts20 cell line that harbors a temperature-sensitive ubiquitin-activating enzyme (E1). E1 ubiquitin-activating enzyme is active at permissive temperature (32°C), whereas inactive at restrictive temperature (39°C) and thereby compromising the ubiquitylation function. In the parent A31N cells, UV irradiation was able to induce XPC modifications at both 32 and 39°C, indicating that UV-induced XPC modifications can also occur in mouse cells (Figure 6A, lanes 1–4). Similarly, UV irradiation also causes XPC modifications in ts20 cells cultured at 32°C, albeit the modifications are weaker than that in A31N cells (Figure 6A, lane 6). In contrast, a very significant decrease of UV-induced XPC modifications could be observed in ts20 cells cultured at 39°C (Figure 6A, lane 8). These results suggest that ubiquitylation is involved in UV-induced XPC modifications. We further examined the involvement of 26S proteasome in UV-induced XPC modifications by using the specific inhibitors of 20S catalytic subunit, MG132 and lactacystin. Interestingly, the treatment of OSU-2 cells with MG132 completely abolished the UV-induced XPC modifications at different times following UV irradiation (Figure 6B, lanes 3–8). Similarly, treatment with another more specific 20S inhibitor, lactacystin, which dramatically decreased the UV-induced XPC modifications (Figure 6C, lane 4 versus lane 2) further strengthened the participatory role of 26S proteasome in UV-induced XPC modification. Taken together, the results demonstrated that ubiquitin–proteasome system is required for XPC modification following UV irradiation.
Figure 6.
Ubiquitin–proteasome system is involved in UV-induced XPC modification. (A) ts20 and its parental cell line A31N were cultured at 32°C or shifted to 39°C for 16 h followed by UV irradiation at 20 J/m2 and further incubation for 1 h at 32°C or 39°C. XPC protein and its modifications were detected by immunoblotting with anti-XPC antibody. (B) OSU-2 cells were treated with 10 μM MG132 or mock treated for 1 h and subjected to UV irradiation at 20 J/m2. The cell lysates were prepared at indicated time points following UV treatment and analyzed. (C) OSU-2 cells were treated with 10 μM lactacystin or mock treated for 1 h and subjected to UV irradiation at 20 J/m2. The cell lysates were prepared after 1 h further incubation and the proteins were detected with anti-XPC antibody.
DISCUSSION
SUMO post-translationally modifies many proteins with demonstrated role in diverse processes including regulation of transcription, chromatin structure and DNA repair [for review, see (19)]. Many factors and enzymes associated with DNA replication and repair including yeast PCNA, the helicase WRN, Topoisomerases I and II, and the thymine-DNA glycosylase enzyme, are post-translationally modified by SUMO [for review, see (19,33)]. SUMO modification has been shown to regulate the subnuclear localization, protein–protein interactions and activity of many factors involved in maintenance of the genome. Ubiquitylation is also believed to be involved in DNA repair, i.e. NER (32,34–36). The NER factors DDB2 and Rad4 have been reported to be ubiquitylated following UV irradiation (13,37). Moreover, while this manuscript was under review, Sugasawa et al. (38) reported that XPC protein, an initial damage recognition factor in NER, is ubiquitylated upon UV irradiation. In this study, we demonstrated that XPC protein is modified not only by ubiquitin, but also by SUMO-1 following UV irradiation. This discovery expands the substrates of SUMO conjugation and for the first time introduces SUMO modification in the phenomenon of NER. In addition, we have also provided direct evidence for XPC ubiquitylation in mammalian cells.
The role of various NER factors in the UV-induced XPC sumoylation and ubiquitylation
While the results described herein provide the very clear evidence for UV-induced XPC sumoylation and ubiquitylation, we do not yet fully understand its significance or role in action mechanism. In our studies, we have shown that several repair factors including DDB2 and XPA are required for UV-induced XPC modification. DDB2, as a subunit of Damaged DNA binding complex (DDB), is thought to be involved in NER [for review, see (39)], transcription, cell cycle (40,41) and p53-mediated apoptosis on exposure to UV radiation (42). During NER, DDB2 is believed to be the very first DNA damage recognition factor prior even to XPC during the GGR of UV-induced CPD and is required for XPC recruitment to DNA damage sites (24–26). Therefore, we speculate that the role of DDB2 in UV-induced XPC modification is to help XPC recruitment and binding to the damage sites, and thereafter enable its sumoylation and ubiquitylation. Several studies have suggested that DDB2 is degraded through ubiquitin-proteasome pathway following binding to the damage sites (43,44). In the absence of ubiquitylation or the degradation function of proteasome, DDB2 could not be eliminated and the recruitment of XPC to damage sites will be compromised along with the accompanying reduction of NER efficiency (32). Thus, the basic prerequisite for XPC modification is that DDB2 would be initially recruited to the damage sites, perhaps to open the local chromatin or distort the local DNA structure, and then be degraded by ubiquitin–proteasome pathway. Thereafter, XPC would be recruited to those damage sites and allow the possibility of its modification. A plausible alternative that DDB2 itself might serve as a SUMO ligase for XPC sumoylation remains to be investigated. For instance, Sugasawa et al. (38) have shown that DDB2 exhibits E3 ligase activity, through a complex with DDB1, Cullin 4A and Roc1, for XPC ubiquitylation. Whether this complex can also sumoylate the interacting XPC protein remains an open question.
It is recognized that DDB2 is not the only factor required for UV-induced XPC modification. Our results showed that even though XPC is localized at the damage sites, it would not be modified in the absence of XPA. It is well known that the assembly of NER factors at the damage sites is sequential and XPC-hHR23B is the principal DNA damage binding protein that is essential for the recruitment of other NER components to the site of DNA damage. The recruitment of XPA to DNA damage requires functional XPC whereas XPA is not needed for the accumulation of XPC at sites of DNA damage (9). However, Sugasawa et al. (38) results did not show any involvement of XPA in UV-induced XPC modification. The apparent discrepancy between these two studies can, at present, only be attributed to the use of different cellular systems. First, two XP-A cell lines are from different individuals. Second, the XP-A cells used in the present study are untransformed, while that used by the Sugasawa et al. study is a SV40 transformed cell line. Like Sugasawa et al. we were also unable to see the modification response in XP12RO, another SV40 transformed cell line (data not shown). We cannot rule out the possibility that some XPA functionality is substituted by other factors during the immobilization step. It may be noted that SV40-transformed cell lines exhibit higher non-specific XPC antibody-cross-reacting bands than in untransformed cell lines (38). Nevertheless, in this study, both the XPA protein restoration experiment and XPA siRNA knockdown experiment show that XPA is absolutely required for UV-induced XPC modification. Therefore, we posit that only when XPA protein is recruited to the damage sites, does XPC get modified.
The role of XPC sumoylation in NER
Based on the current understanding of the involved processes and our results, it is possible to speculate the functional aspects of XPC sumoylation. It has been shown that XPC and its yeast homologue, Rad4, could be degraded through ubiquitin–proteasome pathway (10,13). Our results have shown that UV irradiation could trigger a slight degradation of XPC, which occurs during the very early steps of NER, i.e. 1–2 h following irradiation. However, the level of XPC protein decreased significantly following UV irradiation in cells with little or no XPC sumoylation, e.g. XP-A cells, H460 cells expressing XPA siRNA, and OSU-2 cells transfected with Ubc9 siRNA. It is already clear that one function of SUMO modification of target proteins is to stabilize them by preventing their ubiquitylation via blockage of the lysine residues where ubiquitin would normally attach to the protein (45–48). Therefore, we reason that UV-induced XPC sumoylation serves to inhibit its degradation. After leaving from the NER complex, the XPC protein still remains functionally active to initiate a new round of DNA damage recognition. If, however, XPC is not modified, it will be degraded via ubiquitin–proteasome system. A study in the yeast has already shown that the level of Rad4 is very low in ubc9 knock out cells, indicating that Ubc9 functions in a pathway to stabilize Rad4 possibly through conjugating SUMO modification (49).
The basal amount of XPC protein in the cells is clearly important for the efficient removal of UV-induced photoproducts. Emmert et al. (50) have shown that partially corrected XP-C cells (with detectable but subnormal XPC protein levels) had normal repair of CPD but minimal repair of 6-4PP. A recent study has shown that knocking out both mHR23A and mHR23B genes strongly reduced the steady-state levels of XPC in MEFs which then exhibit the similar repair deficiency as XP-C cells (10). Moreover, the ectopic expression of XPC bypassed the repair defect of these cells. We also tried to determine the role of UV-induced XPC sumoylation in the NER by inhibiting this process through transfecting Ubc9 siRNA into the repair-proficient OSU-2 cells. However, we did not find any difference in the removal of CPD and 6-4PP between the transfected and untransfected cells (data not shown). This can be explained by the fact that siRNA transfection partially knocks down the levels of target protein and the residual expression could be sufficient for normal repair function. As shown by results of Figure 3, Ubc9 siRNA transfection inhibited 60% of the UV-induced sumoylation. However, this level of SUMO modification mediated inhibition caused the degradation of only 50% of cellular XPC. The remaining XPC protein must have been sufficient to undertake a new round of damage recognition and, therefore, not detectably affect repair. In contrast, however, the study in the yeast has shown that the ubc9-1Δ strain (with deletion of Ubc9) had low Rad4 levels and consequently increased UV sensitivity in comparison to the normal strains (49). So, it is obvious that a firm description of the function of UV-induced XPC sumoylation in the mammalian cells will have to await the development of a new and more effective approach to eliminate sumoylation.
Another recognized function of sumoylation is that SUMO can alter substrate interaction with other macromolecules (16). This effect of SUMO on biomolecular interactions also varies for different substrates. For example, sumoylation allows RanGAP1 to bind tightly to the nuclear pore complex protein RanBP2/Nup358 (51,52), whereas SUMO-modified thymine-DNA glycosylase showed a reduced affinity for the DNA substrate (53). Wakasugi and Sancar found that XPC-hHR23B participates in the initial assembly of the excision nuclease, but is then no longer present in the ultimate dual incision complex (54). It is possible that soon after the factors like TFIIH, XPA and/or RPA are gathered, the XPC-hHR23B leaves the complex. As shown by the studies using the cholesterol-derived lesion, the latter segment of the NER reaction can, at least in vitro, be conducted without XPC (55). Therefore, we reckon that one function of XPC sumoylation might be to diminish the affinity of the bound XPC molecules to damaged sites and facilitate its leaving the NER complex after accomplishing the recruitment of subsequent factors, e.g. XPA and ERCC1-XPF. However, this hypothesis will have to be substantiated through future experimentation.
The role of XPC ubiquitylation in NER
In this study, we also showed that XPC is ubiquitylated following UV irradiation in mammalian NER-proficient cells. Usually, the ubiquitylated proteins are targeted to the proteasome for degradation (56). However, ubiquitylated proteins are often known to serve a regulatory function independent of proteolysis. Our results demonstrate an interesting phenomenon in which the repair-proficient OSU-2 cells show a higher level of UV-induced ubiquitylated XPC but lesser XPC degradation than repair-deficient XP-A cells following UV irradiation. It seems that such UV-induced ubiquitylation of XPC is not primarily for degradation, and that ubiquitylated XPC destined for targeted degradation simply cannot be detected due to its rapid turnover. Sugasawa et al. also found that the UV-induced ubiquitylation of XPC did not induce degradation via the 26S proteasome, and suggest that such ubiquitylation alters the DNA binding properties of XPC which could have affected the cell-free NER incision (38).
In summary, we have demonstrated that NER factor XPC can be modified by SUMO-1 and ubiquitin following UV irradiation. The modification requires other functional NER factors, including DDB2 and XPA. A critical interplay of the ubiquitin–proteasome system affects the XPC modification and productive repair at the damage sites within the genome. The function of UV-induced XPC sumoylation might be related to the stabilization of XPC protein and the decreased binding affinity of XPC to damaged DNA. Future work entails the mapping of the sumoylation sites within the XPC protein and clarification of the precise functional role of XPC sumoylation and ubiquitylation in the regulation of NER.
Acknowledgments
We are thankful to Drs Michael Tainsky, Bert Vogelstein, Anatoly Zhitkovich, Fumio Hanaoka, Gilbert Chu, Xin-Hua Feng and Dirk Bohmann for providing valuable cells and reagents. We also thank Dr Robert Snapka for useful discussions and Ahmed S. Jama for helping with experiments. This work was supported by NIH grants ES6074, ES12991 and CA93413 to A.A.W. Funding to pay the Open Access publication charges for this article was provided by National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.De Laat W.L., Jaspers N.G., Hoeijmakers J.H. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 2.Hanawalt P.C. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- 3.Masutani C., Sugasawa K., Yanagisawa J., Sonoyama T., Ui M., Enomoto T., Takio K., Tanaka K., Van der Spek P.J., Bootsma D., et al. Purification and cloning of a nucleotide excision repair complex involving the Xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 1994;13:1831–1843. doi: 10.1002/j.1460-2075.1994.tb06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shivji M.K.K., Eker A.P.M., Wood R.D. DNA repair defect in xeroderma pigmentosum group C and complementing factor from HeLa cells. J. Biol. Chem. 1994;269:22749–22757. [PubMed] [Google Scholar]
- 5.Araki M., Masutani C., Takemura M., Uchida A., Sugasawa K., Kondoh J., Ohkuma Y., Hanaoka F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 2001;276:18665–18672. doi: 10.1074/jbc.M100855200. [DOI] [PubMed] [Google Scholar]
- 6.Sugasawa K., Okamoto T., Shimizu Y., Masutani C., Iwai S., Hanaoka F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugasawa K., Shimizu Y., Iwai S., Hanaoka F. A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair (Amst.) 2002;1:95–107. doi: 10.1016/s1568-7864(01)00008-8. [DOI] [PubMed] [Google Scholar]
- 8.Sugasawa K., Ng J.M.Y., Masutani C., Iwai S., Van der Spek P., Eker A., Hanoaka F., Bootsma D., Hoeijmakers J.H.J. Xeroderma pigmentosum group C complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 9.Volker M., Mone M.J., Karmakar P., Van Hoffen A., Schul W., Vermeulen W., Hoeijmakers J.H., van Driel R., Van Zeeland A.A., Mullenders L.H. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 10.Ng J.M., Vermeulen W., Van der Horst G.T., Bergnik S., Sugasawa K., Vrieling H., Hoeijmakers J.H. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 2003;17:1630–1645. doi: 10.1101/gad.260003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okuda Y., Nishi R., Ng J.M., Vermeulen W., Van der Horst G.T., Mori T., Hoeijmakers J.H., Hanaoka F., Sugasawa K. Relative levels of the two mammalian Rad23 homologs determine composition and stability of the xeroderma pigmentosum group C protein complex. DNA Repair (Amst.) 2004;3:1285–1295. doi: 10.1016/j.dnarep.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Adimoolam S., Ford J.M. p53 and DNA damage-inducible expression of the xeroderma pigmentosum group C gene. Proc. Natl Acad. Sci. USA. 2002;99:12985–12990. doi: 10.1073/pnas.202485699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lommel L., Ortolan T., Chen L., Madura K., Sweder K.S. Proteolysis of a nucleotide excision repair protein by the 26 S proteasome. Curr. Genet. 2002;42:9–20. doi: 10.1007/s00294-002-0332-9. [DOI] [PubMed] [Google Scholar]
- 14.Melchior F. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 15.Hay R.T. Protein modification by SUMO. Trends Biochem. Sci. 2001;26:332–333. doi: 10.1016/s0968-0004(01)01849-7. [DOI] [PubMed] [Google Scholar]
- 16.Johnson E.S. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 17.Saitoh H., Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 18.Tatham M.H., Jaffray E., Vaughan O.A., Desterro J.M., Botting C.H., Naismith J.H., Hay R.T. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 19.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 20.Venkatachalam S., Denissenko M.F., Wani A.A. DNA repair in human cells: quantitative assessment of bulky anti-BPDE DNA adducts by non-competitive immunoassays. Carcinogenesis. 1995;16:2029–2036. doi: 10.1093/carcin/16.9.2029. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds M., Peterson E., Quievryn G., Zhitkovich A. Human nucleotide excision repair efficiently removes chromium-DNA phosphate adducts and protects cells against chromate toxicity. J. Biol. Chem. 2004;279:30419–30424. doi: 10.1074/jbc.M402486200. [DOI] [PubMed] [Google Scholar]
- 22.Chowdary D.R., Dermody J.J., Jha K.K., Ozer H.L. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol. Cell Biol. 1994;14:1997–2003. doi: 10.1128/mcb.14.3.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wani M.A., Zhu Q.Z., El-Mahdy M.A., Wani A.A. Influence of p53 tumor suppressor protein on bias of DNA repair and apoptotic response in human cells. Carcinogenesis. 1999;20:765–772. doi: 10.1093/carcin/20.5.765. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q.E., Zhu Q., Wani G., Chen J., Wani A.A. UV radiation-induced XPC translocation within chromatin is mediated by damaged-DNA binding protein, DDB2. Carcinogenesis. 2004;25:1033–1043. doi: 10.1093/carcin/bgh085. [DOI] [PubMed] [Google Scholar]
- 25.Fitch M.E., Nakajima S., Yasui A., Ford J.M. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J. Biol. Chem. 2003;278:46906–46910. doi: 10.1074/jbc.M307254200. [DOI] [PubMed] [Google Scholar]
- 26.Fitch M.E., Cross I.V., Turner S.J., Adimoolam S., Lin C.X., Williams K.G., Ford J.M. The DDB2 nucleotide excision repair gene product p48 enhances global genomic repair in p53 deficient human fibroblasts. DNA Repair (Amst.) 2003;2:819–826. doi: 10.1016/s1568-7864(03)00066-1. [DOI] [PubMed] [Google Scholar]
- 27.Hwang B.J., Ford J.M., Hanawalt P.C., Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl Acad. Sci. USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Araujo S.J., Tirode F., Coin F., Pospiech H., Syvaoja J.E., Stucki M., Hubscher U., Egly J.M., Wood R.D. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 2000;14:349–359. [PMC free article] [PubMed] [Google Scholar]
- 29.Aboussekhra A., Biggerstaff M., Shivji M.K.K., Vilpo J.A., Moncollin V., Podust V.N., Protic M., Hubscher U., Egly J.-M., Wood R.D. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 30.Guzder S.N., Habraken Y., Sung P., Prakash L., Prakash S. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J. Biol. Chem. 1995;270:12973–12976. doi: 10.1074/jbc.270.22.12973. [DOI] [PubMed] [Google Scholar]
- 31.Mu D., Park C.-H., Matsunaga T., Hsu D.S., Reardon J.T., Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q.E., Wani M.A., Chen J., Zhu Q., Wani G., El-Mahdy M.A., Wani A.A. Cellular ubiquitination and proteasomal functions positively modulate mammalian nucleotide excision repair. Mol. Carcinog. 2005;42:53–64. doi: 10.1002/mc.20065. [DOI] [PubMed] [Google Scholar]
- 33.Muller S., Ledl A., Schmidt D. SUMO: a regulator of gene expression and genome integrity. Oncogene. 2004;23:1998–2008. doi: 10.1038/sj.onc.1207415. [DOI] [PubMed] [Google Scholar]
- 34.Watkins J.F., Sung P., Prakash L., Prakash S. The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol. Cell. Biol. 1993;13:7757–7765. doi: 10.1128/mcb.13.12.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikehata H., Kaneda S., Yamao F., Seno T., Ono T., Hanaoka F. Incubation at the nonpermissive temperature induces deficiencies in UV resistance and mutagenesis in mouse mutant cells expressing a temperature-sensitive ubiquitin-activating enzyme (E1) Mol. Cell. Biol. 1997;17:1484–1489. doi: 10.1128/mcb.17.3.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schauber C., Li C., Tangoankar P., Vega I., Lambertson D., Potts W., Madura K. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 37.Matsuda N., Azuma K., Saijo M., Iemura S., Hioki Y., Natsume T., Chiba T., Tanaka K., Tanaka K. DDB2, the xeroderma pigmentosum group E gene product, is directly ubiquitylated by Cullin 4A-based ubiquitin ligase complex. DNA Repair (Amst.) 2005;4:537–545. doi: 10.1016/j.dnarep.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 38.Sugasawa K., Okuda Y., Saijo M., Nishi R., Matsuda N., Chu G., Mori T., Iwai S., Tanaka K., Tanaka K., et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 39.Tang J., Chu G. Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein. DNA Repair (Amst.) 2002;1:601–616. doi: 10.1016/s1568-7864(02)00052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayes S., Shiyanov P., Chen X., Raychaudhuri P. DDB, a putative DNA repair protein, can function as a transcriptional partner of E2F1. Mol. Cell. Biol. 1998;18:240–249. doi: 10.1128/mcb.18.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiyanov P., Hayes S.A., Donepudi M., Nichols A.F., Linn S., Slagle B.L., Raychaudhuri P. The naturally occurring mutants of DDB are impaired in stimulating nuclear import of the p125 subunit and E2F1-activated transcription. Mol. Cell. Biol. 1999;19:4935–4943. doi: 10.1128/mcb.19.7.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Itoh T., Cado D., Kamide R., Linn S. DDB2 gene disruption leads to skin tumors and resistance to apoptosis after exposure to ultraviolet light but not a chemical carcinogen. Proc. Natl Acad. Sci. USA. 2004;101:2052–2057. doi: 10.1073/pnas.0306551101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fitch M.E., Cross I.V., Ford J.M. p53 responsive nucleotide excision repair gene products p48 and XPC, but not p53, localize to sites of UV-irradiation-induced DNA damage, in vivo. Carcinogenesis. 2003;24:843–850. doi: 10.1093/carcin/bgg031. [DOI] [PubMed] [Google Scholar]
- 44.Rapic-Otrin V., McLenigan M.P., Bisi D.C., Gonzalez M., Levine A.S. Sequential binding of UV DNA damage binding factor and degradation of the p48 subunit as early events after UV irradiation. Nucleic Acids Res. 2002;30:2588–2598. doi: 10.1093/nar/30.11.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 46.Desterro J.M., Rodriguez M.S., Hay R.T. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 47.Lin X., Liang M., Liang Y.Y., Brunicardi F.C., Feng X.H. SUMO-1/Ubc9 promotes nuclear accumulation and metabolic stability of tumor suppressor Smad4. J. Biol. Chem. 2003;278:31043–31048. doi: 10.1074/jbc.C300112200. [DOI] [PubMed] [Google Scholar]
- 48.Lee P.S., Chang C., Liu D., Derynck R. Sumoylation of Smad4, the common Smad mediator of transforming growth factor-beta family signaling. J. Biol. Chem. 2003;278:27853–27863. doi: 10.1074/jbc.M301755200. [DOI] [PubMed] [Google Scholar]
- 49.Ramsey K.L., Smith J.J., Dasgupta A., Maqani N., Grant P., Auble D.T. The NEF4 complex regulates Rad4 levels and utilizes Snf2/Swi2-related ATPase activity for nucleotide excision repair. Mol. Cell. Biol. 2004;24:6362–6378. doi: 10.1128/MCB.24.14.6362-6378.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emmert S., Kobayashi N., Khan S.G., Kraemer K.H. The xeroderma pigmentosum group C gene leads to selective repair of cyclobutane pyrimidine dimers rather than 6–4 photoproducts. Proc. Natl Acad. Sci. USA. 2000;97:2151–2156. doi: 10.1073/pnas.040559697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahajan R., Delphin C., Guan T., Gerace L., Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 52.Matunis M.J., Coutavas E., Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell. Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hardeland U., Steinacher R., Jiricny J., Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wakasugi M., Sancar A. Assembly, subunit composition, and footprint of human DNA repair excision nuclease. Proc. Natl Acad. Sci. USA. 1998;95:6669–6674. doi: 10.1073/pnas.95.12.6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mu D., Hsu D.S., Sancar A. Reaction mechanism of human DNA repair excision nuclease. J. Biol. Chem. 1996;271:8285–8294. doi: 10.1074/jbc.271.14.8285. [DOI] [PubMed] [Google Scholar]
- 56.Hochstrasser M. Ubiquitin-dependent protein degradation. Ann. Rev. Genet. 1992;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]