Abstract
In disease-associated genes, the understanding of the functional significance of deep intronic nucleotide variants may represent a difficult challenge. We have previously reported a new disease-causing mechanism that involves an intronic splicing processing element (ISPE) in ATM, composed of adjacent consensus 5′ and 3′ splice sites. A GTAA deletion within ISPE maintains potential adjacent splice sites, disrupts a non-canonical U1 snRNP interaction and activates an aberrant exon. In this paper, we demonstrate that binding of U1 snRNA through complementarity within a ∼40 nt window downstream of the ISPE prevents aberrant splicing. By selective mutagenesis at the adjacent consensus ISPE splice sites, we show that this effect is not due to a resplicing process occurring at the ISPE. Functional comparison of the ATM mouse counterpart and evaluation of the pre-mRNA splicing intermediates derived from affected cell lines and hybrid minigene assays indicate that U1 snRNP binding at the ISPE interferes with the cryptic acceptor site. Activation of this site results in a stringent 5′–3′ order of intron sequence removal around the cryptic exon. Artificial U1 snRNA loading by complementarity to heterologous exonic sequences represents a potential therapeutic method to prevent the usage of an aberrant CFTR cryptic exon. Our results suggest that ISPE-like intronic elements binding U1 snRNPs may regulate correct intron processing.
INTRODUCTION
Intronic sequences distant from the canonical splice sites are mostly considered functionally neutral regarding pre-mRNA splicing and are thus rarely considered when mapping splicing regulatory elements and are not routinely sequenced in human genetics screening. However, several examples have clearly shown that ‘deep’ intronic mutations may affect the pre-mRNA splicing process in a number of disease-associated genes. The most frequent type of splicing mutation identified in introns induces aberrant splicing by creating or strengthening splice sites (1–6). Because the splice-site consensus motifs are degenerate, the introns contain many matches to each consensus sequence, often with potential exon pairing, which are never selected for splicing (7). In the presence of a particular intronic context, mutations that create a new splice site may define the boundary of the cryptic exon, which is then included in the mature mRNA. For example, a disease-causing intronic 19 C→T substitution (3849 + 10 kb T) ∼10 kb downstream of exon 19 of the CFTR gene has been reported to create a perfect 5′ splice site (5′-ss) consensus. This activates the inclusion of a 84 bp cryptic exon that contains a stop codon (4). However, not all intronic mutations create or strengthen splice sites, suggesting the existence of yet unknown splicing regulatory elements (8,9).
Ataxia-telangiectasia is an autosomal recessive disorder involving cerebellar degeneration, immunodeficiency, chromosomal instability, radiosensitivity and cancer predisposition (10–12). Interestingly, a significant proportion of mutations in the affected ataxia-telangiectasia mutated (ATM) gene (∼48%) cause defects in splicing (13). We have previously identified a new disease-causing mechanism that involves an intronic splicing processing element (ISPE) in ATM intron 20 (9). The sequence of the ISPE (CAG/GTAAGT) resembles adjacent consensus 5′-ss and 3′-ss, and is located in ATM intron 20 approximately 1870 and 570 bp away from neighboring exons 20 and 21, respectively. A 4 bp (GTAA) deletion in the ISPE maintains these potential adjacent consensus 5′-ss and 3′-ss, but the disruption of a non-canonical U1 snRNP interaction with the ISPE activates an aberrant 65 bp exon. Inclusion of this exon in mRNA is directly responsible for ataxia-telangiectasia in an affected patient. The particularity of the ISPE mutation in intron 20 consists in the fact that it does not affect splice sites that defines the boundary of the cryptic exon. In fact, the deletion occurs 12 bp downstream and 53 bp upstream respectively from the two cryptic splice sites, whose sequences were already present in the wild-type intron. We have previously shown that the activation of the cryptic exon occurs through the disruption of a non-canonical interaction of U1 snRNP with the ISPE. Normally, U1 snRNP has a well-known role in 5′-ss definition (14), which does not seem to apply in the case of the ISPE. This particular behavior led us to suggest that this ISPE–U1 snRNP interaction could be involved in resplicing, a mechanism originally identified in the Drosophila gene Ubx and potentially involved in correct intron processing (15,16). To gain greater insight into the basic mechanism involved, we assessed the splicing effect of U1 snRNP binding along the entire cryptic ATM exon, the sequence-specificity of the adjacent consensus 5′-ss and 3′-ss of ISPE, and the pre-mRNA intermediates formed in vivo. At the same time, we assessed the splicing role of consensus ISPE sequences in mouse ATM introns and the artificial recruitment of U1 snRNP on heterologous gene systems. Our results identify cis-acting sequences that can induce aberrant splicing in the context of deep intronic mutations that affect an ISPE sequence and provide a possible general role of non-canonical U1 snRNP binding sites in correct intron processing.
MATERIALS AND METHODS
Construction of hybrid minigenes
pATMΔ minigene has been previously described (9). pATM12T, pATM13A and pATM GT2 were derived from original pATM WT minigene (9) using PCR-mediated site-directed mutagenesis. Genomic mouse DNA was amplified with ATMmo1830dir, CATATGACGAGTAGGTAGGTTAGGATAG, and ATMmo2290rev, GCCAGCTTTATCCACAAATCACTCACTT, primers that contain NdeI restriction site at their 5′ end. Amplified fragments were digested with NdeI and cloned into the pTB NdeI(−) corresponding site (17). pATMmΔ was generated by deletion of the GTAA in mouse ISPE by PCR site-directed mutagenesis. Addition of the 5′-ss to generate pATMm5′ and pATMmΔ5′ was done by cloning the annealed ATM m5′ss dir, CATGGTAAGTAT, and ATM m5′ss rev, CATGATACTTAC, primers in the unique NcoI site. To create pCF-C and pCF-T minigenes corresponding to WT and 3849 + 10 kb C→T mutation (4), respectively, human normal genomic DNA was amplified with CFex19bdir, GGACTTAATCAAAATTCCTCATATGG, and CFex19brev, ATCATATGCTGGTAATGCATGATATCTGA. The amplified fragment of 824 bp that includes the cryptic 84 bp CFTR exon flanked by 377 and 363 bp upstream and downstream intronic sequences, respectively, was cloned in the NdeI site of pTB Nde(−) (17). The 3849 + 10 kb C→T mutation was created by PCR site-directed mutagenesis. pFN-EDA minigene is a pBS derivative of pSVED-A Tot (18) and contains the entire human fibronectin EDA cassette of 3096 bp (that include 92 and 109 bp of preceding and following exonic sequences and the entire introns) cloned in the BstEII site of the α-globin under the SV40 promoter (19). We prepared a pcY7 α-tropomyosin minigene by cloning the 630 bp T2F and T3R amplified fragment derived from pY7 (20), which contains 138 bp of exon 2, 108 bp of intron 2, and 150 bp of exon 3 in the KpnI and Xba sites of pcDNA3. The sequence of the primers is as follows: T2F, AGGGTACCAGCTTGTCGAGGAGGACATCTCAG and T3R, CCTCTAGAGTCGATCGACCTGCAGG. To create the pcY7ATM constructs, the EcoR1–BamHI 165 bp long fragments derived from pATM (WT and Δ), which corresponds to the 65 bp cryptic exon flanked by 52 and 44 bp of intronic sequences, were cloned in the SmaI site of the intron of pcY7 at a distance of 47 and 64 bp from the upstream and downstream α-tropomyosin exons 2 and 3, respectively. To prepare the pATM20 Δ, we amplified human DNA derived from the affected patient with the following primers: ATM J20 Δ, ATGGTGACCTAGAACCTACCAAATCCCTCCACCTGCATATGTTATCTGGCCAGGTG, and ATM Bste II rev, TAGGTCACCGTGGTTTCAGAAGTTCAAGAACATCTTCCATTGG.
The ATM-amplified fragment of 717 bp includes the first 41 bp of exon 20 joined to the ATM Δ cryptic exon, the entire downstream intron and 88 bp of exon 21. The PCR product was digested with BsteII and cloned in the BstEII site of the α-globin under the SV40 promoter (19). All minigene constructs were verified by DNA sequencing. To prepare different U1snRNAs, the sequence between BclI and BglII of the parental U1snRNA clone pG3U1(WT-U1), a derivative of pHU1 (21), was replaced with mutated oligonucleotides as previously described for the U1-Δ (9). The same strategy was used to create U1-32, U1-40, U1-47, U1-66 and their complementary sequences in the ATM cryptic exon are shown in Figure 1B. The complementarity sequences of U1-12T, U1-13A and U1-13A17A are shown in Figure 2A. The tail of U1-CF19, U1-EDA8, U1-EDA19 and U1-α ex2 were created using the following oligonucleotides:
U1-CF19 dir: GATCTCATTCATTACCTTGCAGGGGAGATACCAT;
U1-CF19 rev: GATCATGGTATCTCCCCTGCAAGGTAATGAATGA;
U1-EDA8 dir: GATCTCATTTAGGGCGATGCAGGGGAGATACCAT;
U1-EDA8 rev: GATCATGGTATCTCCCCTGCATCGCCCTAAATGA;
U1-EDA19 dir: GATCTCATGCCAGTCCTGCAGGGGAGATACCAT;
U1-EDA19 rev: GATCATGGTATCTCCCCTGCAGGACTGGCATGA;
U1-αGLO ex2 dir: GATCTCATTGGTGGTGGGGAAGGCAGGGGAGATACCAT;
U1-αGLO ex2 rev: GATCATGGTATCTCCCCTGCCTTCCCCACCACCAATGA.
Figure 1.
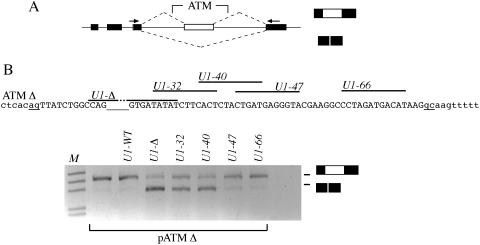
Effect on pre-mRNA splicing of modified U1 snRNAs binding along the entire ATM Δ. (A) Schematic representation of ATM hybrid minigenes used in transient transfection assay. Exons are indicated as black boxes, introns as line and dotted lines represent possible splicing products. Primers used in RT–PCR analysis are shown as black arrows. The position where the ATM intron 20 sequences were cloned is indicated. (B) The upper panel shows the relevant portion of the nucleotide sequences of the ATM Δ intron 20. The cryptic exon is shown in uppercase, intronic sequences are in lowercase and the cryptic acceptor (ag) and donor (gc) sites are underlined. The empty underlined space in the sequence corresponding to the GTAA deletion. The lines above the sequence show the position of modified U1 snRNAs binding site. Lower panel shows the result of the splicing assay. pATM Δ minigene was transfected in Hep3B cells alone or with the indicated U1 snRNAs and the resulting RT–PCR products analyzed on 1.5% agarose gel. The lower and higher MW bands correspond to normal intron processing and inclusion of the 65 bp cryptic exon, respectively.
Figure 2.
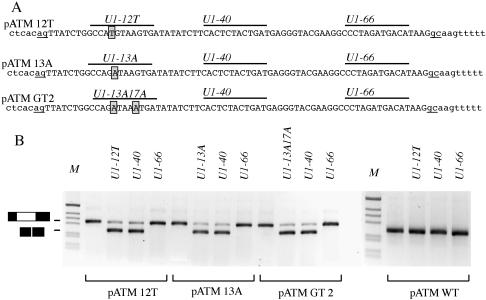
Analysis of mutants at the adjacent consensus 3′-ss and 5′-ss of human ISPE. (A) Nucleotide sequences of ATM intron 20 containing the point mutations (boxed) that were created to disrupt adjacent consensus 3′-ss or 5′-ss at the ISPE at position 12 and 13, respectively and the double GT2 mutant. The cryptic exon is shown in uppercase, intronic sequences are in lowercase and the cryptic acceptor (ag) and donor (gc) sites are underlined. Lines above the sequence show the position of modified U1 snRNAs binding site. (B) Minigene splicing assay. The pATM minigenes were trasfected alone or with the indicated U1 snRNAs in Hep3B cells and the resulting RT–PCR products analyzed on 1.5% agarose gel. The lower and higher MW bands correspond to normal intron processing and inclusion of the 65 bp cryptic exon, respectively. M is the molecular weight marker 1 kb.
Analysis of the hybrid minigene expression and splicing precursors
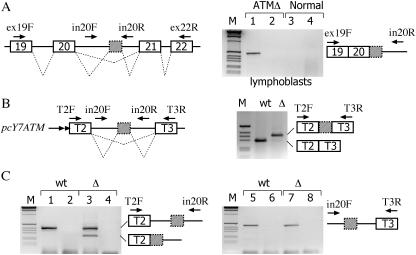
The Hep3B (human hepatocarcinoma) cells were grown in DMEM supplemented with 10% fetal calf serum. Plasmid DNA were purified using Jetstar plasmid purification system by GENOMED. Cells were cotransfected with the DOTAP reagent, with 1.5 μg of each minigene plasmid and when necessary either with 0.5 μg of the control pCMV empty vector, or with the indicated amounts of the U1snRNA codifying plasmids per 35 mm dish of 60–75% confluent cells in the presence of commercial lipofectin. RNA was extracted 48 h later and cDNA was prepared using random primers and reverse transcriptase as previously described (17). Transfections with different pATMs, pCF and pEDA minigenes were amplified with α2-3 and B2 as previously described (17), and visualized on 1.5–2% agarose gel stained with EtBr. The same PCR conditions were applied for the other minigene transfections using the following specific primers: pGLO was amplified with E16DIR CGCACGCTGGCGAGTATGGTGCGG and Glo 395Rev ACAGAAGCCAGGAACTTGTCCAG, pcY7ATMs were amplified with T2F and T3R to detect the mature mRNA, pATM20Δ was amplified with α2-3 and ATM 2550rev CCAGGAAGCTCCTTTAAAAGCATTAGATA. The identity of the resulting splicing products was verified by direct sequence analysis. in20F, GCTCAAGCTCTTAACTGCAACAG, and in20R rev, AATTCGGATCCCAGTACCACAGCCTT, primers were used in combination with the appropriate oligonucleotides to identify the splicing pre-mRNAs both in pcY7ATM minigenes and lymphoblasts. ex19F, GACCGTGGAGAAGTAGAATCAATGG, and ex22R, GTAAGAAACTGTCCTTGAGCATCC, were used for amplification experiments in lymphoblasts. Total RNA, extracted from ATM heterozygous patient's or normal lymphoblasts with RNAwiz RNA isolation reagent, was treated with DNAse RNAse-free and reverse-transcribed with random primer as previously reported (9). The amplification reaction was performed for 40 cycles, 45 s at 94°C, 45 s at 54°C and 45 s at 72°C.
RESULTS
Position-dependent effect of U1 snRNPs binding on the correct ATM intron pre-mRNA processing
Recruiting of U1 snRNA modified to be complementary to the sequence created by the GTAA deletion in the ATM ISPE induced normal pre-mRNA processing by inhibiting cryptic exon inclusion (9). To gain insight into the U1 snRNP functional interaction occurring along the entire cryptic exon, we have now evaluated the effect of several modified U1 snRNAs on the splicing pattern. To test the effect of the position of U1 snRNP association, we prepared snRNAs engineered to interact with unique sequences at consecutive positions along the cryptic ATM exon. Four different U1 snRNAs named U1-32, U1-40, U1-47 and U1-66 were prepared, being complementary to sequences between the GTAA deletion and the cryptic 5′-ss (Figure 1B). These U1 snRNAs and the previously described U1-Δ were cotransfected in Hep3B cells along with the human Δ-ISPE minigene and the pattern of splicing was analyzed by RT–PCR with specific primers (Figure 1). In the ATMΔ minigene (with the GTAA ISPE deletion), the cryptic exon was included in the processed mRNA and, as previously described, cotransfection of U1-Δ restored normal splicing (Figure 1B). Likewise, U1-32 and U1-40 restored normal processing of the intron, whereas the other U1 snRNAs designed to bind further downstream (U1-47 and U1-66) did not affect the normal splicing pattern (Figure 1B). Control experiments, in which the same U1 snRNAs were transfected along with the WT minigene, did not show changes in the splicing pattern (data not shown). These results indicate that correct processing of ATM intron 20 requires binding of U1 snRNP in a sequence that spans between the ISPE and ∼40 nt downstream. Binding of U1 RNAs near the cryptic 5′-ss (U1-47 and U1-66) had no effect. This position-dependent effect indicates that, as previously suggested for normal exons (22), binding of U1 snRNP inside the cryptic exon might produce steric interference between complexes involved in the recognition of splicing signals, and in this case, the acceptor splice site.
Sequence-specificity of adjacent consensus 3′ and 5′ splice site at the ISPE
Interestingly, even if U1 snRNP does not bind the ATM sequence with the GTAA deletion (9), the adjacent CAG/GTAAGT potential consensus 3′-ss and 5′-ss of the original ISPE are not significantly affected. In fact, the GTAA deletion changes the sequence to CAG/GTGATA, which again maintains the ‘core’ consensus AG/GT. We have previously suggested that these consensus splice sites could be potentially used like resplicing signals or ‘zero exons’, as previously found in Drosophila Ultrabithorax gene (15,16). When disrupted, these sequences induce aberrant cryptic exon inclusion. A 5′-ss may be also defined by U1 snRNP binding downstream of the specific cleavage site (23). Thus, it seems important to test if the position-dependent effect of U1 snRNP binding in the ATM context also requires the presence of intact adjacent splice sites at the ISPE. To analyze the possible role of ISPE acting as a resplicing signal, we prepared hybrid minigenes (Figure 2A) where the putative resplicing site AG/GT was destroyed by mutation at the 3′-ss (at position 12 AT/GT) or at the adjacent 5′-ss (at position 13 AG/AT) (Figure 2A, pATM 12T and pATM 13A, respectively). We also prepared a minigene in which two potential GT 5′-ss were inactivated to AT (Figure 2A, pATM GT2). Transfection of these mutants induced aberrant splicing with inclusion of the cryptic exon resembling the splicing pattern observed with the original GTAA ISPE deletion found in the affected patient (Figure 2B). To understand whether the selective disruption of the consensus adjacent splice site is sufficient to induce the aberrant splicing, we prepared additional U1 snRNAs variants, U1-12T, U1-13A and U1-13A17A. These U1 snRNAs were designed to bind 12T, 13A and GT2 ISPE mutants, respectively, through complementarity. Cotransfection of pATM 12T, pATM 13A and pATM GT2 with the corresponding complementary U1 snRNAs showed the disappearance of the cryptic exon (Figure 2). In addition, we tested the effect of two additional U1 snRNA, U1-40 and U1-66 that bound downstream and differentially affected the splicing pattern in the ATM Δ construct (Figure 1). In the three minigenes, cotransfection of U1-40 induces disappearance of the cryptic exon, whereas U1-66 has no effect (Figure 2B). Cotransfection experiments of wild-type ISPE minigene pATM WT with different U1 snRNAs did not show any effect on the splicing pattern (Figure 2B).
Altogether, these results indicate that binding of U1 snRNP at a sequence located within 40 nt downstream of the ISPE prevents aberrant splicing independent of sequence composition of its core AG/GT adjacent consensus 5′-ss and 3′-ss. Thus the function of the ISPE is not dependent on the resplicing signal but on a U1 snRNP complex located at the ISPE or nearby.
Conservation of ISPE sequences in the mouse ATM intron 20 require intronic cryptic splice sites to induce aberrant splicing
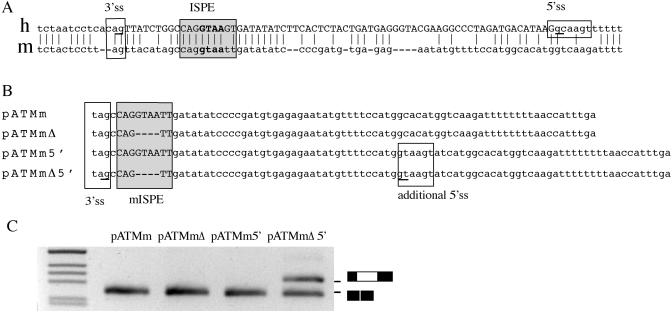
To further evaluate the role of deep intronic elements in splicing regulation, we compared human and mouse intron 20 sequences in the region of the ISPE. The mouse intron has conserved ISPE and cryptic 3′-ss, but no significant homology was observed for the peculiar human cryptic ‘GC’ 5′-ss (Figure 3A) suggesting that this difference may be important in generating the aberrant splicing. To test this hypothesis, we prepared four different mouse minigenes: wild-type mouse ATM intron 20 (pATMm), the same construct with the ISPE deletion (pATMmΔ), the mouse ATM with an artificial 5′-ss (pATMm + 5′-ss), and finally a minigene with the ISPE deletion and the 5′-ss (pATMmΔ + 5′-ss) (Figure 3B). Transfection experiments showed that the wt mouse intron and the mouse intron with the ISPE deletion are normally processed (Figure 3C). However, the deletion of the conserved mouse ISPE activates the cryptic exon in the presence of an artificial 5′-ss located in the corresponding position of the human ‘GC’ 5′-ss (Figure 3C). To study the role of ISPEs as possible targets for disease-causing mutations, we also evaluated two similar sequences in human introns 31 and 47 not flanked by potential cryptic splice sites. Analysis of the corresponding hybrid minigene showed that the ISPE deletions did not affect the splicing pattern (data not shown), strongly suggesting that aberrant splicing due to disruption of a consensus ISPE depends on the context of the intronic sequences where the ISPE is located and in particular, on the presence of flanking cryptic splice sites.
Figure 3.
Functional comparative analysis of the ISPEs in human and mouse ATM intron 20. (A) Alignment of human (h) and mouse (m) ATM intron 20 sequences reveal significant conservation at the cryptic 3′ ss and at the ISPE (boxed). The mouse intron did not show a conserved GC cryptic 5′-ss. The human cryptic splice sites are underlined and the human cryptic exon is in uppercase. (B) Nucleotide sequences of the mouse ATM hybrid minigenes. The conserved 3′-ss and mouse ISPE (mISPE) are boxed. In pATMm 5′ and pATMmΔ5′, the additional sequence that contain the consensus 5′-ss is indicated. Underlined nucleotides are the cryptic splice sites used in pATMmΔ5′ construct. (C) Splicing assay of the mouse ATM hybrid minigenes. Mouse ATM minigenes were transfected in Hep3B cells and the pattern of splicing analyzed with specific primers. RT–PCR amplified fragments were resolved on a 2% agarose gel. The lower MW product correspond to normal intron processing, the higher MW band found in pATMmΔ5′ correspond to inclusion of the cryptic exon. M is the molecular weight marker 1 kb.
U1snRNP recruitment by sequence complementarity to heterologous exonic sequences causes splicing inhibition in some contexts
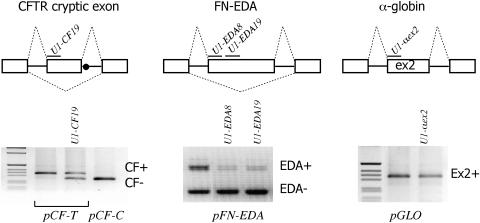
The splicing inhibitory effect of U1 snRNP was further investigated on exonic pre-mRNA sequences. We have previously shown that introduction of the ISPE element near the 3′-ss of CFTR exon 9 induces exon skipping. The same effect was seen by introducing deleted ISPE sequences and cotransfecting the corresponding U1 snRNA (U1 Δ) (9). To evaluate a potential useful tool to modulate exon-specific splicing and to correct aberrant splicing, we now extended our observations by performing complementation experiments with modified U1 snRNAs engineered to specifically bind at the 5′ portion of three exons: a pathological cryptic exon inclusion in CFTR, the EDA fibronectin exon and the second exon of α-globin. The 3849 + 10 kb C→T mutation is known to create a donor site in CFTR intron 19 leading to variable inclusion of a 84 bp long cryptic exon containing a stop codon (24). The EDA exon is a well-characterized example of an alternatively spliced exon subject to tissue-specific and developmental regulation, whereas the α-globin exon is constitutively included. These model systems were selected due to their potentially different exon definition properties. The minigenes corresponding to the three gene systems were cotransfected with different U1 RNAs, each specifically designed to bind to the 5′ portion of the corresponding exon. In the CFTR case, recruitment of modified U1 snRNA partially switched a cryptic exon into the original processed intronic sequence (Figure 4, left panel). Similar results were obtained for the fibronectin exon where two different U1 snRNAs designed to bind 8 and 19 nt downstream of the 3′-ss induced EDA skipping (Figure 4, central panel). On the contrary, binding of U1 at the constitutively included α-globin exon did not modify the splicing patterns (Figure 4, right panel).
Figure 4.
Effect of modified U1 snRNAs on splicing in heterologous exonic sequences. The upper panel shows a schematic representation of the three hybrid minigenes that correspond to the 84 bp cryptic exon in CFTR (pCF), the 270 bp fibronectin alternatively spliced EDA exon (pFN-EDA) and the constitutively included 204 bp exon 2 of α-globin (pGLO). Exons are indicated as boxes, introns as solid line and dotted lines represent the splicing products. The dot in pCF shows the position of the CFTR intron 19 C→T mutation that creates a fully competent 5′-ss. Above each exon are depicted the modified U1 snRNAs binding sites. The lower panel shows the results of the transfection experiments. Hybrid minigenes were transfected alone or with the indicated U1 snRNAs in Hep3B cells. Total RNA was amplified using primers specific for each construct. Resulting products were analyzed on 1.5% agarose gel. The exon inclusion (+) or exclusion (−) forms are indicated on the right of each splicing assay.
Analysis of pre-mRNA splicing intermediates in vivo and in hybrid minigenes
The function of the ISPE is presently not clear, it is possible that the U1 snRNP complex formed in deep intronic sequences may facilitate but may not be essential for efficient removal of the intron. To understand whether the ISPE deletion has a preferential effect on the excision of upstream or downstream intron sequences and thus potentially interferes with the 3′-ss or 5′-ss, we studied the pre-mRNAs formed in vivo in lymphoblast cell lines derived from normal and ATM-affected individuals. We performed in vivo analysis of pre-mRNAs by RT–PCR with primers designed to amplify sequences spanning exon 19 (ex19F) and the downstream portion of intron 20 (in20R) or the upstream segment of intron 20 (in20F) and exon 22 (ex22F) (Figure 5A). In the affected cells, amplification with the ex19F and in20R primers revealed a pre-mRNA form that corresponds to exon 19, exon 20, the cryptic exon and retention of the downstream portion of intron 20 (Figure 5A, lane 1), whereas analysis with in20F and ex22R did not show any intermediate (Figure 5A, lane 2). Analysis of normal cells with either ex19F-in20R or in20F-ex22R primers did not show any amplified band (Figure 5A, lanes 3 and 4, respectively).
Figure 5.
Analysis of ATM pre-mRNA splicing precursors in lymphoblasts and hybrid minigenes. (A) The left panel shows a schematic representation of the human ATM gene and the location of the primers used for RT–PCR analysis. Exons are indicated as boxes, introns as line and dotted lines represent the possible splicing products. The cryptic ATM exon is shown as a grey box with dotted outline. Total RNA, extracted from ATM patient (ATMΔ) and normal lymphoblasts, was treated with DNAse RNAse-free, reverse transcribed and amplified with the indicated primers. Lanes 1 and 3 correspond to amplification with ex19F and in20R, lanes 2 and 4 to amplification with in20F and ex22R. The identity of the amplified precursor product detected in the affected lymphoblasts which contains exon 19, exon 20, the ATMΔ cryptic exon and the downstream intron 20 sequences is schematically represented on the right on the splicing assay along the primers utilized. Control experiments without reverse transcriptase did not reveal any amplified band (data not shown). M is the molecular weight marker 1 kb. (B) Schematic representation of the short α-tropomyosin hybrid minigene (pcY7ATM) used in transient transfection assay. Tropomyosin exons 2 (T2) and 3 (T3) and the ATM cryptic exon (grey) are boxed. The length of the intronic sequences (shown as lines and not in scale) flanking the ATM cryptic exon is of 99 and 108 bases, respectively. Dotted lines represent the possible splicing products. pcY7ATM wt and Δ minigenes were transiently transfected in Hep3B cells and the pattern of splicing analyzed with the indicated primers. Amplification experiments of mature mRNA with T2F and T3R is shown on the right. As expected the GTAA deletion activates the cryptic exon also in the short context of the α-tropomyosin gene. (C) Result of amplification experiments of pcY7ATM wt and Δ with T2F-in20R and in20F-T3R primers on the left and right panel, respectively. The pre-mRNA forms are schematically represented along with the primers utilized. Even lanes correspond to control amplification of RNA samples without reverse transcriptase. All amplified bands were verified by sequencing.
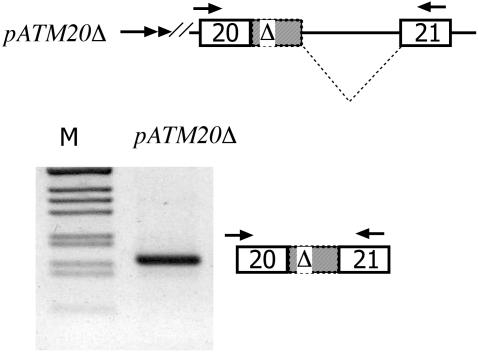
To extend this observation, and to confirm the absence of amplified products with primers located in the upstream portion of intron 20, we searched for the same intermediates in a shorter intron version of the ATM hybrid minigene, which was prepared to improve the RT–PCR detection of pre-mRNA forms (Figure 5B). Control experiments with these minigenes showed that, like the previously reported pTB ATM minigene (9), the Δ ISPE mutation induced cryptic exon inclusion (Figure 5B, right panel). To analyze the pre-mRNA forms produced in the transfected cells, total RNA was amplified with specific primer pairs that span intronic and exonic sequences corresponding to T2F-in20R (Figure 5C, left panel) or in20F-T3R (Figure 5C, right panel). Due to the reduced size of the introns flanking the cryptic exon, amplification experiments in WT and Δ ISPE minigenes showed the full-length unprocessed transcripts with both pairs of primers (Figure 5C, lanes 1, 3, 5 and 7). The partially spliced pre-mRNA form, exclusively present in the Δ ISPE, corresponded to the exclusion of the 3′ portion of intron 20 that precedes the cryptic exon (Figure 5C, lane 3) consistent with the in vivo observations (Figure 5A). To prove that this intermediate, once formed, is processed to the final mature mRNA in vivo in the original context, we prepared the pATM20 Δ minigene (Figure 6A). In this construct, the splicing intermediate lacking the preceding intron is embedded in the natural context with the flanking ATM exon 20, downstream intron sequences, and ATM exon 21. Transfection of pATM20Δ showed a processed transcript similar to that found in vivo in the affected cells, i.e. including the entire cryptic exon (Figure 6B). Altogether, the data show that while in the wild-type situation no intron precursors can be detected, in the ISPE deletion, accumulation of precursors retaining the downstream intron can be seen. Thus, it appears that deletion of the ISPE first causes the removal of the intron 20 sequence upstream of the cryptic exon, followed by the removal of downstream sequences. This process suggests that U1 snRNP binding at the ISPE has a preferential inhibitory effect on the use of the cryptic 3′-ss. The order of aberrant intron removal seems to proceed in a strict 5′–3′ direction as implied by the fact that the 3′ portion of intron 20 is processed before the upstream portion in the ISPE deletion, and the overall result is suggestive of an effect of the ISPE in splicing efficiency.
Figure 6.
The pre-mRNA identified in vivo is correctly spliced in the context of the ATM minigene. The upper panel shows a schematic representation of the pATM20 Δ minigene. ATM exon 20, 21 and the cryptic ATM exon with the GTAA deletion (Δ and grey) are boxed. Dotted lines represent the splicing products and the double arrow at the 5′ end the SV40 promoter. The minigene was transfected in Heb3B cells and the pattern of splicing analyzed with α2-3 and ATM 2550rev primers. The identity of the processed band is shown. M is the molecular weight marker 1 kb.
DISCUSSION
A growing body of evidence indicates that genomic variations at non-canonical splicing regulatory elements may unexpectedly affect the splicing process (25,26). However, their assessment as disease-causing mutations may be a difficult task as the nature of affected splicing regulatory elements is largely unknown and direct RNA analysis is not routinely performed in genomic screenings. We have previously reported a mutation in a deep intronic ISPE element in the ATM gene, which is located far from coding sequences and differs from the majority of the described intronic variants in that it is not directly concerned with changes at splice sites. However, the ISPE ‘CAG/GTAAGT’ sequence contains adjacent consensus 3′-ss and 5′-ss that are never used. A GTAA deletion at ISPE maintains potential adjacent splice sites, disrupts a non-canonical U1 snRNP interaction, and activates an aberrant exon (9). In this paper, we have carefully characterized the composition of cis-acting sequences around the ISPE signal and the role of non-canonical U1 snRNP binding.
To explore the mechanism by which the ISPE–U1 snRNP complex in the ATM intron interferes with the cryptic splice sites, we performed experiments with complementary U1 snRNAs on the ATM Δ minigene. These U1 snRNAs vary in the 5′ end sequence to bind at sequential positions along the cryptic exon (Figure 1). The artificial recruitment of U1 snRNPs at the upstream portion of the cryptic exon induces normal intron processing (U1-Δ, U1-32 and U1-40), whereas further downstream recruitment (U1-47 and U1-66) has no effect (Figure 1). This result identifies a critical region of ∼40 nt downstream the GTAA ISPE deletion in the ATM Δ (or 50 nt downstream the cryptic 3′-ss) where U1 snRNP binding induces normal intron 20 processing. Interestingly, the distance from the 3′-ss adequately corresponds to the suggested minimum size of 50 nt required for internal exon inclusion in vertebrates (22).
Comparison between wild-type and the GTAA deletion mutants (CAG/GTAAGT versus CAG/GTGATA sequences, respectively) shows that the adjacent consensus 3′-ss and 5′-ss AG/GT are maintained. Since these adjacent consensus splice sites might represent a re-splicing signal or ‘zero exon’ that regulates and ensures appropriate intron processing (9,15,16), we evaluated whether the core AG/GT sequence is required for normal processing induced by U1 snRNAs. We created site-directed mutants at either adjacent splice sites that were analyzed alone or in the presence of complementary U1 snRNAs. The results shown in Figure 2 indicate that the position-dependent effect of the U1 snRNA prevents aberrant splicing independent of the sequence composition of its core AG/GT adjacent 5′ and 3′ consensus splice sites. Thus, formation of the U1 snRNP complex in a critical position is sufficient to induce normal ATM intron processing. This effect is independent of the putative resplicing signal contained in the ISPE. On the other hand, amplification experiments in normal cells and in the corresponding hybrid minigene do not show any precursor originating from the usage of the core AG/GT signal (Figure 5). Even if this evidence seems to exclude that the U1 snRNP complex formed on deep ISPE participates in resplicing, the precursors originating from the usage of the resplicing signal might be rapidly processed to the mature mRNA and are not detected in the amplification assay. Additional experiments are necessary to carefully study the kinetics of the splicing reaction of introns containing intronic ISPE–U1 snRNP complexes.
To test the importance of cryptic splice sites around the ISPE, we functionally compared the splicing requirement in the mouse ATM intron 20 sequences. In order to induce aberrant splicing, deletion of the mouse-conserved ISPE requires at least the presence of cryptic splice sites that define the boundaries of the exon (Figure 3) suggesting that additional cis-acting elements adjacent to the ISPE are involved. Actually, deletion of other intronic human ATM ISPE consensus sequences not flanked by cryptic splice sites did not result in aberrant splicing, i.e. activation of a cryptic exon (data not shown).
To analyze the interference with the cryptic splice sites more in detail, we identified the pre-mRNA splicing precursors derived from lymphoblast cells and from hybrid minigene experiments. The ISPE deletion and the resulting disruption of U1 snRNP binding induces the unique formation of a splicing precursor that contains the downstream but not the upstream intron, stimulating a stringent 5′→3′ order of intron removal around the cryptic exon (Figure 5). This obligatory splicing direction induced by the deletion can be observed even in hybrid minigenes with a short intronic context (Figure 5C) and the precursor is properly spliced in vivo in the natural context that includes the flanking ATM exons and natural splice sites (Figure 6). This preferential effect on the excision of the upstream intron suggests that one mechanism of activation of the ATM cryptic exon is an interference of the ISPE–U1 snRNP complex with the cryptic 3′-ss.
We have previously shown that transferring an ISPE sequence into an exon induces its skipping and that this effect is mediated by U1 snRNP (9). We now extended this observation by evaluating the artificial recruitment of U1 snRNAs by complementarity in other heterologous gene systems. We observed that splicing inhibition occurred in the case of the fibronectin EDA alternative splicing and in the 3849 + 10 kb C→T CFTR mutation, but not for the constitutively included α-globin minigene (Figure 4), suggesting that exon-specific elements determine the inhibitory role of U1 snRNP. It is possible that the weaker exon definition present in the alternative spliced EDA and CF cryptic mutation, as compared with the constitutive globin exon, mediates the splicing inhibitory effect of the U1 snRNAs. This reduction in exon definition may not be simply due to weak splice sites, but result from more complex exon-specific interactions that involve both exonic and intronic regulatory elements whose function strongly depend on the context of individual exons. In fact, exonic splicing enhancers have been identified not only in alternatively spliced exons, but also in constitutive globin exons (27). In addition, this result indicates that previously reported exonic splicing silencers identical to ISPE (28) may exert their inhibitory effect through non-canonical binding to U1 snRNP. Interestingly, the increase in the level of correctly spliced RNA transcribed from an endogenous CFTR allele carrying the 3849 + 10 kb T mutation may have therapeutic potential. The specificity provided by the complementarity of the U1 snRNA or by antisense oligoribonucleotides (29) is a preferable therapeutic strategy to correct this splicing defect when compared to overexpression of regulatory splicing factors like SC35 and Htra2-b (30), which have widespread splicing effects in the cell (31,32).
ISPE–U1 snRNP complexes, independent of their interference with cryptic splice sites, could facilitate ATM intron splicing. U1 snRNA has been suggested to regulate transcription initiation by binding to the transcription factor TFIIH (33), and U1 snRNP interaction in the first intron has been shown to enhance PolII transcription (34). Thus, it is possible that the non-canonical interaction of U1 snRNP at the ISPE might affect intron splicing efficiency by regulating the polymerase II processivity or elongation rate due to the close association between transcription and splicing (35,36). Additional experiments are necessary to clarify whether intronic ISPE–snRNP complexes are affecting the kinetics of the splicing reaction.
Non-canonical binding of U1 snRNA has been observed in other gene systems with multiple effects on pre-mRNA processing. Recruitment of U1 snRNP to a pseudo 5′-ss may result in concomitant reduction of U1 snRNP binding to the proper 5′-ss (37). Likewise, U1 snRNA may interfere with the polyadenylation signal (38–40), and targeted loading of modified U1 snRNAs to terminal exons has proved to inhibit gene expression (41,42). In introns, the ISPE–U1 snRNP interaction interfering with the usage of intronic cryptic splice sites, may be a widespread phenomenon not limited to the ATM case.
Acknowledgments
This work was supported by a grant from Associazione Italiana Ricerca Cancro and Italian Research Ministry (FIRB RBNE01 W9PM). We thank C.W.J. Smith for providing the α-tropomyosin clone. Funding to pay the Open Access publication charges for this article was provided by ICGEB.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vervoort R., Gitzelmann R., Lissens W., Liebaers I. A mutation (IVS8 + 0.6kbdelTC) creating a new donor splice site activates a cryptic exon in an Alu-element in intron 8 of the human beta-glucuronidase gene. Hum. Genet. 1998;103:686–693. doi: 10.1007/pl00008709. [DOI] [PubMed] [Google Scholar]
- 2.Chillon M., Dork T., Casals T., Gimenez J., Fonknechten N., Will K., Ramos D., Nunes V., Estivill X. A novel donor splice site in intron 11 of the CFTR gene, created by mutation 1811 + 1.6kbA→G, produces a new exon: high frequency in Spanish cystic fibrosis chromosomes and association with severe phenotype. Am. J. Hum. Genet. 1995;56:623–629. [PMC free article] [PubMed] [Google Scholar]
- 3.Wang M., Dotzlaw H., Fuqua S.A., Murphy L.C. A point mutation in the human estrogen receptor gene is associated with the expression of an abnormal estrogen receptor mRNA containing a 69 novel nucleotide insertion. Breast Cancer Res. Treat. 1997;44:145–151. doi: 10.1023/a:1005753117205. [DOI] [PubMed] [Google Scholar]
- 4.Highsmith W.E., Burch L.H., Zhou Z., Olsen J.C., Boat T.E., Spock A., Gorvoy J.D., Quittel L., Friedman K.J., Silverman L.M., et al. A novel mutation in the cystic fibrosis gene in patients with pulmonary disease but normal sweat chloride concentrations. N. Engl. J. Med. 1994;331:974–980. doi: 10.1056/NEJM199410133311503. [DOI] [PubMed] [Google Scholar]
- 5.Metherell L.A., Akker S.A., Munroe P.B., Rose S.J., Caulfield M., Savage M.O., Chew S.L., Clark A.J. Pseudoexon activation as a novel mechanism for disease resulting in atypical growth-hormone insensitivity. Am. J. Hum. Genet. 2001;69:641–646. doi: 10.1086/323266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christie P.T., Harding B., Nesbit M.A., Whyte M.P., Thakker R.V. X-linked hypophosphatemia attributable to pseudoexons of the PHEX gene. J. Clin. Endocrinol. Metab. 2001;86:3840–3844. doi: 10.1210/jcem.86.8.7730. [DOI] [PubMed] [Google Scholar]
- 7.Sun H., Chasin L.A. Multiple splicing defect in an intronic false exon. Mol. Cell. Biol. 2000;20:6414–6425. doi: 10.1128/mcb.20.17.6414-6425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zavadakova P., Fowler B., Zeman J., Suormala T., Pristoupilova K., Kozich V., Zavad'akova P. CblE type of homocystinuria due to methionine synthase reductase deficiency: clinical and molecular studies and prenatal diagnosis in two families. J. Inherit. Metab. Dis. 2002;25:461–476. doi: 10.1023/a:1021299117308. [DOI] [PubMed] [Google Scholar]
- 9.Pagani F., Buratti E., Stuani C., Bendix R., Dork T., Baralle F.E. A new type of mutation causes a splicing defect in ATM. Nature Genet. 2002;30:426–429. doi: 10.1038/ng858. [DOI] [PubMed] [Google Scholar]
- 10.Sedgwick R.P., Boder E. Ataxia-telangiectasia. In: Jong J.M.B.V., editor. Handbook of Clinical Neurology. Hereditary Neuropathies and Spinocerebellar Atrophie. Vol. 16. The Netherlands: Elsevier Science, Amsterdam; 1991. pp. 347–423. [Google Scholar]
- 11.Lavin M.F., Shiloh Y. The genetic defect in ataxia-telangiectasia. Annu. Rev. Immunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- 12.Gatti R.A., Boder E., Vinters H.V., Sparkes R.S., Norman A., Lange K. Ataxia-telangiectasia: an interdisciplinary approach to pathogenesis. Medicine (Baltimore) 1991;70:99–117. [PubMed] [Google Scholar]
- 13.Teraoka S.N., Telatar M., Becker-Catania S., Liang T., Onengut S., Tolun A., Chessa L., Sanal O., Bernatowska E., Gatti R.A., et al. Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am. J. Hum. Genet. 1999;64:1617–1631. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhuang Y., Weiner A.M. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 15.Burnette J.M., Hatton A.R., Lopez A.J. Trans-acting factors required for inclusion of regulated exons in the Ultrabithorax mRNAs of Drosophila melanogaster. Genetics. 1999;151:1517–1529. doi: 10.1093/genetics/151.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatton A.R., Subramaniam V., Lopez A.J. Generation of alternative Ultrabithorax isoforms and stepwise removal of a large intron by resplicing at exon–exon junctions. Mol. Cell. 1998;2:787–796. doi: 10.1016/s1097-2765(00)80293-2. [DOI] [PubMed] [Google Scholar]
- 17.Pagani F., Buratti E., Stuani C., Romano M., Zuccato E., Niksic M., Giglio L., Faraguna D., Baralle F.E. Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J. Biol. Chem. 2000;275:21041–21047. doi: 10.1074/jbc.M910165199. [DOI] [PubMed] [Google Scholar]
- 18.Muro A.F., Caputi M., Pariyarath R., Pagani F., Buratti E., Baralle F.E. Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol. Cell. Biol. 1999;19:2657–2671. doi: 10.1128/mcb.19.4.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pagani F., Stuani C., Zuccato E., Kornblihtt A.R., Baralle F.E. Promoter architecture modulates CFTR exon 9 skipping. J. Biol. Chem. 2003;278:1511–1517. doi: 10.1074/jbc.M209676200. [DOI] [PubMed] [Google Scholar]
- 20.Gooding C., Roberts G.C., Smith C.W. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated alpha-tropomyosin exon. RNA. 1998;4:85–100. [PMC free article] [PubMed] [Google Scholar]
- 21.Lund E., Dahlberg J.E. True genes for human U1 small nuclear RNA. Copy number, polymorphism, and methylation. J. Biol. Chem. 1984;259:2013–2021. [PubMed] [Google Scholar]
- 22.Hwang D.Y., Cohen J.B. U1 small nuclear RNA-promoted exon selection requires a minimal distance between the position of U1 binding and the 3′ splice site across the exon. Mol. Cell. Biol. 1997;17:7099–7107. doi: 10.1128/mcb.17.12.7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang D.Y., Cohen J.B. Base pairing at the 5′ splice site with U1 small nuclear RNA promotes splicing of the upstream intron but may be dispensable for slicing of the downstream intron. Mol. Cell. Biol. 1996;16:3012–3022. doi: 10.1128/mcb.16.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Highsmith W.E., Jr, Burch L.H., Zhou Z., Olsen J.C., Strong T.V., Smith T., Friedman K.J., Silverman L.M., Boucher R.C., Collins F.S., et al. Identification of a splice site mutation (2789 + 5 G→A) associated with small amounts of normal CFTR mRNA and mild cystic fibrosis. Hum. Mutat. 1997;9:332–338. doi: 10.1002/(SICI)1098-1004(1997)9:4<332::AID-HUMU5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 25.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 26.Pagani F., Baralle F.E. Genomic variants in exons and introns: identifying the splicing spoilers. Nature Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 27.Schaal T.D., Maniatis T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell. Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Friedman K.J., Kole J., Cohn J.A., Knowles M.R., Silverman L.M., Kole R. Correction of aberrant splicing of the cystic fibrosis transmembrane conductance regulator (CFTR) gene by antisense oligonucleotides. J. Biol. Chem. 1999;274:36193–36199. doi: 10.1074/jbc.274.51.36193. [DOI] [PubMed] [Google Scholar]
- 30.Nissim-Rafinia M., Aviram M., Randell S.H., Shushi L., Ozeri E., Chiba-Falek O., Eidelman O., Pollard H.B., Yankaskas J.R., Kerem B. Restoration of the cystic fibrosis transmembrane conductance regulator function by splicing modulation. EMBO Rep. 2004;5:1071–1077. doi: 10.1038/sj.embor.7400273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H.Y., Xu X., Ding J.H., Bermingham J.R., Jr, Fu X.D. SC35 plays a role in T cell development and alternative splicing of CD45. Mol. Cell. 2001;7:331–342. doi: 10.1016/s1097-2765(01)00181-2. [DOI] [PubMed] [Google Scholar]
- 32.Ding J.H., Xu X., Yang D., Chu P.H., Dalton N.D., Ye Z., Yeakley J.M., Cheng H., Xiao R.P., Ross J., et al. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. EMBO J. 2004;23:885–896. doi: 10.1038/sj.emboj.7600054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwek K.Y., Murphy S., Furger A., Thomas B., O'Gorman W., Kimura H., Proudfoot N.J., Akoulitchev A. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nature Struct. Biol. 2002;9:800–805. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- 34.Furger A., O'Sullivan J.M., Binnie A., Lee B.A., Proudfoot N.J. Promoter proximal splice sites enhance transcription. Genes Dev. 2002;16:2792–2799. doi: 10.1101/gad.983602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maniatis T., Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 36.Kornblihtt A.R., de la Mata M., Fededa J.P., Munoz M.J., Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labourier E., Adams M.D., Rio D.C. Modulation of P-element pre-mRNA splicing by a direct interaction between PSI and U1 snRNP 70K protein. Mol. Cell. 2001;8:363–373. doi: 10.1016/s1097-2765(01)00311-2. [DOI] [PubMed] [Google Scholar]
- 38.Ashe M.P., Pearson L.H., Proudfoot N.J. The HIV-1 5′ LTR poly(A) site is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J. 1997;16:5752–5763. doi: 10.1093/emboj/16.18.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lou H., Neugebauer K.M., Gagel R.F., Berget S.M. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol. Cell. Biol. 1998;18:4977–4985. doi: 10.1128/mcb.18.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gunderson S.I., Polycarpou-Schwarz M., Mattaj I.W. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 41.Fortes P., Cuevas Y., Guan F., Liu P., Pentlicky S., Jung S.P., Martinez-Chantar M.L., Prieto J., Rowe D., Gunderson S.I. Inhibiting expression of specific genes in mammalian cells with 5′ end-mutated U1 small nuclear RNAs targeted to terminal exons of pre-mRNA. Proc. Natl Acad. Sci. USA. 2003;100:8264–8269. doi: 10.1073/pnas.1332669100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu P., Kronenberg M., Jiang X., Rowe D. Modified U1 snRNA suppresses expression of a targeted endogenous RNA by inhibiting polyadenylation of the transcript. Nucleic Acids Res. 2004;32:1512–1517. doi: 10.1093/nar/gkh316. [DOI] [PMC free article] [PubMed] [Google Scholar]