Abstract
Regulatory networks involving different cell types control inflammation, morphogenesis and tissue homeostasis. Cell-type-specific transcriptional profiling offers a powerful tool for analyzing such cross-talk but is often hampered by mingling of cells within a tissue. Here, we present a novel method that performs cell-type-specific expression measurements without prior cell separation. This involves inter-species transplantation or chimeric co-culture models among which the human mouse system is frequently used. Here, we exploit the sufficiently divergent transcriptomes of human and mouse in conjunction with high-density oligonucleotide arrays. This required a masking procedure based on transcriptome databases and exhaustive fuzzy mapping of oligonucleotide probes onto these data. The approach was tested in a human–mouse experiment, demonstrating that we can efficiently measure species-specific transcriptional profiles in chimeric RNA samples without physically separating cells. Our results stress the importance of transcriptome databases with accurate 3′ mRNA termination for computational prediction of accurate probe masks. We find that most human and mouse 3′-untranslated region contain unique stretches to allow for an effective control of cross-hybridization between the two species. This approach can be applied to xenograft models studying tumor–host interactions, morphogenesis or immune responses.
INTRODUCTION
Chimeric models have been applied extensively to study tumor–host interactions or embryonic morphogenesis, both of which are controlled by a precisely tuned interplay of different, specialized cell types. For instance, epithelial/mesenchymal interactions have been identified to be essential for formation and patterning of limb buds and epidermal appendages and often involve a complex hierarchy of cross-talk between the two tissues (1–5). Similarly, interactions between carcinoma cells and tumor stroma have been recognized which are causally involved in cancer progression and metastasis (6–9). Attempts to address the complexity of this cross-talk led to the development of cell-type-specific transcriptomics based principally on fluorescent activated cell sorting (FACS) or laser captured micro-dissections (LCM). Known limitations in the FACS approach follow from lengthy dissociation protocols, which can affect the transcriptional program, and from the requirement of appropriate surface markers, which might not be available for all cell types. LCM has been successfully adapted to a variety of experimental settings. However, the isolation of individual cell types based only on morphologic criteria or the isolation of single cells from samples with significant intermingling is limited. In particular, LCM is prone to destroy the immediate interface of adjacent cells that are most important in the analysis of cell–cell communication.
Our approach for cell-type-specific profiling is based on the possibility of studying cell–cell interactions using chimeric models (10,11) and the divergence of genes in their untranslated regions (UTRs). The availability of complete transcriptomes constituted a key resource in the development of our method. In particular, accurate information about 3′-untranslated regions (3′-UTR) of mRNAs is crucial to precisely measure expression of genes in highly similar gene families. This is reflected in the design of GeneChip arrays where probes from UTRs are over-represented. This permitted us to perform species-restricted expression measurements of even highly conserved ortho- and paralogues as sufficient divergence exists in the 3′-UTRs of such genes between different species. We computationally derived masks based on transcriptome databases that allowed us to extend expression profiling to chimeric RNA samples, without loosing the species specificity of the measurements.
Probe masks have been recently used to increase sensitivity/specificity of expression measurements in mammalian species that lack available arrays, but are sufficiently close to humans to permit use of human arrays (11,12). Here, we proceed further in measuring species-specific expression in yet more complex samples, namely mixtures of human and mouse RNAs. Key factors in this method include the generation of probe masks from accurate transcriptomes, together with exhaustive and fuzzy mapping of all oligonucleotide probes onto these transcriptomes. These developments allow us to measure species-specific and hence cell-type-specific expression levels in chimeric RNA samples.
MATERIAL AND METHODS
Expression data
Labeled cRNA was generated from the human colon carcinoma cell line LS174T (HC), human heart (HH; Clontech, Palo Alto, CA) and mouse liver (ML) according to Affymetrix protocols. These cRNAs were mixed in different combinations and hybridized to Human Genome U133 Plus 2.0 GeneChips and Mouse Genome 430 2.0 arrays (details in Table 1). Raw data is available at http://sib-pc27.unil.ch/felix/Chimeric.
Table 1.
Proportions of human colon (HC), human heart (HH) and mouse liver (ML) cRNA in the samples
| Mixtures | Human colon (HC) | Human heart (HH) | Mouse liver (ML) | PolyA spikes | Hybrid. spikes |
|---|---|---|---|---|---|
| 100% HC | 100 | 0 | 0 | 100 | 100 |
| 20% HC + 80% HH | 20 | 80 | 0 | 20 | 100 |
| 50% HC + 50% ML | 50 | 0 | 50 | 100 | 100 |
| 10% HC + 40% HH + 50% ML | 10 | 40 | 50 | 10 | 100 |
| 25% HC + 75% ML | 25 | 0 | 75 | 100 | 100 |
| 100% ML | 0 | 0 | 100 | 100 | 100 |
The first five samples were hybridized onto Human Genome U133 Plus 2.0 arrays and the last sample onto the Mouse Genome 430 2.0 array. The polyA spikes are added prior to cRNA synthesis and are used as controls (cf. Figure 3a, cyan dots). Hybridization spikes are added to the final cRNA samples at equal amounts in each sample.
Masks and normalization
To filter cross-species signals, we identified all individual oligonucleotide probes (25mer) on the Human Genome U133 Plus 2.0 array susceptible to detect spurious signal from mouse mRNA. We used a mouse transcriptome as defined by the union of two databases, RefSeq6 (http://www.ncbi.nlm.nih.gov/RefSeq) and tromer (13) (ftp://ftp.licr.org/pub/databases/trome). To exhaustively find all sequence similarities with a given number of mismatches (MMs), gapless global Smith–Waterman alignments were performed using GeneMatcher hardware (Paracel, Pasadena, CA). The whole procedure was repeated by mapping the probes of the Mouse Genome 430 2.0 array onto the human transcriptome in order to allow measurements of mouse transcripts from chimeric RNA samples (mask files can be accessed at http://sib-pc27.unil.ch/felix/Chimeric). Statistics about the number of probes masked for various stringencies can be found in Table 2. Probes were classified as coding or non-coding if unambiguously mapped onto the coding sequence (CDS) or 3′-UTR part of RefSeq sequences. Probes that mapped to several RefSeq sequences were considered only if they mapped to the 3′-UTR in all cases, or vice versa for the CDS. Probe masking is implemented by modifying the cdf environment in BioConductor (14) and standard RMA signal estimation (15) was subsequently applied to the truncated probe sets (PSs). CEL files were quantile normalized as part of the standard RMA signal estimation procedure.
Table 2.
Masked probes and discarded probe sets (PS) of the Human Genome U133 Plus 2.0 and Mouse Genome 430 2.0 arrays after masking all probes with a fixed maximal number of MMs onto the transcriptome of mouse or human, respectively
| Maximal number of mismatches | Human array | Mouse array | ||
|---|---|---|---|---|
| Discarded PSs (total 54 675) | Probes masked (total 604 258) | Discarded PSs (total 45 101) | Probes masked (total 501 592) | |
| 0 | 81 (0.14%) | 11 159 | 94 (0.17%) | 9718 |
| 1 | 410 (0.75%) | 31 611 | 355 (0.64%) | 27 925 |
| 2 | 1432 (2.6%) | 61 028 | 1301 (2.3%) | 55 048 |
| 3 | 3448 (6.3%) | 102 306 | 3230 (5.9%) | 92 308 |
| 4 | 7568 (13.8%) | 231 692 | 6933 (12.7%) | 200 856 |
PSs were discarded when the number of probes per PS was <4.
SAGE data
A test set of genes differentially expressed in heart or colon was identified using SAGE (cf. Figure 2). For this, we used SAGE Genie (http://cgap.nci.nih.gov/SAGE) and compared libraries from colon cancer cell lines with heart tissue (parameters used: F = 2, P = 0.05).
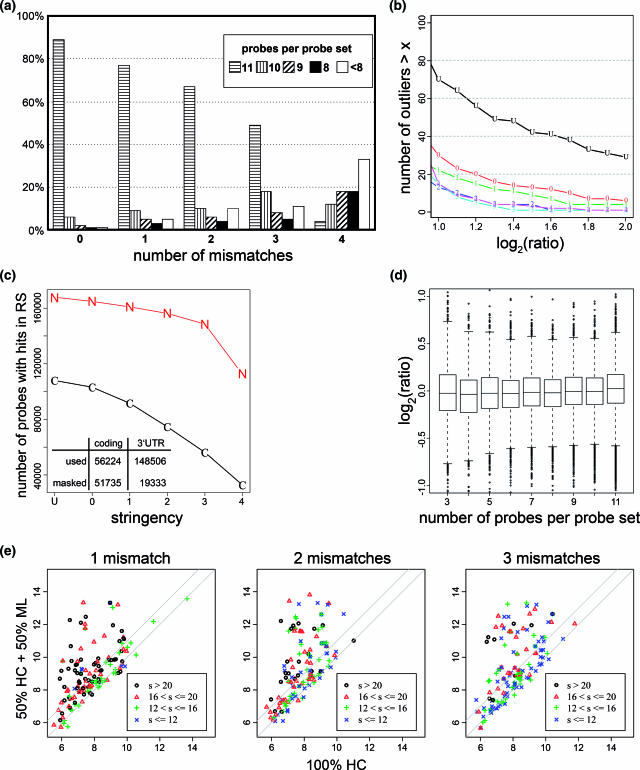
Figure 2.
Oligonucleotide probe masks. (a) Distribution of probes per probe set (PS) of the Human Genome U133 Plus 2.0 array after masking all probes onto the mouse transcriptome with a fixed maximal number of MMs. (b) Masking efficiency. Outlier counts according to Figure 1b as a function of log-ratio thresholds. ‘U’ is the unmasked results, ‘0’ masks only probes with perfect matches, ‘1’ with up to one mismatch, etc. (c) Number of probes in the coding (C) or non-coding (N) part of mRNAs after masking up to a given number of MMs (x-axis). The number of used or masked probes with 3 MMs in relation to its localization in the coding or UTR part of mRNAs is shown in the insert. Only probes with matches on RefSeq are considered. (d) Accuracy of expression signals for truncated PSs in the 50% HC + 50% ML versus 100% HC comparison. Analysis is stratified according to the number of probes (NP) left after masking. Only pairs with mean intensity >7 (unit and scale according to RMA output) are used. Boxplots show uniform behavior in function of NP, and tight correlation between diluted and undiluted signal estimates: 50% of the data are reproducible within a factor of 20.15∼1.1. (e) All individual probes belonging to the 100 most outlying PSs in Figure 1a are stratified according to the number of MMs to their predicted target and the longest perfectly matching stretch (s).
Raw data, masks, scripts for signal estimation are implemented as an R package available at http://sib-pc27.unil.ch/felix/Chimeric.
RESULTS
To establish and verify our approach, RNA was isolated from the human colon carcinoma cell line LS174T (HC), human heart (HH) and mouse liver (ML). These RNAs were labeled, mixed in different combinations and hybridized to Affymetrix GeneChips (Table 1). The transcriptional profile of the HC sample was determined and compared to the profiles of HC diluted 1:1 by ML prior to hybridization. This design enabled us to analyze both the effects of cross-species hybridization and the decrease in signal strength induced by diluting the human sample with mouse (Figure 1a and b). Cross-species hybridization was evident as outliers above the diagonal in Figure 1a. The profile of the mouse liver sample was measured separately on a mouse array and confirms that the outliers can be explained by highly expressed mouse transcripts hybridizing to human probes. As expected, most of these genes increase the expression measurements of the corresponding human orthologue, however cross-hybridization between non-orthologous genes also occur (data not shown).
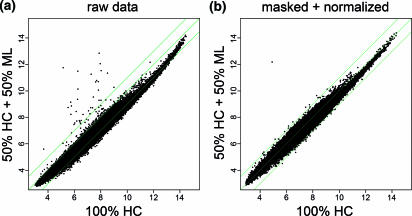
Figure 1.
Raw and masked expression signals. (a) and (b) compare expression signals of a 1:1 human colon (HC) and mouse liver (ML) mixture versus pure HC RNA. +/− 2-fold lines are indicated in green. (a) Standard RMA algorithm16 without masking or normalization. (b) Masked and normalized data. Oligonucleotide probes with three or fewer MMs to a mouse transcript in RefSeq or tromer were masked, and PSs with <4 probes left were discarded.
Masking cross-species hybridization
To control cross-species hybridization, we defined masks with increasing stringency by discarding probes with zero up to four mismatches (MMs) to any documented transcript in the competing species (Table 2). A consequence of increased stringency was the rapid decrease in the number of probes left in each probe set (PS) (Figure 2a) when MMs beyond three were considered. For example, the number of PSs with the original number of eleven probes drops from 49 to 4% when four instead of three MMs were considered. Thus, discarding up to three MMs provided the maximal masking stringency compatible with sufficient number of probes per PS. Signals for each transcript were re-calculated using the reduced PSs and the remaining number of outliers was determined in function of the maximal number of MMs (Figure 2b). These statistics confirm that three MMs provide an optimal masking stringency, as the 3 MM line lies systematically below the 2 MM line. As expected, probes which had to be masked are strongly enriched in the coding part of mRNAs (Figure 2c) thus reflecting evolutionary constraints. Sequence divergence and hence species specificity is larger in the 3′-UTR, as was found from all probes with unambiguous matches in RefSeq (cf. Methods). Association between masked oligos and their location in 3′-UTRs was highly significant for any masking stringency (χ2 statistics, P ∼ 0, 2 × 2 contingency table for 3 MMs is shown in Figure 2c).
We further tested whether we could restrict masking only to those probe–target pairs with long, perfectly aligned stretches. However, we found no straightforward and generally applicable criteria. In Figure 2e, all individual probes belonging to the 100 most outlying PSs in Figure 1a were stratified according to two criteria: the number of MMs to their predicted target and the longest, perfectly matching stretch. It is seen that relatively short stretches in the range of 12–16 often sense mouse signals. Such lengths correspond to stretches found in single mismatch probes (MM), known to measure a fair amount of specific signal (11). Therefore, the length criteria cannot be used alone. On the other hand, existing free energy models of DNA/RNA hybrids have not been developed to precisely predict annealing of short oligonucleotide sequences with several MMs and cannot be used reliably either (16). Since the masking performance was satisfactory and the fraction of lost PSs was small, we opted for a stringent masking based only on the number of MMs.
Next, we established the minimum number of probes per PS that still provide sufficient precision for differential expression measurements. For this, we calculated the variability in expression ratios between the 100% HC and 50% HC + 50% ML profiles as a function of the minimal number of used probes (Figure 2d). This showed that the inter-quartile range was not very sensitive to the number of probes used, and indicated that using four probes per PS provided a precision comparable to the full set. Consequently, only 6.3% of all human PSs with fewer than four remaining probes were discarded from our analyses (counts for other stringencies in Table 2).
Signals for each transcript were re-calculated from the reduced PSs using the standard RMA algorithm (15). This enabled us to eliminate virtually all cross-species hybridization (Figure 1b). This is reflected in the lack of outliers in Figure 1b; the one clear outlier remaining represents a PS consisting of repetitive sequences with no sequence match on either the human or mouse genome. Such PSs can easily be identified and filtered out a posteriori.
Sensitivity/specificity assessment of chimeric samples
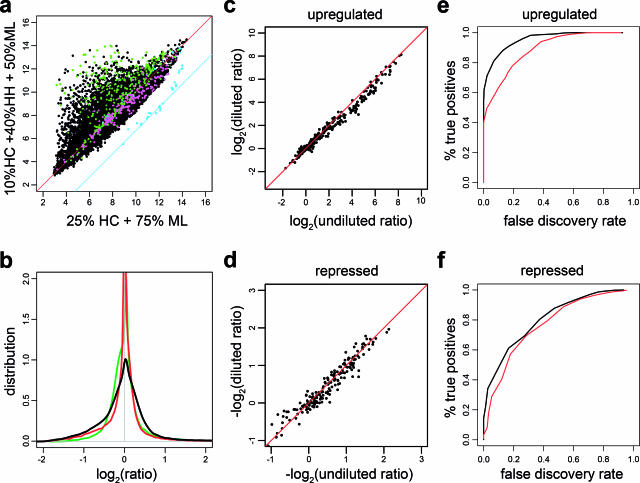
A mixture of human colon and heart RNA was used to evaluate the sensitivity of our approach in detecting gene expression changes after dilution with mouse RNA (Figure 3). When comparing the human colon/heart mixture to the pure human colon sample, we expect to see an entire range of induced genes (theoretical ratio is unbound for heart-restricted genes) while the most repressed, colon-restricted genes are reduced by 5-fold (or log2 (1/5) = −2.32). This asymmetry is visible in Figure 3a, with genes above the diagonal showing stronger induction than the ones below. Notice that this comparison uses different dilutions with mouse RNA on the two axes (x-axis 1:3 and y-axis 1:1) indicating the robustness of our method. To assess our method, a test set of differentially expressed genes was identified independently by comparing published SAGE libraries from colon cancer cell lines and heart tissue (http://cgap.nci.nih.gov/SAGE). Notice that about half of the genes predicted by SAGE do not show clear differential expression in our samples even in the undiluted comparison, indicating partial overlap between the two technologies. Nevertheless, in transcripts that are heart-restricted in both our measurements and the SAGE analysis, the measured inductions after dilution with mouse RNA correlate very well with the measurements in pure human samples (Figure 3c and d). This demonstrated that neither the dilution with mouse RNA nor the masking hinders measuring differential expression. Expectedly, higher dilutions induced progressive compression in the dynamic range, as seen in the nested distributions of log2 ratios (Figure 3b). To refine our evaluation, we defined a larger set of differentially expressed genes as positives, which consisted of 2% of the most up- and down-regulated genes in the undiluted comparison. These genes were then monitored in the presence of 1:1 and 1:3 human/mouse dilutions, and the fraction of recovered positives was calculated as the function of the false discovery rate (FDR) (Figure 3e and f). Although dilution of samples pose an intrinsic signal to noise problem, the obtained results indicate that our procedure can recover high percentages of positives with acceptable FDRs, e.g. >80% of the upregulated positives can be recovered after 1:1 dilution with a FDR rate of 20% (Figure 3e). The higher FDR observed for repressed genes (Figure 3f) follows from the asymmetric distribution of differentially expressed genes in our experimental design.
Figure 3.
Assessment of human-specific differential expression after dilution with mouse RNA. All samples were masked and normalized as in Figure 1b. (a) (10% HC + 40% HH + 50% ML) versus (25% HC + 75% ML). Genes differentially expressed in heart (magenta) or colon (green) according to SAGE (P < 0.05). The cyan polyA spikes should fall onto the indicated 10-fold line (Table 1). (b) Compression induced by dilution. The density of log2 ratios is narrowest for 1:3 human:mouse mixture (green) and widens for smaller dilutions (red is 50%, and black 0% mouse). (c and d) Comparison of the expression changes for genes in the test set (green and magenta in panel a) in the pure human mixture (80% HH + 20% HX) versus the human mixture diluted 1:1 with mouse. (e and f) Fraction of positives recovered after addition of mouse RNA plotted against the FDR for the 1:1 (black) and 1:3 (red) human:mouse mixtures. Here, ‘positives’ were defined as the 2% most induced or repressed genes in the undiluted 20% HC 80% HH versus 100% HC comparison.
DISCUSSION
Our study has shown that species-specific, and hence cell-type-specific transcriptional profiles of chimeric tissues can be obtained. We have developed a robust approach, which allows accurate measurements of differential expression even in experimental settings with changing species proportions. More precisely, our data showed that dilutions, with RNA from different species in the range of 1:3–3:1 and possibly larger, did not significantly affect the sensitivity and specificities of the profiling method. Such ranges will be sufficient for most applications.
It is interesting that the number of probes homologous to transcripts in the other species increases rapidly when matches with >3 MMs out of 25 nt are considered (cf. Table 2). More precisely, the number of masked probes increases by a factor more than two between 3 and 4 MMs, at which level more than a third of all oligonucleotides potentially cross-hybridize. At 5 MMs, we found that the entire array would be masked, presumably reflecting the overall level of conservation between mouse and human transcripts. This indicates that the degree of divergence between human and mouse 3′-UTRs, together with the stringency of short oligonucleotide hybridization allows for an effective control of cross-hybridization between human and mouse. Since we found that oligonucleotides with 3 MMs to an RNA molecule in the target can easily pick up signals under the current hybridization conditions (cf. Figure 2b), this approach would be difficult for more closely related species.
Additionally, our results stress the importance of transcriptome databases with accurate 3′ mRNA termination for computational prediction of accurate probe masks. Indeed, use of RefSeq alone was unable to provide optimal masking. Several reasons contribute to this: first, RefSeq sequences often do not contain complete 3′-UTRs (17), which is crucial since oligonucleotide probes are located primarily in these regions. Second, the tromer database used in this study compiles available expressed sequence tags and mRNA, and therefore presents a broader coverage of human or mouse transcriptomes than RefSeq. It would be interesting to explore whether similar masking can be exploited for better control of cross-hybridization in standard single-species experiments. Since hybridization is essentially governed by mass-action kinetics (18), it is almost unavoidable that highly expressed genes lead to spurious signals onto probes corresponding to lowly expressed transcripts. Therefore, one might consider identifying highly expressed genes with unmasked signal estimation, then predict the probes susceptible to contamination and mask these before re-computing the expression signals. Our masking strategy considered every possible transcript and did not incorporate expression data for the analyzed tissues. This is essential to correctly identify upregulated genes which are normally not expressed in the analyzed cell types but might be induced in the course of the experiment, e.g. during metastatic colonization of liver tissue. Nevertheless, this very generalized masking strategy allowed precise measurement of 94% of PSs, and we consider that the coverage under the proposed stringency is sufficient for most screening applications.
We have demonstrated that cell-type-specific, transcriptional profiles of chimeric tissues can be obtained by combining GeneChip arrays with probe masks. This new approach will facilitate the study of reciprocal interactions in a variety of chimeric systems either by co-cultures in vitro or after transplantation in vivo. Future applications of our profiling method can be envisaged in the field of tumor/stroma interactions in cancer progression and metastasis (19,20), studies of the hematopoietic system (21) or of tissue interactions during organogenesis (22) and homeostasis.
Acknowledgments
We would like to thank Otto Hagenbuechle for advice on the experimental design, Josiane Wyniger for excellent technical support, Dorota Retelska for advice on the manuscript, and the DNA Array Facility Lausanne for financial support. This work was supported in part by grants from the NCCR ‘Molecular Oncology’ to F.N. and J.H. Funding to pay the Open Access publication charges for this article was provided by the NCCR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pispa J., Thesleff I. Mechanisms of ectodermal organogenesis. Dev. Biol. 2003;262:195–205. doi: 10.1016/s0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- 2.Tickle C. Patterning systems—from one end of the limb to the other. Dev. Cell. 2003;4:449–458. doi: 10.1016/s1534-5807(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 3.Jahoda C.A., Oliver R.F., Reynolds A.J., Forrester J.C., Gillespie J.W., Cserhalmi-Friedman P.B., Christiano A.M., Horne K.A. Trans-species hair growth induction by human hair follicle dermal papillae. Exp. Dermatol. 2001;10:229–237. doi: 10.1034/j.1600-0625.2001.100402.x. [DOI] [PubMed] [Google Scholar]
- 4.Isogawa N., Terashima T., Nakano Y., Kindaichi J., Takagi Y., Takano Y. The induction of enamel and dentin complexes by subcutaneous implantation of reconstructed human and murine tooth germ elements. Arch. Histol. Cytol. 2004;67:65–77. doi: 10.1679/aohc.67.65. [DOI] [PubMed] [Google Scholar]
- 5.Mitsiadis T.A., Cheraud Y., Sharpe P., Fontaine-Perus J. Development of teeth in chick embryos after mouse neural crest transplantations. Proc. Natl Acad. Sci. USA. 2003;100:6541–6545. doi: 10.1073/pnas.1137104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liotta L.A., Kohn E.C. The microenvironment of the tumour–host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 7.Mueller M.M., Fusenig N.E. Friends of foes—bipolar effects of the tumour stroma in cancer. Nat. Rev. Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 8.Liu S., Tian Y., Chlenski A., Yang Q., Zage P., Salwen H.R., Crawford S.E., Cohn S.L. Cross-talk between Schwann cells and neuroblasts influences the biology of neuroblastoma xenografts. Am. J. Pathol. 2005;166:891–900. doi: 10.1016/S0002-9440(10)62309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vosseler S., Mirancea N., Bohlen P., Mueller M.M., Fusenig N.E. Angiogenesis inhibition by vascular endothelial growth factor receptor-2 blockade reduces stromal matrix metalloproteinase expression, normalizes stromal tissue, and reverts epithelial tumor phenotype in surface heterotransplants. Cancer Res. 2005;65:1294–1305. doi: 10.1158/0008-5472.CAN-03-3986. [DOI] [PubMed] [Google Scholar]
- 10.Bonnet D. Haematopoietic stem cells. J. Pathol. 2002;197:430–440. doi: 10.1002/path.1153. [DOI] [PubMed] [Google Scholar]
- 11.Davis P.H., Stanley S.L. Breaking the species barrier: use of SCID mouse–human chimeras for the study of human infectious diseases. Cell. Microbiol. 2003;5:849–860. doi: 10.1046/j.1462-5822.2003.00321.x. [DOI] [PubMed] [Google Scholar]
- 12.Khaitovich P., Muetzel B., She X., Lachmann M., Hellmann I., Dietzsch J., Steigele S., Do H.H., Weiss G., Enard W., et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sperisen P., Iseli C., Pagni M., Stevenson B.J., Bucher P., Jongeneel C.V. trome, trEST and trGEN: databases of predicted protein sequences. Nucleic Acids Res. 2004;32:D509–D511. doi: 10.1093/nar/gkh067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugimoto N., Nakano S., Katoh M., Matsumura A., Nakamuta H., Ohmichi T., Yoneyama M., Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 17.Iseli C., Stevenson B.J., de Souza S.J., Samaia H.B., Camargo A.A., Buetow K.H., Strausberg R.L., Simpson A.J., Bucher P., Jongeneel C.V. Long-range heterogeneity at the 3′ ends of human mRNAs. Genome Res. 2002;12:1068–1074. doi: 10.1101/gr.62002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hekstra D., Taussig A.R., Magnasco M., Naef F. Absolute mRNA concentrations from sequence-specific calibration of oligonucleotide arrays. Nucleic Acids Res. 2003;31:1962–1968. doi: 10.1093/nar/gkg283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiter D., Bogenrieder T., Elder D., Herlyn M. Melanoma–stroma interactions: structural and functional aspects. Lancet Oncol. 2002;3:35–43. doi: 10.1016/s1470-2045(01)00620-9. [DOI] [PubMed] [Google Scholar]
- 20.Edlund M., Sung S., Chung L. Modulation of prostate cancer growth in bone microenvironments. J. Cell. Biochem. 2004;91:686–705. doi: 10.1002/jcb.10702. [DOI] [PubMed] [Google Scholar]
- 21.Lensch M., Daley G. Origins of mammalian hematopoiesis: in vivo paradigms and in vitro models. Curr. Top. Dev. Biol. 2004;60:127–196. doi: 10.1016/S0070-2153(04)60005-6. [DOI] [PubMed] [Google Scholar]
- 22.Fontaine-Perus J. Mouse–chick chimera: an experimental system for study of somite development. Curr. Top. Dev. Biol. 2000;48:269–300. doi: 10.1016/s0070-2153(08)60759-0. [DOI] [PubMed] [Google Scholar]