Abstract
We have developed a novel three-primer, one-step PCR-based method for site-directed mutagenesis. This method takes advantage of the fact that template plasmid DNA cannot be efficiently denatured at its reannealing temperature (Tra), which is otherwise a troublesome problem in regular PCR. Two flanking primers and one mutagenic primer with different melting temperatures (Tm) are used together in a single PCR tube continuously without any intervention. A single-stranded mutagenic DNA (smDNA) is synthesized utilizing the high Tm mutagenic primer at a high annealing temperature, which prevents the priming of the low Tm primers (i.e. the two flanking primers). A megaprimer is then produced using this smDNA as the template at a denaturing temperature that prevents wild-type template DNA activity. The desired mutant DNA is then obtained by cycling again through these first two steps, resulting in a mutagenic efficiency of 100% in all tested cases. This highly automated method not only eliminates the necessity of any intermediate manipulation and accomplishes the mutagenesis process in a single round of PCR but, most notably, enables complete success of mutagenesis. This novel method is also both cost and time efficient and fully automated.
INTRODUCTION
Site-directed mutagenesis based on PCR is used to test the role of particular residues in the structure, catalytic activity and ligand-binding capacity of a protein by introducing the desired mutation into target DNA sequences (1). Of the many reported variants of PCR-based mutagenesis, the megaprimer method, which uses three oligonucleotide primers and two rounds of PCR, appears to have great promise because of its potential simplicity and low cost (2,3). However, these original megaprimer methods are time-consuming and require a laborious purification step in order to remove the residual primers from the megaprimer synthesized in the first PCR step, which makes these methods limited in their application (1).
The megaprimer method modified by Ke and Madison (4) is the most popular method because it does not require purification of DNA between subsequent rounds of PCR due to innovative primer design. However, this method cannot be completely automated because after the completion of the first PCR step, a high Tm flanking primer has to be added to the reaction tube, along with the megaprimer for the second PCR step. In addition, it is generally necessary to select six colonies for DNA sequencing to verify the presence of the desired mutation(s) (1) because this method yields mutants with an efficiency of only ∼82%. Also, Ke and Madison (4) report no examples of mutagenesis involving insertions or deletions. Consequently, these drawbacks prevent the megaprimer strategy from being widely employed.
Although various versions of the megaprimer method have been introduced frequently, there is not a three-primer, single PCR-based method for mutagenesis that can create base substitutions, insertions and deletions. Here, we have developed, tested and validated a novel and highly efficient PCR strategy that uses the reannealing temperature (Tra) of wild-type template DNA as a denaturing temperature. At this temperature, the wild-type template DNA cannot be efficiently denatured. A novel protocol has been established successfully based on the hypothesis that when two types of templates (i.e. double-stranded and single-stranded) coexist in the reaction mixture during PCR, the double-stranded template (i.e. the wild-type template) will not be amplified at the Tra. All primers (i.e. one mutagenic and two flanking primers) can be added simultaneously and used together in a single PCR tube continuously without any intervention, such as stopping the reaction and/or the addition of other reagents. This single step PCR-based method will be widely used for mutagenesis involving base substitutions, insertions and deletions.
MATERIALS AND METHODS
Preparation of materials and reagents
PCR experiments were performed in an Eppendorf Mastercycler gradient thermal cycler. DNA polymerase, restriction enzymes and DNA ligase were obtained from TakaRa and all primers were from Sangon. Several DNA templates, i.e. CNA (calcineurin A subunit); PP1 (protein phosphatase catalytic subunit); CNB (calcineurin B subunit); OB (human obese); CAM (calmodulin), were used in the gradient PCR (shown in Figure 1 and Table 1) and inserted into pET-21a system by Nde1 and BamH1, except for OB by BamH1 and EcoR1 (Figure 1e and k) and CNB by EcoRV into pIRES1neo (Figure 1g); and OB into pGEX-1 Lambda T by BamH1 and EcoR1 (Figure 1m). However, only CNA, PP1 and CNB DNA templates were used in 17 mutagenesis experiments to create base substitutions, insertions or deletions.
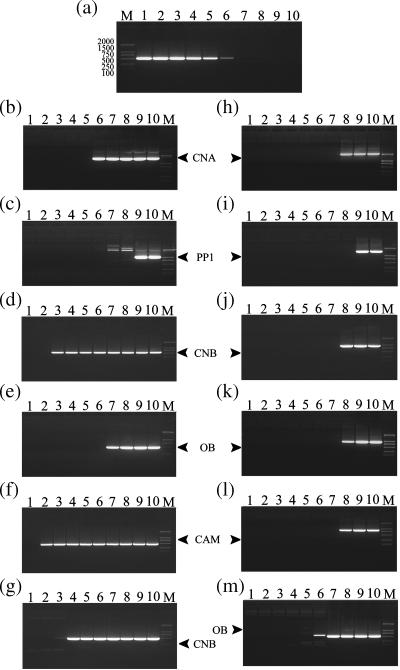
Figure 1.
The determination of denaturing temperatures, T1 and T2. The electrophoretic analysis of gradient PCR products is shown on a 1.2% agarose gel. Lane M, DNA molecular weight marker; fragment lengths (in bp) are indicated (DL2000 TaKaRa). (a) DNA products of PCR using a gradient of annealing temperatures; lane 1, 53.5°C; lane 2, 55.3°C; lane 3, 57.5°C; lane 4, 60.1°C; lane 5, 62.8°C; lane 6, 65.5°C; lane 7, 68°C; lane 8, 70.1°C; lane 9, 71.6°C; lane 10, 72.5°C. From (b) to (m) DNA products of PCR using a gradient denaturing temperature, lane 2, 79.8°C; lane 3, 81.1°C; lane 4, 82.4°C; lane 5, 84.0°C; lane 6, 85.9°C; lane 7, 87.9°C; lane 8, 89.9°C; lane 9, 91.5°C; lane 10, 94.1°C, except for lanes 1, 85.6°C (b); 90.0°C (c); 80.1°C (d); 87.6°C (e); 79.5°C (f); 80.7°C (g); 88.4°C (h); 89.7°C (i); 88.1°C (j); 88.2°C (k); 88.3°C (l); 87.7°C (m), respectively.
Table 1.
Results of mutagenesis experiments using the novel three-primer, one-step PCR protocol
| Mutant name | Original sequences/template (bp) | Mutagenic primer with mutant sequences | Mutagenesis efficiencya (%) |
|---|---|---|---|
| CNB/K134H | AAA/CNB (554) | GCAGATTGTAGACCACACCATAATAAACG | 7/7 = 100 |
| CNB/G120GGb | GGC/CNB (554) | GCGTATCTTTCAGGTTGTTGCCGCCCACCATCATCTTCAACAC | 7/7 = 100 |
| CNB/G120c | GGC/CNB (554) | GAAGATGATGGTGAACAACCTGAAAGATACGCAGTTACAG | 5/5 = 100 |
| CNB/K134R | AAA/CNB (554) | AGATTGTAGACCGCACCATAATAAACGCA | 1/1 = 100 |
| CAN/RY112-3LT | CGCTAC/CNA (1577) | CCCCTAAGAAGAGTGTCAAAGTGTTGGCAGGAGAT | 1/1 = 100 |
| CAN/V314H | GTG/CNA (1577) | GCACCAAATTACTTAGATCATTACAATAATAAAGC | 1/1 = 100 |
| PP1/L88L | CTG/PP1 (1034) | CACGTAGTCACCCAGGAATAGGTAGTTGCTCTC | 1/1 = 100 |
| CNB/M118N | ATG/CNB (554) | TTGAAGATGAATGTGGGCAACAACCTGAAAGATACGC | 2/2 = 100 |
| CNB/M118K | ATG/CNB (554) | TGAAGATGAAAGTGGGCAACAAC | 1/1 = 100 |
| CNB/M118W | ATG/CNB (554) | TTGAAGATGTGGGTGGGCAACAACCTGAAAGATACG | 1/1 = 100 |
| CNB/M118G | ATG/CNB (554) | TTGAAGATGGGCGTGGGCAACAACCTGAAAGATAC | 1/1 = 100 |
| CNB/M118H | ATG/CNB (554) | TTGAAGATGCATGTGGGCAACAACCTGAAAGATACG | 1/1 = 100 |
| CNB/M118E | ATG/CNB (554) | TTGAAGATGGAAGTGGGCAACAACCTGAAAGATACGC | 1/1 = 100 |
| CNB/V119R | GTG/CNB (554) | TTGAAGATGATGCGTGGCAACAACCTGAAAGATA | 1/1 = 100 |
| CNB/G120Y | GGC/CNB (554) | TTGAAGATGATGGTGTATAACAACCTGAAAG | 1/1 = 100 |
| CNB/G120R | GGC/CNB (554) | TTGAAGATGATGGTGCGTAACAACCTGAAAG | 1/1 = 100 |
| CNB/L123R | CTG/CNB (554) | ATGGTGGGCAACAACCGCAAAGATACGCAGT | 1/1 = 100 |
aMutagenesis efficiency was calculated as the number of confirmed mutants out of a number of randomly selected clones.
bCNB/G120GG represents bases insertions.
cCNB/G120 represents bases deletions.
Gradient PCR system
Each reaction mixture (200 μl) included 10 experimental PCR sample mixtures; each sample was subjected to a different annealing (Figure 1a) or denaturing (Figure 1b–m) temperature following a gradient with the mean temperature of 62°C (Figure 1a) and 87°C (Figure 1b–m). The reaction mixture (200 μl) contained 5 U pyrobest™ DNA polymerase (TakaRa), 16 μl of 2.5 mM of each dNTP (stock concentration), 20 μl 10× Pyrobest Buffer (supplied by the manufacturer), 200–300 ng plasmid DNA template (quantified by GBC cintra 10e), 120 pmol forward flanking primer and 120 pmol reverse flanking primer. Flanking primers were designed according to the sequence of the corresponding vector: R1 and F2 (Figure 1a–f); R1 and F2′ (Figure 1h–l); MP1 and MP2 (Figure 1m), except for GP1 and GP2 which were complementary to the CNB sequence (Figure 1g). The PCR protocol, programmed into an Eppendorf Mastercycler gradient thermal cycler, was 30 cycles of amplification using one of the following reaction conditions: (i) a gradient annealing temperature of 53.5–72.5°C during PCR (Figure 1a) with 94°C for 45 s (except for a 4 min first cycle), 62°C (G = 10) for 1 min and 72°C for ‘N’ min with a final extension step for 8 min at 72°C; (ii) a gradient denaturing temperature of 79.8–94.1°C during PCR (Figure 1b–m) with 87°C (G = 7) for 45 s (except for a 4 min first cycle), 42°C for 1 min and 72°C for ‘N’ min with a final extension step for 8 min at 72°C.
The ‘N’ min is calculated according to the amplification length of the target DNA, e.g. 1 min is needed when the amplification length is 1000 bases.
Mutagenesis reaction system
These reaction mixtures (100 μl) contained 2.5 U pyrobest™ DNA polymerase (TakaRa), 8 μl of 2.5 mM of each dNTP (stock concentration), 10 μl of 10× Pyrobest Buffer (supplied by the manufacturer), 100–150 ng of plasmid DNA template (quantified by GBC cintra 10e), 10 pmol of mutagenic primer, 60 pmol forward flanking primer (F2) and 60 pmol reverse flanking primer (R1). Amplification cycling parameters (Figure 3b) were programmed in Eppendorf Mastercycler gradient thermal cycler as follows: Step 1 with six sub-cycles of 45 s at 94°C (except for the 4 min first cycle), and 100 s at 72°C; Step 2 with three sub-cycles of 80°C for 45 s (except for 94°C for the 45 s first cycle), 42°C for 1 min, 72°C for 45 s, and a final extension step for 2 min at 72°C; Step 3 with six cycles from Step1 to Step 2. Electrophoresis analysis of each mutagenesis product (or ‘each of the mutagenesis products’) using this protocol was performed on a 1.2% agarose gel. If necessary (Figure 3d), ∼10 rounds of linear amplifications with 45 s at 94°C (except for the 4 min first cycle), 2 min at 72°C were added to the program prior to Step 1 (Figure 3c).
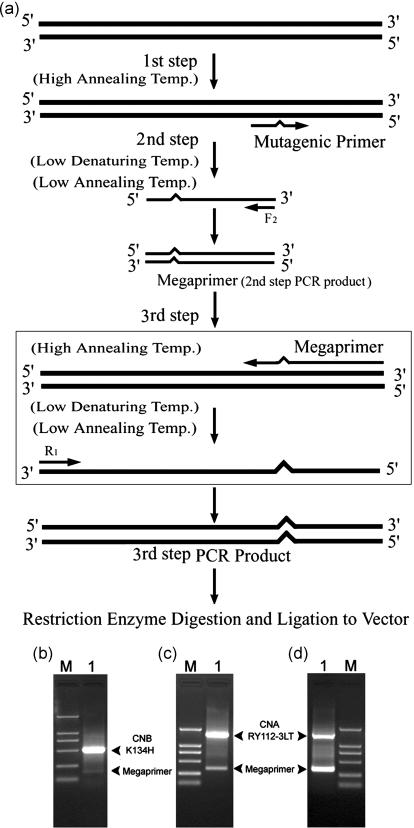
Figure 3.
(a) A schematic outline of the mutagenesis protocol: Step 1, a single-stranded mutagenic DNA (smDNA) was amplified with the high Tm mutagenic primer and a high annealing temperature of 72°C, at which the activities of two low Tm flanking primers is wholly restrained. Step 2, the megaprimer is synthesized using the smDNA template and one of the flanking primers such as F2 at a low annealing temperature (usually 36–46°C) and a low denaturing temperature (i.e. the Tra), where the wild-type DNA template restores its double-stranded status. Step 3, by running the two procedures above in the same system, an entire mutagenic single-stranded template DNA can be first produced with the megaprimer at a high annealing temperature (usually 72°C) and then the desired mutagenic DNA will be obtained by using the other low Tm flanking primer such as R1 under the conditions of Step 2. Electrophoretic analysis of mutagenesis products using this protocol is shown on a 1.2% agarose gel. For instance (b) lane M, Marker DL2000; lane 1, PCR product during CNB/K134H mutagenesis (554 bp) with its megaprimer (151 bp); (c) and (d) lane M, Marker DL2000; lane 1, PCR product during CNA/RY112-3LT mutagenesis (1577 bp) with its megaprimer (365 bp).
DNA sequencing
The DNA product of the mutagenesis experiments was purified by agarose gel electrophoresis, digested with Nde1 and Sal1, ligated into a pET-21a vector and transformed into competent Escherichia coli DH5α. Colonies from each mutagenesis reaction were selected and sequenced to confirm the presence of the desired mutation and the absence of any additional mutations. Mutagenesis efficiency was calculated as the number of confirmed mutants out of a number of randomly selected clones.
Calculation of base composition and correlation coefficients (r)
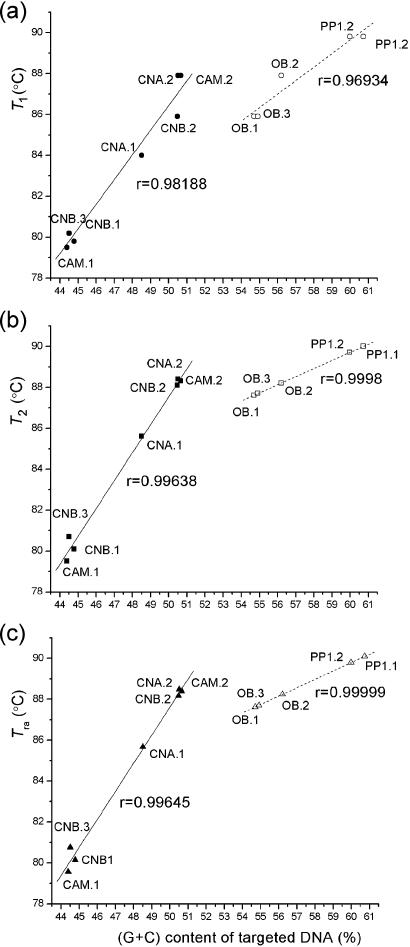
(G + C) content of each targeted DNA and its recombinant plasmid DNA were calculated (using BioEdit Version 4.7.2): 44.4 and 52.33, CAM.1; 44.51 and 51.30, CNB.3; 44.77 and 52.28, CNB.1; 48.51 and 52.02, CNA1; 50.47 and 52.28, CNB.2; 50.5 and 52.02, CNA.2; 50.66 and 52.33, CAM.2; 54.7 and 53.20, OB.1; 54.91 and 50.47, OB.3; 56.21 and 53.20, OB.2; 59.99 and 54.30, PP1.2; 60.74 and 54.30, PP1.1 (Figure 2).
Figure 2.
A comparison of correlations among the denaturing temperatures: (a) T1; (b) T2; and (c) the highest reannealing temperature (Tra), relative to (G + C) content of each targeted DNA. The numbers 1, 2 and 3 after the gene name represent the gene itself shown in Figure 1b–f, the gene with a part of the vector sequence shown in Figure 1h–l and the gene in other vector shown in Figure 1g and m, respectively. The values of T1 and T2 are shown in Figure 1. When (G + C) content of each targeted DNA is less than that of its recombinant plasmid, the solid line and solid symbols (circles, squares and triangles) represent the values of its T1 (a), T2 (b) and Tra (c), respectively; otherwise, they are represented by dashed lines and open symbols.
The correlation between the base composition and reannealing temperature of the target DNA were calculated by both SPSS.10.0 and Origin 7.0 software packages. The calculated correlation coefficients (r) are shown in Figure 2.
The sequences are available on GenBank: X57115 (CNA); X14832 (PP1); L03554 (CNB); U18915 (OB); and U89673 (pIRES1neo); U13849 (pGEX-1λT) and http://www.merckbiosciences.com/docs/docs/PROT/pET-21a-gb.html (pET-21a).
RESULTS
Primer design
At 68°C, low Tm primers cannot be efficiently annealed to template DNA (Figure 1a and shown in Materials and Methods). Noting this fact, we designed two flanking primers and a mutagenic primer with a different Tm (Figure 3a). All flanking primers (R1 and F2) were 14–16 bases with a calculated Tm between 36 and 46°C. Usually, the desired mutation should be located in the middle of the mutagenic primer with ∼8–10 bases of correctly matched sequences on each side of the mismatched region (1). Note that with >10 bases of correctly matched sequences at the 3′ end of the mutagenic primer, the PCR can be initiated effectively. Usually, the mutagenic primer excluding the mismatched region has a calculated Tm ∼62°C. The Tm (in °C) of oligonucleotide primers can be estimated using the standard formulas (5): Tm = 4(G + C) + 2(A + T).
PCR with a gradient of denaturation temperatures
PCR was conducted on a number of different DNA templates (contained in PET-21a vectors) using flanking primers designed according to the pET-21a vector sequence: R1: 5′-CGGAGATATACATATG-3′; F2: 5′-ATTGTCGACGGAGCT-3′ (Figure 1b–f, lanes 2–10). Each PCR was conducted with a gradient of denaturation temperatures of 79.8–94.1°C. Each template showed an initial and approximate denaturation temperature, which we shall call T1. T1 values vary depending on the sequence, ranging from 79.8°C (Figure 1d, lane 2) and 84.0°C (Figure 1b, lane 5) to 89.8°C (Figure 1c, lane 8) in this study. Above T1, the target DNA can be amplified well; below this temperature, the target DNA was not amplified (Figure 1b–f). In order to quantify this phenomenon, we repeated the experiment by changing the amplification length of the target DNA using primer F2′: 5′-ACCATCACCCTAATC-3′ instead of F2 (Figure 1h–l), and then used other vector systems with their corresponding primers [such as pIRES1neo with GP1: 5′-GTAGGATCCATGGGAAATGAGGCGAG-3′ and GP2: 5′-TGCCGTCGACTCACACATCTACCACC-3′ (Figure 1g) and pGEX-1λT with MP1: 5′-TTCCGCGTGGATCC-3′ and MP2: 5′-TCAGTCAGTCACGAT-3′ (Figure 1m)]. Similar amplification results were observed following these modifications (Figure 1g–m). Based on these results, we found a statistical correlation between the content of guanine and cytosine bases in each targeted DNA and the initial denaturation temperature (T1) (Figure 2a), irrespective of the vector and primers (Figures 1 and 2). As indicated in the Materials and Methods, the initial denaturation temperature (T1) observed in the gradient PCR, may represent the reannealing temperature of the template DNA amplified. In other words, the target DNA cannot be amplified when the denaturation temperature during PCR is at or below T1 because the target DNA is not denatured.
Nature and function of reannealing temperature
Upon determination of approximate T1 and Tm (at Tm, denaturation occurs in PCR and target DNA is amplified), we were interested to find out whether we can further obtain a temperature (T2), which more closely approaches Tm than T1. T2 would retain the characteristics of T1, at which target DNA denaturation has still not occurred. Thus, we carried out more detailed experiments by using fine gradient denaturation temperatures in a narrow window. In the case of CNA, for example, T1 is 84.0°C (Figure 1b, lane 5) and Tm is 85.9°C (Figure 1b, lane 6). By fine-tuning the temperature in the narrow window of 84.0–85.9°C, we obtained a T2 of 85.6°C for CNA (Figure 1b, lane 1). Based on the results obtained from many experiments including various DNA targets, variation of amplification length of the target DNA and primers as well as type of plasmids, we analyzed the relationships between T2 and Tm (5,6), and found an excellent correlation. When (Gi + Ci)% is less than recombinant plasmid (G + C)%, the difference between Tm and T2 (Tm − T2) is equivalent to 100 times the difference between (A + T)% in the amplification part of the recombinant plasmid and that in the recombinant plasmid (Figure 2). The existence of such a correlation evidently indicates that (A + T) content plays an important role in reannealing between the double-stranded DNA templates, as (G + C) content does in denaturation.
Essentially, T2 is very close to the highest reannealing temperature (Tra) of the template DNA. Tra is the critical reannealing temperature of the template DNA; the target DNA cannot be amplified when the denaturation temperature during PCR is at Tra. Here, we advanced a novel formulation based on the original one developed by Marmur and Doty (5) to calculate the Tra as follows:
Here Ai, Ti, Gi and Ci represent bases of each targeted DNA and A, T, G and C represent bases of each recombinant plasmid DNA.
To validate the novel formulation, 12 different amplification fragments with different (Gi + Ci) content ranging from 44.4 to 60.7% were denatured at Tra during PCR. No band of the desired product was observed in the agarose gel. It is suggested that the wild-type template DNA is not efficiently denatured at Tra. We can hypothesize that when two types of templates (i.e. double-stranded and single-stranded) coexist in the reaction mixture during PCR, the double-stranded templates (i.e. wild-type template) will not be amplified at the Tra. The denaturation temperature is critically <94°C, which is the typical denaturation temperature of regular PCR, but at this temperature, single-stranded type can be amplified well.
A novel PCR protocol for mutagenesis
Taking advantage of the fact that the wild-type template DNA could not be efficiently denatured at Tra, we developed a novel PCR strategy for mutagenesis. This method, outlined in Figure 3a, begins with a high annealing temperature in first step that allows the mutagenic primer, but not the flanking primers, to anneal to the template DNA. Next, a low denaturing temperature and a low annealing temperature in the second step of the protocol allows one of the flanking primers such as F2 to generate a complementary megaprimer, without amplifying the wild-type template DNA. Finally, repeating these temperature cycles amplifies the desired mutant DNA sequence using a high annealing temperature, the megaprimer and the wild-type template DNA, followed by a low denaturing temperature, a low annealing temperature, the other flanking primers such as R1 and the megaprimer PCR product. Obviously, the final step can be carried out by cycling again through the first two steps. Note that one mutagenic and two flanking primers can be added simultaneously in a single continuous reaction tube owing to their distinctly different Tm. Without any intervention such as reaction stoppage and/or addition of other reagents, the PCR product of this reaction is directly cloned to screen for mutants.
The step-by-step protocol of the mutagenesis method is described below:
Design and synthesize the flanking primers (R1 and F2) and mutagenic primer based on the known sequence of the DNA, as outlined in the primer design of the Results section.
Dissolve the flanking primers (R1 and F2) and mutagenic primer in H2O at a concentration of 10 μM (10 pmol/μl).
The Tra (in °C) of the target DNA (the wild-type template DNA) can be estimated using the formula and/or can be obtained using the gradient denaturing reaction conditions (see Materials and Methods).
In a sterile 0.5 ml microfuge tube or amplification tube (on ice), set up PCR by mixing the following reagents: 100–150 ng template DNA, 10 μl 10× amplification buffer, 8 μl of 2.5 mM dNTP solution, 6 μl of 10 μM primer R1 (60 pmol), 6 μl of 10 μM primer F2 (60 pmol), 0.8 μl of 10 μM mutagenic primer (8 pmol), 2.5 U thermostable DNA polymerase, and 100 μl H2O. An access of flanking primers compared to the mutagenic primer is required for optimal results. In this study, a molar ratio of 15:2 (flanking primers: mutagenic primer) was used.
Place the tubes in a thermocycler, such as Eppendorf Mastercycler gradient, and program the thermocycler as follows: (a) 4 min at 94°C; (b) 45 s at 94°C; (c) ‘1 + N’ min at 72°C; (d) go to (b), repeat six sub-cycles; (e) 45 s at 94°C; (f) 45 s at Tra°C; (g) 1 min at 42°C; (h) ‘N’ min at 72°C; (i) go to (f), repeat three sub-cycles; (j) 2 min at 72°C; (k) go to (b), repeat six cycles; (l) hold at 4°C. Note that the polymerization for ‘N’ min is every 1000 bp of length of the target DNA per minute.
Examine 10% of the PCR on an agarose or polyacrylamide gel and estimate the concentration of amplified target DNA.
Verify the complete sequence of the amplified DNA fragment after cloning to ensure that no mutations other than those in the mutagenic primer were introduced during these manipulations.
To test this novel mutagenesis strategy, three different DNA templates, inserted into pET-21a plasmid between Nde1 and BamH1 sites, were used in 17 mutagenesis experiments to create base substitutions, insertions or deletions with flanking primers (i.e. R1 and F2). All mutagenic primers and corresponding mutagenic efficiencies are summarized in Table 1. These data show that, in every single case, a successful mutated sequence was generated with very high efficiency.
DISCUSSION
Our experiments show that generating pure, high-yield mutant PCR product requires the following:
First, the two flanking primers should have identically low Tm values compared to the mutagenic primer, rather than having significantly different Tm values [as in a previous protocol described by Ke and Madison (4)]. Moreover, we found that a molar excess of the flanking primers compared to the mutagenic primer was required for optimal results. The advantages of adopting this ratio include (i) depleting mutagenic primers quickly in Step 1; (ii) producing megaprimers rapidly in Step 2; (iii) avoiding potential interference caused by the mutagenic primers to ensure normal priming of megaprimers at the high annealing temperature in Step 3; and (iv) expediting annealing between the other flanking primer and the entire mutagenic template in Step 3 (Figure 3a). Usually, the PCR is initiated most effectively when not less than 13 bases of correctly matched sequences are at the 3′ end of the mutagenic primer (Table 1). Furthermore, to expedite the depletion of the mutagenic primer in limited sub-cycles at the high annealing temperature, a high molar concentration of the wild-type template may be used (500 ng in 100 μl reaction mixtures). This high concentration can be used without the risk of generating copies of the wild-type DNA since the combined action of the high annealing and the low denaturing temperatures in Steps 1 and 2, respectively, limit the ability of the flanking primers to anneal to the wild-type plasmid DNA template.
Second, the accuracy of temperature values is very important. Most mutagenesis methods involving megaprimers need isolation and purification steps, and therefore cannot be automated without amplifying wild-type template. We found that using the Tra, we can selectively prevent the amplification of wild-type templates, hence ensuring that only the mutagenesis template is amplified. This result demonstrates that fully automated, high purity and high yield mutagenesis is possible. Obviously, this method depends on the accuracy of the chosen Tra. Moreover, if the low denaturation temperature is too far below Tra, then the third step in the PCR protocol may not work well, most likely because the full-length single-stranded template anneals to itself. Thus explaining why after using T1 we still needed to find T2 through further experiments to generate the formula. That is also why we cannot use a fixed general temperature. Since Tra becomes very close to Tm, during the PCR if the low denaturation temperature is higher than Tra, then the wild-type template will also be amplified. Note that if the low denaturation temperature in Step 3 is too far below the Tra, the yield would probably decrease, suggesting that an annealing effect may happen in the entirely mutagenic single-stranded DNA originating from the megaprimer.
Third, the optimal number of sub-cycles in each PCR step must be used to achieve exponential amplification (Figure 3a). Generally, the optimal number of sub-cycles is six or seven in the first PCR step and two or three in the second PCR step due to the high concentration of flanking primer. In addition, judging from the fluorescence intensity of the megaprimer band in the agarose gel analysis, we can find that the megaprimer is at a very low level relative to a high yield of the desired PCR product (Figure 3b and c); otherwise (Figure 3d), ∼10 rounds of additional linear amplification are necessary to deplete the mutagenic primer specifically using a high annealing temperature (usually 72°C), prior to the first step (shown in Materials and Methods). Consequently, the yield and purity of the desired product can be improved greatly (Figure 3c), indicating that avoiding potential interference caused by the mutagenic primers to ensure normal priming of megaprimers is helpful for maximizing the efficiency of this method. Generally speaking, by agarose gel analysis, a good PCR is reflected by a dim megaprimer band and a bright target band (Figure 3b and c), although sometimes it can be the other way around (Figure 3d). In the latter case, the problem can be overcome by addition of ∼10 cycles of linear amplification at the beginning of the protocol (i.e. Step 1). This will facilitate the consumption of the mutagenic primer and give rise to better results as the interference caused by the mutagenic primer is reduced.
Unlike the previous megaprimer protocols (1–4), this novel method uses one mutagenic and two flanking primers added simultaneously in a single reaction tube by creative use of Tra. Consequently, no human intervention (including stoppage of the reaction, addition of other reagents and/or intermediate purification) is required between steps during this single PCR. This method is cheap, simple, efficient and automatable. Most importantly, the success rate of mutagenesis has been 100% for all tested cases. Furthermore, the specific amplification or the differential display of the genomic DNA differing in the thermal stability seems to be allowed by means of Tra, an idea that is worthy of experimental study.
Acknowledgments
We thank co-workers in our department for testing our protocol. We are greatly indebted to Mr Lianwen Zhang and Mr Yong Lin for their critical comments on the manuscript. Useful discussion with Dr Vinay Singh is appreciated. This work was supported in part by a grant from the National Natural Science Foundation of China, the Research Fund for the Doctoral Program of High Education and the National Important Basic Research Project. Funding to pay the Open Access publication charges for this article was provided by the grant from the National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sambrook J., Russell D.W. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 2.Kammann M., Laufs J., Schell J., Gronenborn B. Rapid insertional mutagensis of DNA by polymerase chain reaction (PCR) Nucleic Acids Res. 1989;17:5404. doi: 10.1093/nar/17.13.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giebel L.B., Spritz R.A. Site-directed mutagenesis using a double-stranded DNA fragment as a PCR primer. Nucleic Acids Res. 1990;18:4947. doi: 10.1093/nar/18.16.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ke S.H., Madison E.L. Rapid and efficient site-directed mutagenesis by single-tube ‘megaprimer’ PCR method. Nucleic Acids Res. 1997;25:3371–3372. doi: 10.1093/nar/25.16.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marmur J., Doty P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J. Mol. Biol. 1962;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- 6.Marmur J., Doty P. Heterogeneity in deoxyribonucleic acids. I. Dependence on composition of the configurational stability of deoxyribonucleic acids. Nature. 1959;183:1427–1429. doi: 10.1038/1831427a0. [DOI] [PubMed] [Google Scholar]