Abstract
A number of missense mutations in the Na,K-ATPase α2 catalytic subunit have been identified in familial hemiplegic migraine with aura. Two alleles (L764P and W887R) showed loss-of-function, whereas a third (T345A) is fully functional but with altered Na,K-ATPase kinetics. This study describes two additional mutants, R689Q and M731T, originally identified by Vanmolkot et al. [Vanmolkot, K. R., et al. (2003) Ann. Neurol. 54, 360-366], which we show here to also be functional and kinetically altered. Both mutants have reduced catalytic turnover and increased apparent affinity for extracellular K+. For both R689Q and M731T, sensitivity to vanadate inhibition is decreased, suggesting that the steady-state E1 ↔ E2 poise of the enzyme is shifted toward E1. Whereas the K′ATP is not affected by the R689Q replacement, the M731T mutant has an increase in apparent affinity for ATP. Analysis of the structural changes effected by T345A, R689Q, and M731T mutations, based on homologous replacements in the known crystal structure of the sarcoplasmic reticulum Ca-ATPase, provides insights into the molecular bases for the kinetic alterations. It is suggested that the disease phenotype is the consequence of lowered molecular activity of the α2 pump isoform due to either decreased K+ affinity (T345A) or catalytic turnover (R689Q and M731T), thus causing a delay in extracellular K+ clearance and/or altered localized Ca2+ handling/signaling secondary to reduced activity in colocalized Na+/Ca2+ exchange.
Keywords: sodium pump kinetics, missense mutations, ATP1A2 gene
The Na,K-ATPase catalyzes the ATP-driven exchange of three intracellular Na+ ions for two extracellular K+ ions across the plasma membrane of virtually all animal cells and is essential to the maintenance of the electrochemical alkali cation gradients that are dissipated by ion channels in the propagation of action potentials (for recent reviews, see refs. 1 and 2). The catalytic cycle of this P type ion pump involves phosphorylation and dephosphorylation of a conserved aspartate residue in its catalytic α subunit and conformational transitions of phospho- and dephosphoenzyme, commonly referred to as E1P ↔ E2P and E1 ↔ E2 conformational transitions, respectively (see Scheme 1). Four isoforms of α and three isoforms of β have been described thus far. All are distributed in a tissue- and developmentally dependent manner. In adult mammals, α2 is located principally in skeletal muscle and brain, in particular in glial cells, and, to a lesser extent, in heart, adipocytes, and the eye (see refs. 3-6).
Scheme 1.
Abbreviated Na,K-ATPase model. ATPL represents ATP binding with low affinity. EXT, extracellular; CYT, cytoplasmic.
The discovery of the association of missense mutations in the ATP1A2 gene on chromosome 1q23 that encodes the Na,K-ATPase α2 isoform with familial hemiplegic migraine (FHM) has been an important breakthrough in that it reflects a disease caused by a kinetically altered sodium pump and is therefore an important lead in migraine pathophysiology. Although a migraine is a common polygenic disorder, FHM is a rare autosomal dominant form of migraine with aura and is usually additionally associated with hemiparesis and other clinical features ranging from ataxia to epileptic seizures. Thus far, at least two subtypes of FHM have been described, those associated with missense mutations in the CACNA1A gene, encoding the α1 subunit of the voltage-dependent neuronal (P/Q type) calcium channel, which account for >50% of all FHM families (FHM1), and the more recent association of FHM2 with missense mutations of ATP1A2. Together, they suggest that the cause of FHM, and perhaps other forms of migraines, lies in a disruption of normal cation transport.
To date, there are at least nine well documented alleles of the α2 gene in families segregating FHM2, although several other candidate alleles, albeit from smaller families, await validation. No deletions, frameshifts, nonsense, or splice site alleles have been clearly identified thus far. In the case of the first two of the reported mutations, L764P and W887R, DeFusco et al. (7) suggested that the disease is the result of haploinsufficiency because the mutant enzyme does not support the growth of cells in culture. We have since shown that in contrast to cells bearing mutants L764P and W887R, those bearing the T345A replacement had normal growth but showed altered (decreased) K+ affinity, thus likely accounting for the disease (8).
In this study, we show that mutants R689Q and M731T first identified by Vanmolkot et al. (9), like T345A, are also functional enzymes that support cell growth. Kinetic alterations effected by these three replacements and their relevance to the disease mechanism are presented together with a model of the consequences of mutations of the analogous residues in the closely homologous regions of the sarcoplasmic reticulum Ca-ATPase (SERCA) crystal structures reported by Toyoshima et al. (10-13).
Materials and Methods
Mutagenesis, Transfection, and Cell Culture. The R689Q and M731T mutations of the rat α2 cDNA were derived from the ouabain-insensitive rat α2*∥ cDNA developed by Jewell and Lingrel (14), and mutagenesis was carried out as described in ref. 8. The pCDNA-α2 mutant constructs were transfected into HeLa cells. Cells expressing the relatively ouabain-resistant rat α2* enzymes were selected, amplified, and subsequently grown in culture medium containing 1 μM ouabain as described in refs. 14 and 15.
Membrane Preparation and Enzyme Assays. NaI-treated microsomal membranes were prepared as described in refs. 14 and 15. For the determination of maximal Na,K-ATPase activity, assays were carried out with (final concentrations) 1 mM ATP/100 mM NaCl/10 mM KCl/4 mM MgSO4/30 mM Tris·HCl, pH 7.4/5 mM EGTA, pH 7.4/5 μM ouabain. Components of Na,K-ATPase activity were measured as the release of 32Pi from [γ-32P]-ATP as described in ref. 16. Baseline hydrolysis was determined by using 5 mM ouabain. Catalytic turnover was estimated from the ratio of Vmax to maximal phosphoenzyme (EPmax) as in ref. 17, except that 50 mM NaCl (or KCl for baseline EP) was included during the room temperature incubation with oligomycin. Transport assays were carried out on cells grown in 24-well plates as described in ref. 18. Kinetic constants were determined by fitting the data to either a simple one-site Michaelis-Menten model (K′ATP), or a two-site (K0.5(K)) or 3-site (K0.5(Na)) cooperative model (v = Vmax[cat]n/(K + [cat]n) where n = 2 or 3 for the cation (cat), either K+ or Na+, respectively. Curve fitting was carried out by using kaleidagraph (Synergy Software, Reading, PA).
Polyacrylamide Gel Electrophoresis and Western Blotting. SDS/PAGE was carried out as described in ref. 8, and densitometry was performed in the linear range of sample addition and film exposure. α2* protein was immunoblotted with monoclonal antibodies (McB2), kindly provided by Kathleen Sweadner (Harvard Medical School Neuroscience Center, Charlestown, MA).
All measurements were carried out concurrently on mutant and control (WT) α2* enzymes, performing each assay in triplicate on at least two separate clones. Each value shown is the mean ± SD.
Sequence Alignment and Structural Analysis of Mutations Based on SERCA. The sequence of Na,K-ATPase α2 isoform (GenBank accession no. P50993) was manually aligned to SERCA according to Sweadner and Donnet (19). SERCA crystal structures relative to states E1.2Ca (PDB ID code 1SU4, ref. 10), E1-ATP.2Ca (PBD ID code 1VFP, ref. 12), E2-Pi.2Ca (PDB ID code 1WPG, ref. 13), and E2 (PDB ID code 1IWO, ref. 11) were used. These structures correspond, in the Na,K-ATPase catalytic cycle, to states E1.3Na, E1-ATP.3Na, E2-Pi.[2K] and E2[2K], respectively. For the α2* protein mutant T345A, the corresponding residue G322†† in SERCA was mutated into T, then into A, by using sybyl6.9.1 (Tripos Associates, St. Louis). For the mutant R689Q, the corresponding residue R678 was mutated to Q, and the loop 201-NQDK-204 was replaced by α2* loop 228-NPLET-232 by using the protein loops module implemented in sybyl6.9.1. For mutant M731T, the corresponding residue M720 was mutated to T. For each WT/mutant couple, the protein was subjected to energy minimization to a root-mean-square gradient of 0.01 kcal/(mol·Å) by using the amber all-atom force-field (20), a distance-dependent dielectric function (4rij) and an 8-Å nonbonded cutoff. Homology modeling of the entire protein was not necessary for the purposes of this study because of the high sequence conservation between Na,K-ATPase and SERCA around the regions of interest. These “locally homology-modeled” structures can suggest the structural implications of the point mutations of interest in the Na,K-ATPase.
Results
Growth Rates and α2* Subunit Expression. The full-length cDNA of the rat α2 enzyme containing the R689Q and M731T substitutions were cloned and introduced into WT HeLa cells. To distinguish exogenous rat α2 pumps from the endogenous HeLa enzyme, the Q116R and N127D mutations were introduced at the border of the first extracellular loop of the mutant enzymes to render them relatively ouabain insensitive. As in earlier studies, this ouabain-resistant WT control is referred to as α2*. Thus, cells expressing the exogenous pumps were selected by growth in culture medium containing low (1 μM) concentrations of ouabain. As mentioned in ref. 8 and shown in Fig. 1, cells expressing R689Q and M731T survive in 1 μM ouabain. Although Na pump activities of the mutant cells were generally lower in the clones expressing these mutations, particularly R689Q (see legends to Figs. 3C and 4), clone-to-clone variation in expression (≈2-fold between clones of the same WT or mutant pumps) may reflect gene dosage in the transformant rather than functional alterations effected by the residue replacements, per se.
Fig. 1.
Comparison of α2*- and mutant α2*-transfected HeLa cell growth rates and α2* protein expression. Growth curves for WT- and mutant-transfected HeLa cells were generated as described in Materials and Methods. Inset shows Western blot analysis for α2* protein expression for samples with the units [(μmol/mg per min) x 10-12] of Na,K-ATPase activity shown above each lane. Symbols and lines are as follows: ○, α2*; □, R689Q; ⋄, M731T.
Fig. 3.
ATP, Na+, and K+ activation of Na,K-ATPase activity. Data shown are means ± SD of the number of experiments shown in parentheses, each carried out in triplicate. Symbols are as in Fig. 1. (A) ATP hydrolysis was assayed in the presence of 100 mM NaCl/10 mM KCl/4 mM MgSO4 and varying ATP concentrations as described in Materials and Methods and normalized to 100% Vmax. Data were fitted to a one-site Michaelis-Menten model. K′ATP values were 158.3 ± 19.5 (6), 142.8 ± 21.3 (4), and 67.31 ± 15.1 μM (3) for α2*, R689Q, and M731T, respectively. P < 0.0001 for M731T compared with α2*. (B) Na+ activation of Na,K-ATPase was determined as described in Materials and Methods, maintaining a constant KCl concentration of 10 mM. The data were fitted to a three-site cooperative model and normalized to 100% Vmax. K0.5(Na) values were 5.43 ± 0.88 (10), 5.77 ± 1.43 (7), and 6.48 ± 1.46 mM (5) for α2*, R689Q, and M731T, respectively. P > 0.2 for R689Q and M731T. (C) K+ activation of Na,K-ATPase was determined as described in Materials and Methods, maintaining a constant NaCl concentration of 100 mM. Data were fitted to a two-site cooperative model and normalized to 100% vmax. K0.5(K) values were 1.69 ± 0.21 (6), 0.34 ± 0.01 (3), and 0.56 ± 0.06 mM (3) for α2*, R689Q, and M731T respectively. P < 0.0001 for R689Q and M731T, compared with α2*.
Fig. 4.
K+ activation of (86Rb+)K+ influx. Assays were carried out as described in Materials and Methods with cells equilibrated and assayed in the presence of 20 mM Na+, in medium containing varying concentrations of KCl as indicated. Data were fitted to a two-site cooperative model and normalized to 100% Vmax. K0.5(KEXT) values were 0.48 ± 0.11 (3), 0.25 ± 0.03 (3), and 0.27 ± 0.02 mM (3) for α2*, R689Q, and M731T respectively. P < 0.05 for R689Q and M731T, compared with α2*.
Western blot analysis of membranes isolated from the cells (Fig. 1 Inset) shows that density/unit activity is notably higher (5.6-fold) in membranes isolated from the R689Q- and M731T-transfected cells compared with WT α2* cells. Similar results were obtained with intact cells in which ouabain-sensitive
86Rb(K+) influx and α2* expression (Triton X-100 solubilized cells) were assayed concurrently (density/activity ratio ≥ 10-fold higher in the mutants, data not shown) raising the possibility that these mutations decrease catalytic turnover of the α2* pump.
Catalytic Turnover. To test whether catalytic turnover is indeed decreased, estimates of the ratio of maximal Na,K-ATPase activity (Vmax) to maximal phosphoenzyme (EPmax) were carried out by using permeable membranes; for the measurement of EPmax, oligomycin was added to trap the phosphoenzyme in the E1P state. Thus, the ratio of Vmax/EPmax in WT is 4,619 ± 426 min-1, similar to that reported in ref. 21 and is reduced to 2,042 ± 374 and 880 ± 247 min-1 for R689Q and M731T replacements, respectively.
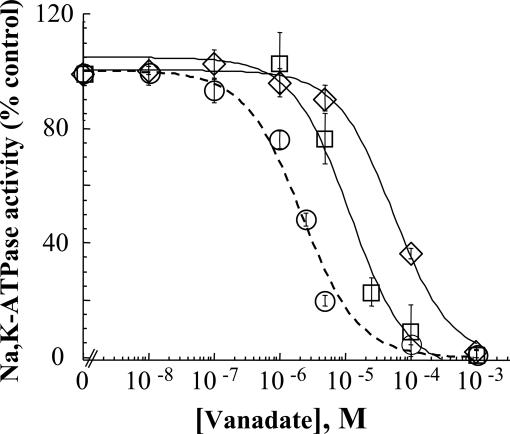
Effects on the E1/E2 Poise. To determine whether the reduced catalytic turnover reflects a shift in steady-state E1 ↔ E2 poise of enzyme intermediates, sensitivity of Na,K-ATPase to inhibition by vanadate was tested as in earlier studies (22). Inorganic orthovanadate is a transition state analog of inorganic phosphate that binds to the enzyme in the E2 conformation. Accordingly, sensitivity to vanadate inhibition is a measure of the E1/E2 conformational equilibrium. The representative experiment shown in Fig. 2 indicates that both mutations decrease sensitivity to vanadate inhibition. However, the effect of the R689Q replacement is notably less, i.e., the increase in I50 for R689Q is ≈6-fold compared with ≈25-fold for the M731T (see Table 1), suggesting a larger shift in E1 ↔ E2 poise in favor of E1 for M731T.
Fig. 2.
Vanadate sensitivity of α2*, R689Q, and M731T. ATP hydrolysis was determined in medium containing 100 mM NaCl, 10 mM KCl, and 1 mM ATP and with varying vanadate concentrations as described in Materials and Methods. Data presented as percent of the control Na,K-ATPase measured in the absence of vanadate were fitted to a one-compartment model by using a nonlinear least-square analysis of a general logistic function (23). Values shown are the mean ± SD of triplicate determinations from a representative experiment. I50 vanadate values obtained from replicate experiments with two clones were 2.12 ± 0.17, 13.17 ± 3.43, and 57.83 ± 11.61 μM for α2*, R689Q and M731T, respectively. Symbols are as in Fig. 1. For R689Q and M731T, P < 0.05, compared with α2.
Table 1. Effects of mutations on kinetic parameters.
| K′ ATP† | K0.5(Na)† | K0.5(K)† | K0.5(KEXT)‡ | l50† (vanadate) | Catalytic turnover† (Vmax/EPmax) | |
|---|---|---|---|---|---|---|
| α1 | 3.0(24) | 1.04(14) | 1.05(14) | 1.10(25) | 0.03(17) | 1.67(17) |
| α2* | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| T345A | 0.30 | 1.11 | 1.92 | 1.79 | 3.13 | 1.00 |
| R689Q | 0.90 | 1.06 | 0.21 | 0.52 | 6.22 | 0.44 |
| M731T | 0.42 | 1.19 | 0.33 | 0.57 | 27.3 | 0.19 |
Values shown are normalized to a value of 1.0 for the WT α2* enzyme. Numbers in superscript refer to references from which the data (shown in italics) for α1 were taken.
Kinetic parameters are from assays of Na,K-ATPase hydrolytic activity.
Kinetic parameters is from the assay of ouabain-sensitive K+ influx, whereby K0.5(KEXT) refers to the K0.5 for extracellular K+.
Apparent ATP Affinity. Fig. 3A shows that the M731T mutant has a 3-fold lower K′ATP for low affinity ATP binding, presumably in the E2(K) + ATP → ATP.E2.K step, compared with α2*. In contrast, the R689Q replacement does not alter K′ATP.
Na+- and K+-Activation Kinetics. The experiments shown in Fig. 3 B and C describe the Na+- and K+-dependent activation profiles of Na,K-ATPase activity of the R689Q and M731T mutants compared with WT α2*. The results indicate that the two mutations have little, if any, effect on apparent Na+ affinity (Fig. 3B), whereas they increase K+ affinity >3-fold (Fig. 3C).
The increase in apparent affinity for extracellular K+ is also seen under the more physiological conditions prevailing in intact cells. Thus, assays of ouabain-sensitive 86Rb(K+) influx into intact cells equilibrated with 20 mM NaCl (Fig. 4) indicate that both mutations cause a 2-fold decrease in K0.5(K). A similar 2-fold decrease in K0.5(K) was seen at 80 mM Na+ (data not shown).
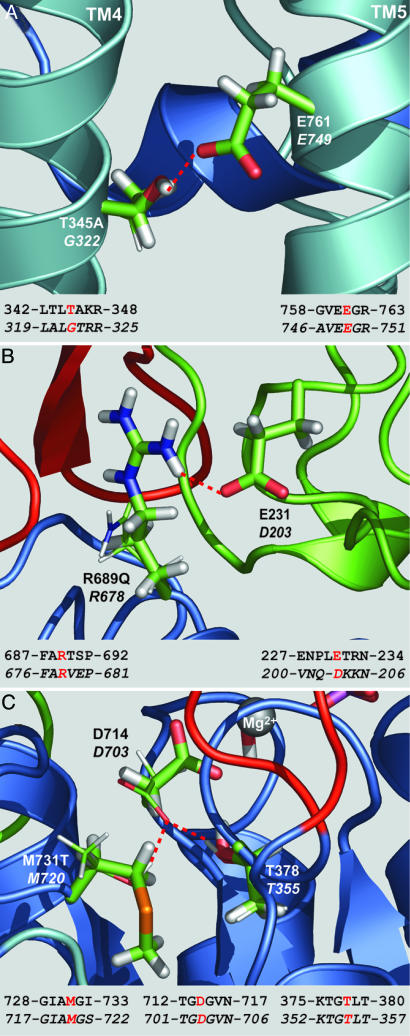
Structural Changes Effected by T345A, R689Q, and M731T Mutations Based on the Homologous Replacements in SERCA. In all four states in which crystal structures are available, SERCA residue G322 of TM4 faces E749 (E761 in α2) of TM5, and water, likely present in the ion channel, can hydrate E749. In the WT α2* protein, residue T345, corresponding to SERCA G322, does not allow water to approach E761 but rather is hydrogen-bonded to E761 Oε 1 (2.9 Å, 123°), thus preserving an electrostatically favorable environment (Fig. 5A). In the mutant T345A, the side-chain methyl group, not a hydrogen bond donor, cannot form a hydrogen bond with E761 Oε 1 and, because of its size and hydrophobicity, additionally prevents water from approaching E761 (Fig. 5A).
Fig. 5.
Structural analysis of the mutants by comparison with SERCA. Color convention is in agreement with Sweadner and Donnet (19): A domain in green, P domain in blue, N domain in red, and TMs in pale blue. Residues of the WT enzyme are in stick representation. The mutated residues and the WT residues that change conformation upon mutation are displayed as thin sticks. Na,K-ATPase and SERCA corresponding residues of interest are labeled, the latter in italics. Below each panel, the local alignment is shown with the displayed residues in red and SERCA sequence in italics. (A) T345A mutant (SERCA E2 state, PDB ID code 1IWO, ref. 11). (B) R689Q mutant (SERCA E2-Pi.2Ca state, PDB ID code 1WPG, ref. 12). (C) M731T mutant (SERCA E1-ATP.2Ca state, PDB ID code 1VFP, ref. 13). Red dashed lines indicate hydrogen bonds. Figure designed with pymol 0.93(DeLano Scientific, South San Francisco, CA).
In SERCA state E1-ATP.2Ca (PDB ID code 1VFP, ref. 12) R678 forms a hydrogen bond with O3′ of the ribose of ATP analog AMPPCP through side chain Nη 1H (3.0 Å, 136°). This hydrogen bond is likely present in the corresponding state E1.ATP.3Na of the WT α2* protein but not in the mutant R689Q, where the side chain of the glutamine is too short (Fig. 5B). More importantly, from the analysis of SERCA E2-Pi.2Ca structure (PDB ID code 1WPG, ref. 13), R678 (P domain, R689 in α2) may be involved in the docking of the A on to the P domain of SERCA by forming hydrogen bonds with Oδ2of D203 (3.2 Å, 141°) and with backbone carbonyl of Q202 (3.3 Å, 134°), both part of the A domain. Such bonds are not present in the other forms of the enzyme whose structures are available because of the larger distance between the A and P domains. By modeling α2 loop 228-NPLET-232 in place of loop 201-NQDK-204, we verified that one hydrogen bond between R689 (P domain) and E231 (corresponding to D203, A domain) is present (3.0 Å, 142°) (Fig. 5B).
By analogy with SERCA E1-ATP.2Ca state (PBD ID code 1VFP, ref. 12), in the mutant M731T (M720 in SERCA), the D714 side chain may rotate toward T731 (torsion angle NH-Cα-Cβ-Cγ passes from 63° to 270°) to form two hydrogen bonds, one between T731 Oγ1-H and D714 Oδ2 (2.7 Å, 166°) and the other between T378 (T355) Oγ1-H and D714 Oδ2 (2.8 Å, 162°) (Fig. 5C). These two hydrogen bonds compete with Mg2+ in the sharing of one lone pair of D714 Oδ2, thus likely affecting Mg2+ binding and catalysis.
Discussion
At least nine families with missense mutations in the ATP1A2 gene, which encodes the α2 isoform of the Na,K-ATPase, have been identified thus far. Four (T345A, R689Q, M731T, and L759P) are located in the catalytic loop between M4 and M5. The initial finding that two alleles, L764P and W887R, lead to expression of nonfunctional protein was based on their inability to support growth of HeLa cells (7). Whereas the disease phenotype of these two alleles was attributed to haploinsufficiency, missense replacements R689Q and M731T, like T345A described in ref. 8, result in active pump enzymes. Thus, the latter three all support growth of the analogous rat mutant α2* pump transformants after transfection into HeLa cells whose endogenous pump activity was suppressed by growth in ouabain. A comparison of the kinetic characteristics of the three mutants to those of the WT α2* enzyme are presented in Table 1. As noted in Table 1, compared with the ubiquitous α1 isoform, α2* has a higher apparent ATP affinity, a lower catalytic turnover, and a 25-fold lower sensitivity to vanadate inhibition, suggesting (in addition to the higher ATP affinity) that its E1 ↔ E2 conformational poise is E1 shifted (see refs. 17 and 21). The higher affinity for ATP and maintenance of activity under acidic conditions (17) may be important for maintaining pump function under energy-compromised conditions, particularly anoxia.
Structural Alterations Effected by the T345A, R689Q, and M731T Replacements and Their Impact on the Kinetic Behavior of α2. The notable alteration effected by the T345A mutation is the decrease in apparent affinity for K+ (8). In that report, we argued that increased hydrophobicity created by the T345A replacement may interfere with the normal displacement of M4, thereby weakening the coordination of K+ ions at “site II” (26) and, consequently, decrease the intrinsic binding affinity for extracellular K+. As depicted in Fig. 5A, the SERCA structure that was used for structural comparison shows that the hydrogen bond between T345 and E761 is missing when A345 replaces T345, thus altering the K+ entry path involving the cytoplasmic stalk regions of the M4 and M5 helices.
In contrast to the alterations effected by the T345A mutation, the R689Q and M731T replacements cause a decrease in the catalytic turnover and increase the apparent affinity for K+, with either no change or increased ATP affinity caused by the R689Q and M731T mutations, respectively.
In the case of R689Q, it is evident from Fig. 5B that the R → Q replacement has an important effect on the structural transitions involving interactions between the phosphorylation (P), Actuator (A), and nucleotide binding (N) domains, which likely have long-range effects on the enzyme's catalytic turnover and apparent K+ affinity as discussed below.
As seen in Fig. 5C, in the T731 mutant, an important competition between Mg2+, T731 Oγ1-H, and T378 Oγ1-H for the lone pair of D714 Oδ2 would be expected to decrease the Mg2+ affinity for Mg2+-dependent phosphoryl transfer reactions and, thus, decrease overall catalytic turnover. (For a discussion of the role of Mg2+ in the Na,K-ATPase reaction steps, see ref. 27 and reviews by Jorgensen and coworkers (2, 28).
Interestingly, there is a relevant paradigm in the work of Jorgensen et al. (28), whose mutational analysis of residues in the N and P domains indicates that structural changes of various residues important for N/A/P domain interactions have diverse long-range consequences for ligand interactions. Thus, mutations of residues in pig α1 corresponding to D709A and N712A in rat α2 shift the conformation toward E2, yet the former, but not latter, mutation alters occlusion of the K+ congener Tl+. In an analogous way, the complexity of long-range effects of the α2 mutations analyzed in this study are underscored by several findings (see Table 1), namely: (i) the distinct changes in catalytic turnover (decreased for R689Q and M731T, but not T345A) and (ii) the different extents of conformational shifts toward E1, evidenced by the decreased sensitivity to vanadate inhibition (greatest for M731T and least for T345A), yet (iii) the increase in ATP affinity seen with T345A and M731T, but not R689Q, whereas (iv) K0.5(K) is decreased for R689Q and M731T, but increased for T345A. Interestingly, none of the alleles appear to alter the sodium affinity of the pump.
How Do the Functional Changes Explain the FHM2 Disease Phenotype? Because the three mutant enzymes T345A, R689Q, and M731T are all functional, but kinetically altered, it is evident that the ouabain-resistant growth phenotype, per se, is no longer a valid measure of allele pathogenicity. As discussed in ref. 8, a decrease in K+ affinity and, thus, raised extracellular K+ may explain the cortical spreading depression that could be the basis for the disease phenotype caused by T345A (for a comprehensive review of spreading depression, see ref. 29). It is also plausible that excessive clearing of extracellular K+ caused by a higher apparent affinity of the mutant R689Q and M731T pumps for K+ can induce spontaneously appearing waves of spreading depression as shown by Dahlem et al. (30). Those authors showed that the more the extracellular K+ concentration differed from the physiological level, the greater the number of spontaneous spreading depression waves in a given period. Perhaps such aberrant K+ handling described herein may be the basis for FHM, or diverse symptoms associated with FHM2 seen in the different alleles, for example, benign familial infantile convulsions in the R689Q family (9).
Overall, functional studies of mutations linked with FHM2 suggest that the functional alteration common to all of the five alleles analyzed to date is the reduction of pump activity. Reduction of activity is thus the result of either haploinsufficency, as appears to be the case for the L764P and W887R mutations, decreased affinity for extracellular K+, as seen with the T345A mutation, or decreased catalytic turnover, as seen with the R689Q and M731T mutants. In the cases of R689Q and M731T, it is also plausible that their lower catalytic turnover overrides the effect of the increase in K+ affinity, at least in the range of elevated K+ concentrations present extracellularly during the depolarization phase of the action potential, with the result that extracellular K+ clearance is, in fact, slowed. Although this study does not specifically address the possibility that R689Q and M731T mutations alter trafficking to the cell surface, there is no indication of a reduction in the fraction of mutant and WT pumps associated with the cell surface, i.e., the ratio Vmax(pump)/Vmax(Na,K-ATPase) is not reduced by either mutation (see legends to Figs. 3C and 4). On the other hand, the decrease in ratio of pump activity/Na,K-ATPase protein caused by both mutations (≥10-fold decrease as mentioned in Results) is greater than the decrease in catalytic turnover, which raises the possibility that these mutations affect protein stability. Future studies should address the issue of altered trafficking and/or stability of the R689Q and M731T mutant pumps.
At face value, it is difficult to explain the FHM2 disease phenotype, given the evidence that the α2 isoform constitutes a relatively minor component of the total Na,K-ATPase isoforms in the brain, e.g., ≈20% α2 and ≈80% α1 in astrocytes. In terms of activity, the contribution of α2 is even lower (≈10%), given the ≈50% lower catalytic turnover of α2 compared with α1 (17, 31). In fact, in the heterozygote α2 knockout mouse, bulk intracellular Na+ in the astrocyte is not altered (32). The most likely explanation for an important role of α2 in the function of cells of the neuromuscular system is that they are confined to microdomains of the plasma membrane, leading to changes in cation concentrations in localized regions at the mouth of ion channels and exchangers, as first shown in the immunocytochemistry studies of Juhasova and Blaustein (33). Colocalization of the α2 Na,K pump and the plasma membrane Na/Ca exchanger provides an attractive basis for proposing that a common functional change leads to FHM caused by either a missense mutation in the CACNA1A gene that encodes the α1 subunit of the voltage-dependent neuronal (P/Q) type calcium channel (FHM1) or the Na,K-ATPase α2 pump (FHM2). Both would lead to elevated intracellular Ca2+ in a critical microdomain involved in calcium signaling. In the case of the α2 missense mutations, the reduction in pump activity would raise the local intracellular Na+ concentration which, in turn, would lead to an increase in local Ca2+ entry and replenishment of the endoplasmsic reticulum calcium store. More recently, Golovina et al. (32) provided evidence in support of the notion that this localized increase in Ca2+ occurs adjacent to the ER Ca2+ stores. As a result, Ca2+ signaling is elevated without any change in bulk intracellular Na+, as proposed by Juhasova and Blaustein (33). In support of this notion is a recent study comparing immortalized optic nerve astrocytes derived from Na,K-ATPase α2 homozygous knockout mice with those of WT littermates (34). The results provided strong evidence in support of a critical role of α2, but not α1, in calcium signaling and capacitative calcium entry in these cells. It remains to be determined whether astrocytes derived from animals engineered to have FHM2 missense mutations have similarly altered Ca2+ responses.
Another perspective of the importance of such microdomains in pathogenesis is hinted at by the LQT4 alleles of ankyrin B and the surprising finding that defects in a protein without transporter function can produce the Long QT channelopathy syndrome, apparently solely because such assemblies of transporters and channels such as the Na,K pump, Na/Ca exchanger and, in that case, inositol trisphosphate receptor, are disrupted (35, 36).
Acknowledgments
We thank Drs. Jerry B. Lingrel (University of Cincinnati College of Medicine, Cincinnati) for the α2* cDNA and Kathleen Sweadner for the gift of McB2 monoclonal antibodies. This work was supported by Canadian Institutes for Health Research Grant MT-3876 (to R.B.). This work is National Research Council of Canada publication no. 47473.
Author contributions: L.S. and R.B. designed research; L.S. and R.S. performed research; A.M. and E.P. contributed new reagents/analytic tools; A.M., R.S., and E.P. analyzed data; and L.S., J.J.G., and R.B. wrote the paper.
Abbreviation: FHM, familial hemiplegic migraine.
Footnotes
α2* denotes the relatively ouabain-insensitive rat α2 enzyme (14).
Residues in italics refer to residues in SERCA homologous to those in the Na, K-ATPase α2.
References
- 1.Kaplan, J. H. (2002) Annu. Rev. Biochem. 71, 511-535. [DOI] [PubMed] [Google Scholar]
- 2.Jorgensen, P. L., Hakansson, K. O. H. & Karlish, S. J. D. (2003) Ann. Rev. Physiol. 65, 817-849. [DOI] [PubMed] [Google Scholar]
- 3.Sweadner, K. J. (1989) Biochim. Biophys. Acta 988, 185-220. [DOI] [PubMed] [Google Scholar]
- 4.Moseley, A. E., Dean, W. L. & Delamere, N. A. (1996) Invest. Ophthalmol. Visual Sci. 37, 1502-1508. [PubMed] [Google Scholar]
- 5.Orlowski, J. & Lingrel, J. B. (1988) J. Biol. Chem. 263, 10436-10442. [PubMed] [Google Scholar]
- 6.Wetzel, R. K. & Sweadner, K. J. (2001) Invest. Ophthalmol. Visual Sci. 42, 763-769. [PubMed] [Google Scholar]
- 7.De Fusco, M., Marconi, R., Silvestri, L., Atorino, L., Rampoldi, L., Morgante, L., Ballabio, A., Aridon, P. & Casari, G. (2003) Nat. Genet. 33, 192-196. [DOI] [PubMed] [Google Scholar]
- 8.Segall, L., Scanzano, R., Kaunisto, M. A., Wessman, M., Palotie, A., Gargus, J. J. & Blostein, R. (2004) J. Biol. Chem. 279, 43692-43696. [DOI] [PubMed] [Google Scholar]
- 9.Vanmolkot, K. R., Kors, E. E., Hottenga, J. J., Terwindt, G. M., Haan, J., Hoefnagels, W. A., Black, D. F., Sandkuijl, L. A., Frants, R. R., Ferrari, M. D. & van den Maagdenberg, A. M. (2003) Ann. Neurol. 54, 360-366. [DOI] [PubMed] [Google Scholar]
- 10.Toyoshima, C., Nakasako, M., Nomura, H. & Ogawa, H. (2000) Nature 405, 647-655. [DOI] [PubMed] [Google Scholar]
- 11.Toyoshima, C. & Nomura, H. (2002) Nature 418, 605-611. [DOI] [PubMed] [Google Scholar]
- 12.Toyoshima, C. & Mizutani, T. (2004) Nature 430, 529-535. [DOI] [PubMed] [Google Scholar]
- 13.Toyoshima, C., Nomura, H. & Tsudo, R. (2004) Nature 432, 361-368. [DOI] [PubMed] [Google Scholar]
- 14.Jewell, E. A. & Lingrel, J. B. (1991) J. Biol. Chem. 266, 16925-16930. [PubMed] [Google Scholar]
- 15.Lane, L. K., Feldmann, J. M., Flarsheim, C. E. & Rybczynski, C. L. (1993) J. Biol. Chem. 268, 17930-17934. [PubMed] [Google Scholar]
- 16.Segall, L., Lane, L. K. & Blostein, R. (2002) J. Biol. Chem. 277, 35202-35209. [DOI] [PubMed] [Google Scholar]
- 17.Segall, L., Daly, S. E. & Blostein, R. (2001) J. Biol. Chem. 276, 31535-31541. [DOI] [PubMed] [Google Scholar]
- 18.Garty, H., Lindzen, M., Scanzano, R., Aizman, R., Fuzesi, M., Goldshleger, R., Farman, N., Blostein, R. & Karlish, S. J. (2002) Am. J. Physiol. 283, F607-F615. [DOI] [PubMed] [Google Scholar]
- 19.Sweadner, K. J. & Donnet, C. (2001) Biochem. J. 356, 685-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiner, S. J., Kollman, P. A., Nguyen, D. T. & Case, D. A. (1986) J. Comput. Chem. 7, 230-252. [DOI] [PubMed] [Google Scholar]
- 21.Daly, S. E., Lane, L. K. & Blostein, R. (1996) J. Biol. Chem. 271, 23683-23689. [DOI] [PubMed] [Google Scholar]
- 22.Boxenbaum, N., Daly, S. E., Javaid, Z. Z., Lane, L. K. & Blostein, R. (1998) J. Biol. Chem. 273, 23086-23092. [DOI] [PubMed] [Google Scholar]
- 23.DeLean, A., Munson, P. J. & Rodbard, D. (1978) Am. J. Physiol. 235, E97-E102. [DOI] [PubMed] [Google Scholar]
- 24.Daly, S. E., Lane, L. K. & Blostein, R. (1994) J. Biol. Chem. 269, 23944-23948. [PubMed] [Google Scholar]
- 25.Munzer, J. S., Daly, S. E., Jewell-Motz, E. A., Lingrel, J. B. & Blostein, R. (1994) J. Biol. Chem. 269, 16668-16676. [PubMed] [Google Scholar]
- 26.Ogawa, H. & Toyoshima, C. (2002) Proc. Natl. Acad. Sci. USA 99, 15977-15982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patchornik, G., Munson, K., Goldshleger, R., Shainskaya, A., Sachs, G. & Karlish, S. J. D. (2002) Biochemistry 41, 11740-11749. [DOI] [PubMed] [Google Scholar]
- 28.Jorgensen, P. L. (2001) J. Bioenerg. Biomembr. 33, 367-377. [DOI] [PubMed] [Google Scholar]
- 29.Somjen, G. G. (2001) Physiol. Rev. 81, 1065-1096. [DOI] [PubMed] [Google Scholar]
- 30.Dahlem, Y. A., Dahlem, M. A., Mair, T., Braun, K. & Muller, S. C. (2003) Exp. Brain Res. 152, 221-228. [DOI] [PubMed] [Google Scholar]
- 31.Crambert, G., Hasler, U., Beggah, A. T., Yu, C., Modyanov, N. N., Horisberger, J. D., Lelievre, L. & Geering, K. (2000) J. Biol. Chem. 275, 1976-1986. [DOI] [PubMed] [Google Scholar]
- 32.Golovina, V. A., Song, H., James, P. F., Lingrel, J. B. & Blaustein, M. P. (2003) Am. J. Physiol. 284, C475-486. [DOI] [PubMed] [Google Scholar]
- 33.Juhaszova, M. & Blaustein, M. P. (1997) Ann. N.Y. Acad. Sci. 834, 524-536. [DOI] [PubMed] [Google Scholar]
- 34.Hartford, A. K., Messer, M. L., Moseley, A. E. & Delamere, N. A. (2004) Glia 45, 229-237. [DOI] [PubMed] [Google Scholar]
- 35.Mohler, P. J., Splawski, I., Napolitano, C., Bottelli, G., Sharpe, L., Timothy, K., Priori, S. G., Keating, M. T. & Bennett, V. (2004) Proc. Natl. Acad. Sci. USA 101, 9137-9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohler, P. J., Schott, J. J., Gramolini, A. O., Dilly, K. W., Guatimosim, S., duBell, W. H., Song, L. S., Haurogne, K., Kyndt, F., Ali, M. E., et al. (2003) Nature 421, 634-639. [DOI] [PubMed] [Google Scholar]