Abstract
Because of the extreme impact of genome sequencing projects, protein sequences without accompanying experimental data now dominate public databases. Homology searches, by providing an opportunity to transfer functional information between related proteins, have become the de facto way to address this. Although a single, well annotated, close relationship will often facilitate sufficient annotation, this situation is not always the case, particularly if mutations are present in important functional residues. When only distant relationships are available, the transfer of function information is more tenuous, and the likelihood of encountering several well annotated proteins with different functions is increased. The consequence for a researcher is a range of candidate functions with little way of knowing which, if any, are correct. Here, we address the problem directly by introducing a computational approach to accurately identify and segregate related proteins into those with a functional similarity and those where function differs. This approach should find a wide range of applications, including the interpretation of genomics/proteomics data and the prioritization of targets for high-throughput structure determination. The method is generic, but here we concentrate on enzymes and apply high-quality catalytic site data. In addition to providing a series of comprehensive benchmarks to show the overall performance of our approach, we illustrate its utility with specific examples that include the correct identification of haptoglobin as a nonenzymatic relative of trypsin, discrimination of acid-d-amino acid ligases from a much larger ligase pool, and the successful annotation of BioH, a structural genomics target.
Keywords: enzymes, function prediction, EC, psi-blast
Assigning function to protein sequences continues to be of key importance (1). Currently, most approaches to protein function prediction rely on searching sequence databases to identify homologous sequences with prior annotation. The most widely used search tools are blast and psi-blast (2); at the National Center for Biotechnology Information alone, >70,000 blast searches are performed each day for the general public. It is certainly no coincidence that the blast algorithm was the most highly cited paper of the last decade, surpassing all biology publications (3). psi-blast is an iterative method that uses results from a blast search to create a profile (position-specific scoring matrix). The profile is used to search the database for additional homologues, and these results can be used to further improve the profile. A profile captures family-specific information, including functionally and structurally important residue positions, and can therefore identify distant homologues not recognized by alignment to a single sequence.
psi-blast and powerful fold recognition methods such as genthreader (4) are good at identifying distantly related proteins, even down to 10% sequence identity. However, recent studies have shown that, simply on the basis of overall similarity, it is generally impossible to infer the function of one protein from another below 40% sequence identity (5). More pessimistically, a study by Tian and Skolnick (6) found that precise function diverges below identities of 60%, which decreases the value of iterative database search methods because confident functional assignment cannot be achieved.
In this way, the utility of popular curated databases such as Pfam (7), CDD (8), PRINTS (9), and PROSITE (10) is restricted by the ability to correlate protein relationships with a similarity in function. This situation is particularly severe for methods that use profiles to perform the search stage, because they regularly find relationships below 40% sequence identity. More recently, the electronic transfer of Gene Ontology (11) terms onto proteins of known structure has been investigated (12). Although the overall approach appears promising, it is likely to face the same generic problems. As methods develop to recognize ever more distant homologues, the requirement for appropriate strategies for functional assignment becomes critical.
One indication of the scale of the problem is that 39% of the sequences in the nonredundant sequence database (13) do not even have a Swiss-Prot (14) homologue from which to infer function above 40% sequence identity. Furthermore, although function is best differentiated at the domain level, the majority of proteins in Swiss-Prot have multiple domains that can occur in many different combinations, which means that a simplistic approach of transferring information from one multidomain protein to another will inevitably lead to erroneous annotation (15). A typical example to illustrate this problem is the Swiss-Prot entry Q12792, twinfilin 1/protein tyrosine kinase 9, which has no kinase domain.
A further example is our own annotation of nicastrin, a protein in the Alzheimer's related complex, where we identified a clear relationship to the aminopeptidase/transferrin receptor superfamily (16). The problem in that instance was determining whether nicastrin was likely to be an aminopeptidase-like enzyme or merely a peptide-binding protein in the transferrin receptor mold. The absence of critical zinc-binding residues led us to conclude that nicastrin is not enzymatic. Making incorrect functional predictions can have far-reaching consequences in terms of erroneous and costly experimental validation (17) and proliferation of incorrect assignments in the sequence databases (18).
Identifying conserved residues can be useful in assigning function to previously uncharacterized sequences. Residues that are important for the structure and function of a protein are conserved through evolution. Many groups are working on ways to identify these residues in an attempt to assign function. For example, evolutionary trace methods work by identifying conserved residues in branches of phylogenetic trees (19–21). Our work differs in that we begin with the knowledge of which residues in a protein are important for its function.
In this study, we have analyzed the extent to which enzymes with equivalent function can be correctly identified amongst homologues well below the 40% sequence identity threshold by using knowledge of catalytic residue data. The key to our approach is to filter homologues detected by using psi-blast searches into two sets: those with conserved function and those where function is expected to differ. Only homologues with a completely conserved catalytic site are considered to be functionally equivalent. Data on catalytic residues are taken from >480 functional sites in the Catalytic Site Atlas (CSA) (22) (www.ebi.ac.uk/thornton-srv/databases/CSA). These data have been carefully derived from experimental tertiary structures in the Protein Data Bank (23) (PDB) and combined with associated literature to provide a unique resource. Entries in the CSA have broad coverage and comprise 58% of all current three-digit EC numbers (24). Our filtering process is general and automated and will benefit from ongoing CSA curation efforts. We will refer to the complete filtering process as the iCSA (Inpharmatica CSA) filter.
Methods
The iCSA Filter. Each enzyme in the CSA is used as a query to retrieve sequence homologues over four psi-blast iterations. The parameter settings for psi-blast are an E value (e) for hit acceptance of 10–3, an E value for inclusion of a hit into the next profile (h) of 3 × 10–3 and a fixed search space (Y) of 9 × 10–9. These parameters are in line with the recommended approaches for running psi-blast (25). These homologues are extracted from release 14 of the Biopendium [a proteome-scale annotation resource (26)].
The iCSA filter then realigns each homologue with its query sequence and checks the known catalytic site positions for conservation of residue type. However, because it is now critical that the small number of catalytic positions is aligned accurately, we use a number of different alignment methods (where necessary) to overcome the deficiencies in any individual approach. So, in detail, we start by using psi-blast and assess whether the required catalytic residues have been conserved. In cases where the residues are not found to be conserved, we repeat the process by using clustalw (27) and, failing that, Smith–Waterman (28). If all three methods produce a negative result, we conclude that the homologue either is a nonenzyme or operates by a different mechanism.
A functional assignment is made when a homologue is found to conserve all catalytic residues. In cases where the main chain of a residue has been annotated as the functional group instead of the side chain, then any residue is allowed to match the query residue unless the query residue is glycine (glycine residues have to align to glycine residues to be accepted). Any homologues that do not conserve all of the site residues are recorded as being homologues with a potentially different function. Various manual checks of the homologues have been made, on both those assigned a function by iCSA and those not, to verify the automatic procedures being used.
Benchmarking the iCSA Filter by Using Swiss-Prot/EC Data. To benchmark our approach, we first required independently assigned function data for a wide range of sequences. For this purpose, we used proteins with EC assignments from the well annotated Swiss-Prot sequence database, now incorporated into UniProt (29). We assume that these sequences have correct EC annotations (although later we shall highlight some exceptions). Some EC numbers are partially complete; for example, the enzyme class is known, but a subclass has not been assigned and is denoted by a dash instead of a number. Any sequence with an EC number that was not complete to the third digit or had the term “probable” or “putative” in its annotation was removed from the sequence test set.
Function assignments were deemed to be in agreement when the first three levels of the EC hierarchy (e.g., EC 1.1.1) assigned by our iCSA filter matched those recorded in the Swiss-Prot entry. The fourth level of the EC hierarchy is often used to describe substrate specificity; curated sites in the CSA describe purely catalytic residues and not those responsible for substrate binding.
We used the literature-annotated CSA enzymes as queries in psi-blast searches of the Swiss-Prot database. Homologues that were accepted by the iCSA filter were assigned the EC number of the query, and this EC number was compared with the annotation in the Swiss-Prot database. To compare the iCSA filter with sequence homology alone, all Swiss-Prot homologues were given the EC number of the query, regardless of whether catalytic residues were found to be conserved, and again compared with the Swiss-Prot annotation. In both cases, we calculated what proportion of the EC numbers assigned was correct (i.e., agreed with the Swiss-Prot annotation).
Results
The results for the iCSA filter are presented in Tables 1 and 2, and a measure of the improvement seen with the iCSA filter is provided in Fig. 1. As mentioned in Methods, it is important to remember that although the iCSA filter categorizes homology-based hits into two separate groups (essentially conserved function and nonconserved function), the simplistic homology search generates just a single group of hits. For the purposes of this work, we have assumed that all hits from the simplistic homology search would be given the same function as the query, in line with the way a researcher would typically transfer function information between proteins.
Table 1. Accuracy of EC prediction by iCSA filter.
| PSI-BLAST iteration | Swiss-Prot homologues identified by 482 queries | Swiss-Prot homologues with same EC as query | Total EC predictions using iCSA filter | Correct EC predictions using iCSA filter | Accuracy of EC prediction by iCSA filter, % |
|---|---|---|---|---|---|
| 1 | 27,072 | 22,941 | 19,024 | 17,335 | 91.1 |
| 2 | 11,567 | 4,386 | 1,844 | 1,048 | 56.8 |
| 3 | 9,471 | 2,738 | 395 | 185 | 46.8 |
| 4 | 7,439 | 2,254 | 176 | 105 | 59.7 |
| Overall | 55,549 | 32,319 | 21,439 | 18,673 | 87.1 |
Results are shown for the iCSA prediction of EC number for Swiss-Prot homologues detected at each PSI-BLAST iteration, up to four, using 482 nonredundant CSA enzymes as queries. “Overall” figures use the results from all four interations in one calculation. EC predictions are deemed correct if they match to the third digit of the EC annotation in the Swiss-Prot database. Accuracy is calculated using the number of correct predictions divided by the number of predictions made. (Note that each Swiss-Prot may occur more than once because it may have been identified by separate queries. The totals include all predictions in order to provide an unbiased calculation of accuracy.)
Table 2. Coverage of EC prediction by iCSA filter.
| Swiss-Prot homologues
|
||||
|---|---|---|---|---|
| psi-blast iteration | Identified by 482 queries | With same EC as query | With same EC as query and correct iCSA filter assignment | Coverage, % |
| 1 | 17,450 | 15,966 | 13,792 | 86.4 |
| 2 | 3,882 | 2,029 | 764 | 37.7 |
| 3 | 1,483 | 657 | 125 | 19.0 |
| 4 | 961 | 388 | 62 | 16.0 |
| Overall | 23,776 | 19,040 | 14,743 | 77.4 |
Results are shown for the iCSA prediction of EC number for Swiss-Prot homologues detected at each psi-blast iteration, up to four, using 482 nonredundant CSA enzymes as queries. “Overall” figures use the results from all four iterations in one calculation. EC predictions are deemed correct if they match to the third digit of the EC annotation in the Swiss-Prot database. Coverage is calculated as the number of homologues correctly assigned the same EC by iCSA divided by the number with the same EC as the queries. The figures given are for a nonredundant set of Swiss-Prot homologues (i.e., each Swiss-Prot appears only once because we need to record only whether an assignment has or has not been made in order to calculate coverage).
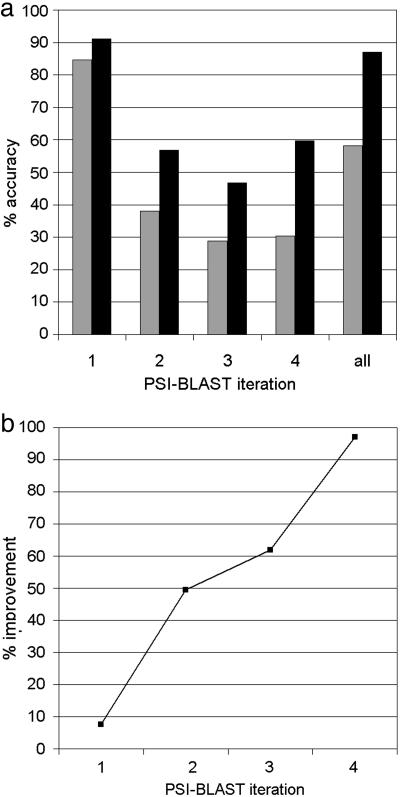
Fig. 1.
Improvement in function assignment accuracy of iCSA compared with sequence homology alone. (a) Results are shown for the accuracy of EC assignment to Swiss-Prot homologues obtained by using the iCSA filter (black bars) and compared, at each psi-blast iteration up to four, with results obtained by using just psi-blast (gray bars). (b) Percentage improvement in function assignment accuracy with iCSA compared with psi-blast only.
Fig. 1 shows that when an EC assignment is made by the iCSA filter, it is more likely to be correct than an assignment using sequence homology alone. The improvement is 8% at the first iteration of psi-blast and rises to 50% at the second iteration, 62% at the third iteration, and 97% at the fourth iteration. Importantly, the benefits of using the iCSA filter become more significant as the homologues identified become more distant.
In theory, an improvement in assignment accuracy (selectivity) could be achieved without any decrease in coverage (sensitivity), because all proteins with the requisite catalytic site would be accepted. In practice, however, the iCSA filter is limited by its reliance on an alignment that is correct over all of the key catalytic residues. We therefore decided to look at what proportion (coverage) of the Swiss-Prot homologues with the same three-digit EC number as the CSA queries was correctly identified by the iCSA filter (Table 2).
After four iterations of psi-blast and at the third level of EC, the simplistic homology search made the correct assignment of EC only 58% of the time. In contrast, the iCSA filter achieved 87% success while still covering 77% of the total set. Because the simplistic homology search can make only positive assignments, it successfully found all of the real enzymes. However, it also found many other proteins. Because it is unable to distinguish between these two sets, its application to real prediction work is restricted. In contrast, the iCSA filter neatly segregates the results into two pools. Of the 23% of homologues classified as having an alternative function by the iCSA filter, more than half (58%) actually have a different function; the remainder have alternative catalytic sites not yet recorded in the CSA database, misaligned sequences, or an incorrect EC assignment in Swiss-Prot. For example, Swiss-Prot protein P25036, incorrectly annotated as a metalloendopeptidase (EC 3.4.24) by Swiss-Prot, is actually a serine endopeptidase (EC 3.4.21) as correctly identified by iCSA. (The error was reported and has been corrected by Swiss-Prot.) For these reasons, coverage of 100% is unlikely to be achieved by the iCSA filter.
When the equivalent figures are calculated for all four levels of EC, the accuracy when using the iCSA filter decreases to 55%, with a slightly higher coverage of 87%, compared with an accuracy of only 26% when using sequence homology alone. The differing results when using the third and fourth digit of the EC number as a benchmark suggest that, by ensuring that the catalytic residues are conserved, we capture proteins with the same “reaction” but ignore the specificity that is broadly described at the fourth EC level. Further understanding of the residues responsible for the specificities of an enzyme reaction will add to the success of functional assignment.
Swiss-Prot is trivially nonredundant, so to check for bias toward a particular sequence family, we also reduced the Swiss-Prot set to ensure that no sequence had >40% sequence identity to any other sequence with the same EC. The results for this condensed set are very similar to those presented for the full set, with an accuracy of 83.4% and coverage of 70.3%.
Although a distant homologue alone would not be trusted to confer function, this possibility becomes real when using the iCSA filter and is particularly valuable in the many cases where no well annotated, close homologues are available. However, it should also be considered an important step even when using information from close homologues, because, in principle, a single point mutation in the appropriate residue could destroy catalytic activity. Here we use psi-blast to show that even distant relationships can confer high-quality function annotation when combined with the knowledge of the catalytic residues. The iCSA filter is not limited to psi-blast but can be applied as a postfilter with any homology detection tool.
d-alanine:d-alanine Ligase (EC 6.3.2.4). One of the largest differences in performance between the two methods is for d-alanine:d-alanine ligase. EC assignment accuracy is 100% when using the iCSA filter with the d-alanine:d-alanine ligase [PDB ID code 2DLN (30)] annotated site, compared with 19.2% when using sequence homology alone. d-alanine:d-alanine ligase, which is responsible for the biosynthesis of bacterial cell walls, belongs to the large ATP-grasp superfamily (31). Members of this superfamily have many specific physiological functions, which leads to difficulty when assigning function to previously uncharacterized sequences (32).
The iCSA filter correctly identifies 56 of the 64 homologues with the same function at the third level of EC (EC 6.3.2: acid-d-amino acid ligases), two-thirds of which have <40% sequence identity to the query. Four of the eight unidentified enzymes have conservative substitutions at the catalytic site. More significantly, iCSA successfully leaves unannotated 210 of the 218 homologues picked up by the homology search that have different functions at the third level of EC. These functions include EC 6.3.4 (other carbon-nitrogen ligases) and EC 6.3.5 (carbon-nitrogen ligases with glutamine as the amido-N-donor), which belong to the ATP-grasp superfamily.
Trypsin-Like Serine Proteases (EC 3.4.21). For this family, with a clearly defined catalytic triad (His-Asp-Ser, shown in trypsin in Fig. 2a), the accuracy of both the iCSA filter and homology alone is 100% in our benchmark. However, the benefits of the iCSA filter are to be seen in the homologues not assigned the trypsin EC by iCSA.
Fig. 2.
Trypsin catalytic site and trypsin sequence aligned to haptoglobin sequence. (a) Trypsin 3D structure (PDB ID code 1A0J33, chain A). Catalytic residues are shown in red (His-57), green (Asp-102), and blue (Ser-195). (b) Trypsin (PDB ID code 1A0J, chain A, residues 1–196) aligned to haptoglobin (Swiss-Prot sequence P19006, residues 85–303). Catalytic residue positions in trypsin are marked by an asterisk. 1A0J retrieved P19006 with a blast-level search; the sequences have 26% pairwise sequence identity.
Ninety-five percent of the Swiss-Prot homologues with EC 3.4.21 are identified by iCSA, leaving 21 Swiss-Prot entries with the same three-digit EC as the trypsin query that do not conserve the catalytic site and have therefore not been assigned this function. Seventeen of these homologues do not overlap completely with the trypsin catalytic domain and therefore do not contain all of the necessary residues to be active. Two of the remaining four have an arginine residue instead of the histidine and have a caution notice in the full Swiss-Prot entry (O13057 and O13060), which suggests that the iCSA is correct to reject these homologues and that researchers should indeed view these particular entries with caution.
In addition to the enzymes included in the current benchmark, there are also trypsin homologues that are known to be nonenzymatic, such as haptoglobin. The iCSA filter correctly identifies these proteins as homologues lacking the catalytic triad required and classifies them as nonenzymatic (see Fig. 2b). In contrast, sequence similarity alone, which is used by many popular annotation systems such as Pfam and CDD, incorrectly assigns a serine protease function to haptoglobin. Such annotation mistakes can be costly because the information is frequently used to guide the design of subsequent biological assays.
Use of the iCSA Filter in Structural Genomics. Structural genomics groups have varying rationale for prioritizing protein targets for structure determination. In general, two approaches receive the majority of attention: finding sequences with distant similarities to known structures or identifying sequences with no detectable similarity to any known structure. Although these approaches are pragmatic, large numbers of candidates remain. Further prioritization could be usefully pursued by using the iCSA filter.
One such case is the protein BioH, selected by the Midwest Center for Structural Genomics as a protein of unknown structure and unknown function. Although structure determination revealed BioH to be a member of the α/β hydrolase family with a conserved Ser-Asp-His site (PDB ID code 1M33) (34), applying psi-blast to data available before the BioH structure was determined shows that BioH could have been placed in the α/β hydrolase family without structure determination. Furthermore, hand analysis by an expert would have revealed conservation of the Ser-Asp-His triad, indicating probable hydrolase activity. Repeating the search today with the iCSA filter (while ignoring information from the available BioH structure) rapidly identifies three curated entries, all with Ser-Asp-His active sites, allowing a prediction of function from sequence alone.
Applying the iCSA Filter to Unannotated Swiss-Prot Entries. The iCSA filter has identified a conserved catalytic site in 883 Swiss-Prot sequences that do not have an EC number, including 58 human sequences. Although these proteins may not have a catalytic function, it seems likely that their function either has not been identified experimentally or simply has not been annotated in the source database. In contrast, a further 4,814 homologues without EC annotation have no conserved site according to the iCSA filter. Rapidly providing this sort of information for unannotated proteins is clearly of benefit, particularly with the rise of poorly annotated sequences from high-throughput genome sequencing.
Annotating the Human Genome. Genomes are now sequenced routinely by major centers, resulting in an exponential increase in primary data. In contrast, experimental approaches for identifying encoded proteins and, more importantly, assigning function have not kept pace. As a result, researchers now rely primarily on computational approaches. The ability to apply a detailed functional annotation method uniformly across all sequences is a particular strength of our approach.
Taking human ensembl (35) data as our example (from release 14 of Biopendium), the iCSA filter annotated 2,064 homologous sequences with an EC number and rejected a further 2,257 homologues as not having the complete complement of catalytic residues necessary for function. We identified entries within the 2,257 set that had very close homologues in Swiss-Prot (at least 90% pairwise sequence identity and a difference in length of <10% of the shorter sequence). For 73% of these, the Swiss-Prot entry had either an EC number that was different from the EC number of the CSA query or no recorded EC number. For this 73%, the system is working as designed, suggesting that there will be benefits from increasing the EC coverage of the CSA. The remaining 27% that had the same EC as the CSA query fall into a number of categories: some will be incomplete sequences or splice variants, and others will be correct sequences that are misaligned, have a catalytic mechanism different from the one defined in the CSA, or are miscurated in Swiss-Prot.
Extended Assignment Using Conservative Substitutions. Function assignment by iCSA is strict, requiring that every catalytic residue is found in a homologue. Ultimately, coverage is best extended by increasing the range of curated entries in the CSA. However, a more immediate approach would be to accept conservative substitutions observed in Swiss-Prot homologues with the same EC in the sites identified by each CSA query. Using this approach, each CSA catalytic residue has a set of allowed substitutions. The coverage greatly improves, because we are now assigning function to sequences that were previously rejected. What is surprising, however, is that the accuracy of assignment remains high, with 84.9% accuracy and 91.9% coverage overall.
Discussion
Our results clearly show that the iCSA filter can substantially improve function assignment accuracy, especially when homologues are distant, and will be very useful in annotation of previously uncharacterized sequences. Equally important, it shows that only half of the homologues returned after four iterations of psi-blast are likely to have a similar function to the query protein (there is a total of 69,151 Swiss-Prot homologues returned after four iterations of psi-blast searches with each CSA query; 36,832 of these homologues have either no EC annotation or a different EC annotation from their respective query), highlighting the need to use more sophisticated knowledge-based approaches to function annotation.
As increasingly large numbers of enzymes are discovered in genome sequencing projects, the number of enzymes without an EC designation continues to rise. It is possible that Swiss-Prot sequences without an EC number that pass the iCSA filter criteria are newly discovered enzymes that fit into well known catalytic mechanisms but may work on alternative substrates to the known enzymes. A key advantage of the iCSA filter is that it can be applied directly to any sequence detected using any database search tool. This feature is particularly important for high-throughput genome sequencing pipelines where almost no hand annotation is possible in the time available before release, as well as for databanks such as GenBank (36), the European Molecular Biology Laboratory Nucleotide Sequence Database (37), and the DNA Data Bank of Japan (38), where annotation depends on each depositor.
Structural biology is an increasingly popular tool for obtaining functional clues for a protein. Over the past few years, several structural genomic initiatives have been established worldwide to increase the throughput of structural elucidation. However, despite these efforts, structure determination remains a relatively low-throughput approach and is costly to implement. Our technique helps to prioritize proteins for structure determination by quickly estimating the likelihood of a protein having a function similar to a previously determined structure. Furthermore, because the data for our approach are derived from 3D structures, there is a natural synergy with structural genomics initiatives. Using our method, efforts can be concentrated on those family members with no clear functional mechanism.
These results clearly show that the iCSA filter correctly distinguishes between homologues with alternative functions and highlight the merits of using a key residue-based system in combination with homologue detection. When predicting the function of a previously uncharacterized sequence by using homology, the closest well annotated homologue will usually be chosen. The iCSA filter becomes more useful as the closest available homologue becomes more distant. The CSA is continually being updated at the European Bioinformatics Institute with new catalytic sites, which can only lead to improved function assignment by iCSA. Although this study has concentrated on catalytic function, the approach is generic, and it would be possible to extend it into any areas where invariant residues could be confidently assigned. Antibody loops could be particularly well suited to this, given their high degree of conservation (39). Application to protein–protein interaction surfaces would also be extremely fertile ground, should data emerge to show examples of residue invariance.
Acknowledgments
We thank Michael Stevens (European Bioinformatics Institute) for helpful discussions and John Overington and Felix Sheinerman for drawing our attention to haptoglobin and twinfilin, respectively. We are grateful for technical advice from Ian Carruthers and Richard Bickerton (both of Inpharmatica). This work was supported by the Department of Trade and Industry LINK program in Applied Genomics funded by the Biotechnology and Biological Sciences Research Council (R.A.G., R.V.S., and C.T.P.) and the BioSapiens project, which is funded by the European Commission within its FP6 Program under the thematic area “Life Sciences, Genomics, and Biotechnology for Health,” contract no. LHSGCT-2003-503265 (J.M.T.).
Author contributions: R.A.G., R.V.S., B.A.-L., J.M.T., and M.B.S. designed research; R.A.G. and R.V.S. performed research; R.A.G., R.V.S., G.J.B., A.G., M.W.M., and C.T.P. contributed new reagents/analytic tools; R.A.G. and R.V.S. analyzed data; and R.A.G. and R.V.S. wrote the paper.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 29, 2003.
Abbreviations: CSA, Catalytic Site Atlas; iCSA, Inpharmatica CSA.
See accompanying Profile on page 12296.
References
- 1.Whisstock, J. C. & Lesk, A. M. (2003) Q. Rev. Biophys., 36, 307–340. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous (1999) Scientist 13, 15. [Google Scholar]
- 4.Jones, D. T. (1999) J. Mol. Biol. 287, 797–815. [DOI] [PubMed] [Google Scholar]
- 5.Todd, A. E., Orengo, C. A. & Thornton, J. M. (2001) J. Mol. Biol. 307, 1113–1143. [DOI] [PubMed] [Google Scholar]
- 6.Tian, W. & Skolnick, J. (2003) J. Mol. Biol. 333, 863–882. [DOI] [PubMed] [Google Scholar]
- 7.Bateman, A., Coin, L., Durbin, R., Finn, R. D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E. L., et al. (2004) Nucleic Acids Res. 32, D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchler-Bauer, A., Anderson, J. B., Cherukuri, P. F., DeWeese-Scott, C., Geer, L. Y., Gwadz, M., He, S., Hurwitz, D. I., Jackson, J. D., Ke, Z., et al. (2005) Nucleic Acids Res. 33, D192–D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Attwood, T. K., Bradley, P., Flower, D. R., Gaulton, A., Maudling, N., Mitchell, A. L., Moulton, G., Nordle, A., Paine, K., Taylor, P., et al. (2003) Nucleic Acids Res. 31, 400–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hulo, N., Sigrist, C. J., Le Saux, V., Langendijk-Genevaux, P. S., Bordoli, L., Gattiker, A., De Castro, E., Bucher, P. & Bairoch, A. (2004) Nucleic Acids Res. 30, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis, S. E. (2005) Genome Biol. 6, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pal, D. & Eisenberg, D. (2005) Structure 13, 121–130. [DOI] [PubMed] [Google Scholar]
- 13.Altschul, S. F., Boguski, M. S., Gish, W. & Wootton, J. C. (1994) Nat. Genet. 6, 119–129. [DOI] [PubMed] [Google Scholar]
- 14.Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M. C., Estreicher, A., Gasteiger, E., Martin, M. J., Michoud, K., O'Donovan, C., Phan, I., et al. (2003) Nucleic Acids Res. 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegyi, H. & Gerstein, M. (2001) Genome Res. 11, 1632–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fagan, R., Swindells, M. B., Overington, J. & Weir, M. (2001) Trends Biochem. Sci. 26, 213–214. [DOI] [PubMed] [Google Scholar]
- 17.Iyer, L. M., Aravind, L., Bork, P., Hofmann, K., Mushegian, A. R., Zhulin, I. B. & Koonin, E. V. (2001) Genome Biol. 2, RESEARCH0051.1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galperin, M. Y. & Koonin, E. V. (1998) In Silico Biol. 1, 55–67. [PubMed] [Google Scholar]
- 19.Livingstone, C. D. & Barton, G. J. (1993) Comput. Appl. Biosci. 9, 745–756. [DOI] [PubMed] [Google Scholar]
- 20.Lichtarge, O., Bourne, H. R. & Cohen, F. E. (1996) J. Mol. Biol. 257, 342–358. [DOI] [PubMed] [Google Scholar]
- 21.Mihalek, I., Res, I. & Lichtarge, O. (2004) J. Mol. Biol. 336, 1265–1282. [DOI] [PubMed] [Google Scholar]
- 22.Porter, C. T., Bartlett, G. J. & Thornton, J. M. (2004) Nucleic Acids Res. 32, D129–D133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000) Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.International Union of Biochemistry and Molecular Biology (1992) Enzyme Nomenclature: Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes (Academic, New York).
- 25.Jones, D. T. & Swindells, M. B. (2002) Trends Biochem Sci. 27, 161–164. [DOI] [PubMed] [Google Scholar]
- 26.Swindells, M. B., Rae, M., Pearce, M., Moodie, S., Miller, R. & Leach, P. (2002) Philos. Trans. R. Soc. London A 360, 1179–1189. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith, T. F. & Waterman, M. S. (1981) J. Mol. Biol. 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 29.Bairoch, A., Apweiler, R., Wu, C. H., Barker, W. C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R., Magrane, M., et al. (2005) Nucleic Acids Res. 33, D154–D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan, C., Moews, P. C., Walsh, C. T. & Knox, J. R. (1994) Science 266, 439–443. [DOI] [PubMed] [Google Scholar]
- 31.Galperin, M. Y. & Koonin, E. V. (1997) Protein Sci. 6, 2639–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, H., Xu, H., Graham, D. E. & White, R. H. (2003) Proc. Natl. Acad. Sci. USA 100, 9785–9790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schroder, H. K., Willassen, N. P. & Smalas, A. O. (1998) Acta Crystallogr. D 54, 780–798. [DOI] [PubMed] [Google Scholar]
- 34.Sanishvili, R., Yakunin, A. F., Laskowski, R. A., Skarina, T., Evdokimova, E., Doherty-Kirby, A., Lajoie, G. A., Thornton, J. M., Arrowsmith, C. H., Savchenko, A., et al. (2003) J. Biol. Chem. 278, 26039–26045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hubbard, T., Andrews, D., Caccamo, M., Cameron, G., Chen, Y., Clamp, M., Clarke, L., Coates, G., Cox, T., Cunningham, F., et al. (2005) Nucleic Acids Res. 33, D447–D453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J. & Wheeler, D. L. (2005) Nucleic Acids Res. 33, D34–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanz, C., Aldebert, P., Althorpe, N., Baker, W., Baldwin, A., Bates, K., Browne, P., van den Broek, A., Castro, M., Cochrane, G., et al. (2005) Nucleic Acids Res. 33, D29–D33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tateno, Y., Saitou, N., Okubo, K., Sugawara, H. & Gojobori, T. (2005) Nucleic Acids Res. 33, D25–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lesk, A. M. & Chothia, C. (1988) Nature 335, 188–190. [DOI] [PubMed] [Google Scholar]