Abstract
Background
Carotid atherosclerotic plaque is the primary cause of cardiovascular and cerebrovascular diseases. It is closely related to oxidative stress and immune inflammation. This bioinformatic study was conducted to identify key oxidative stress-related genes and key immune cell infiltration involved in the formation, progression, and stabilization of plaques and investigate the relationship between them.
Results
We show that the up-regulation of oxidative stress-related genes such as IDH1 and CD36 in resident-like macrophages and foam macrophages play a key role in the formation and progression of carotid atherosclerotic plaques.
Conclusions
We discuss the role of oxidative stress and immune inflammation in the formation, progression, and stabilization of plaques by combining predictive models with analysis of single-cell data. It introduced novel insights into the mechanisms underlying carotid atherosclerosis formation and plaque progression and may assist in identifying potential therapeutic targets for their treatment.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13062-025-00600-7.
Keywords: Oxidative stress, Immune inflammation, Single-cell sequencing, Machine learning
Background
According to the World Health Organization, an estimated 17.9 million people die from cardiovascular diseases (CVDs) each year, accounting for 32% of global mortalities and remaining the leading cause of death [1]. Notably, carotid atherosclerosis is the principal cause of cerebrovascular diseases. Indeed, it can lead to partial or complete narrowing of the lumen of the blood vessels, thus limiting blood flow to the brain and resulting in symptoms such as blurred and transient loss of vision. Furthermore, acute rupture of carotid atherosclerotic plaques can lead to local thrombosis and blood blockage [2, 3], which is the primary cause of stroke and the second leading cause of death worldwide [4, 5]. While improvements in living standards and medical care, as well as health management practices, have significantly lowered the incidence and mortality rates of carotid atherosclerosis [6], it remains a public health concern in most countries.
Decades ago, atherosclerosis was regarded as a disease induced by dyslipidemia [7, 8]. At present, mounting evidence suggests that atherosclerosis is a chronic immune inflammatory disease [9–11]. Oxidative stress is closely related to inflammation and the mutual promotion between them play a vital role in the occurrence and progression of atherosclerotic plaques [12]. Oxidative stress is essentially triggered by the excessive production of reactive oxygen species (ROS) and dysregulation of the antioxidant system [13, 14]. Previous studies have reported that excessive ROS production can trigger apoptosis of endothelial cells and vascular responses to external stimuli, thus leading to inflammation. Subsequently, macrophages, T cells, B cells, tumor necrosis factor (TNF), other immune cells and inflammatory cytokines infiltrate the vascular wall, thereby promoting the occurrence of atherosclerosis [15–17]. Meanwhile, some inflammatory cytokines (such as TNF and Interleukin-1) secreted in response to the inflammatory reactions can result in inhibitory kappa B kinase (IKK) phosphorylation-dependent activation of the NF-κB essential modulator (NEMO) complex. These interactions eventually activate the nuclear factor-κB (NF-κB) pathway, which contributes to more ROS generation. For example, enzymes such as NADPH oxidase (Nox), xanthine oxidase, inducible nitric oxide synthase (iNOS), or neuronal nitric oxide synthase (nNOS) are regulated by the NF-κB pathway, facilitating the production of ROS and peroxynitrites [18–20]. Eventually, the disbalance between ROS production and the anti-oxidant mechanisms induces atherosclerosis. Therefore, the mutual promotion between inflammation and oxidative stress may play an indispensable role in the initiation and development of atherosclerosis. More importantly, studies have described that oxidative stress is closely associated with plaque rupture and intraplaque hemorrhage, which play a critical role in the prognosis of carotid atherosclerosis [21–24].
Therefore, exploring the interactions between key oxidative stress genes and key inflammatory cells, as well as the mechanisms underlying the formation of atherosclerotic plaques, may assist in the management, prevention, and development of innovative therapeutic strategies for atherosclerosis.
Methods
Data collection, collation, and oxidative stress-related gene acquisition
Four carotid atherosclerosis-related datasets were retrieved from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/), namely GSE28829 [25], GSE163154 [26], GSE43292 [27] and GSE159677 [28], as listed in Table 1. In brief, we searched the GEO database using ‘atherosclerosis’ as the keyword, restricted the sample type to Homo sapiens, and selected GSE28829 (early and advanced carotid plaques), GSE43292 (carotid plaques and control group), and GSE163154 (intra-plaque hemorrhage and non-intra-plaque hemorrhage carotid plaques) to explore the relationship between oxidative stress and the occurrence, progression, and rupture of carotid atherosclerotic plaque samples (CAS). Meanwhile, GSE159677 (calcified atherosclerotic core plaques and the patient-matched proximal adjacent portions of carotid artery) was selected to conduct cluster analysis performed on single-cell data to identify key oxidative stress-related genes and key immune cell infiltration. A total of 444 genes related to oxidative stress were identified and acquired according to gene ontology annotation at AmiGO 2 tool (https://amigo.soybase.org/amigo/amigo/landing). The details of genes are presented in Supplementary Table 1. The main clinical characteristics of the patients of each dataset were displayed in Supplementary Table 2.
Table 1.
The details of four carotid atherosclerosis-related datasets
| Plaque (series) | Platform | Samples |
|---|---|---|
| 28829 | GPL570 | Carotid plaque 29 [advanced 16(thin or thick fibrous cap atheroma);early 13(pathological intimal thickening and intimal xanthoma)] |
| 163154 | GPL6104 |
Carotid plaque 43 (non-IPH 16;IPH 27) IPH:intra-plaque hemorrhage |
| 43292 | GPL6244 | Carotid plaque 32;Control group(Macroscopically intact carotid tissue adjacent to the atheroma plaque) 32(they are all from media and neo-intima) |
| 159677 | GPL18573 | Calcified atherosclerotic core (AC) plaques and patient-matched proximal adjacent (PA) portions of carotid artery were collected from three patients |
Single sample gene set enrichment analysis (ssGSEA)
Using "GSVA" (version 1.34.0) [29] in R software (4.1.2), single-sample gene set enrichment analysis (ssGSEA) was performed on the three datasets to analyze oxidative stress characteristics related to the identified oxidative stress-related genes. In brief, we organized three datasets into a gene expression matrix format, with genes as rows and samples as columns. We used oxidative stress-related genes as the target gene set and calculated the enrichment of this gene set in each sample to generate an ssGSEA score. This score represents the enrichment level of the target gene set in each sample. We then analyzed the ssGSEA results to examine the scores of the target gene set across different samples.
Differential analysis and screening of key oxidative stress-related genes
Oxidative stress-related genes were identified from the three datasets, and differentially expressed genes were identified using the "limma" package [30] in R software (4.1.2) using the screening criteria of corrected p-value < 0.05 (Benjamini-Hochberg, to control the false positive rate) and false discovery rate of 0.05 (Used for large-scale multiple hypothesis testing, it can more effectively balance the relationship between discovering truly significant results and controlling the proportion of false positives). Oxidative stress-related genes differentially expressed across the three datasets were regarded as key oxidative stress-related genes.
Consistency cluster analysis, principal component analysis (PCA), and immune cell infiltration analysis
Consensus Cluster Plus [31] in R software (4.1.2) was utilized to conduct consensus clustering analysis of samples based on key oxidative stress-related genes, and different clusters were distinguished according to the results of consensus clustering matrix analysis. PCA analysis of different clusters was performed using the "limma" package [30] in R software (4.1.2). Then, GSVA (version 1.34.0) [29] in R software (4.1.2) was employed to perform ssGSEA, and the enrichment scores of 23 immune-related cells and functions in the CAS group, control group, and different clusters were determined [32]. Lastly, we analyzed the clusters to identify differentially expressed genes.
Gene ontology (GO) and kyoto encyclopedia of genes and genomes (KEGG) enrichment analyses
GO and KEGG enrichment analyses of differentially expressed genes in different clusters were performed using the cluster Profiler 4.0 package [33].
Construction and validation of oxidative stress prediction models
The "randomForest" and "kernlab" packages [34, 35] in the R software (4.1.2) were used to analyze the data. We used the random forest method (to handle classification problems in a nonlinear model) and the support vector machine (SVM) method (to handle classification problems in a nonlinear model with a polynomial kernel) to construct the predictive models. For the random forest method, we used fivefold cross-validation to select appropriate hyperparameters, setting n_estimators to 500, min_samples_split to 2, and max_depth to 20. For the SVM model, we applied a polynomial kernel of degree 3, selected gamma = 0.1 as the kernel function parameter, and tuned the regularization parameter (C) using cross-validation to determine the optimal configuration. The reliability of the predictive models constructed using the two methods was compared by comparing the residual values and the reverse cumulative distribution of residuals using receiver operating characteristic (ROC) curve analysis. The method with higher confidence was selected to construct the predictive model based on the top 5 genes identified by importance scores of oxidative stress-related genes calculated using the corresponding method. Finally, the accuracy of the predictive model was validated by comparing the accuracy curve of the predictive model with the ideal curve and the bias correction curve.
Decision curve analysis and construction of nomogram
DCA was used to assess the clinical decision utility of the predictive model. We used predictive models to calculate the predicted probabilities for each patient, apply decision curve analysis (DCA) to assess the net benefit of the model at different thresholds, and plot the decision curves by using "dcurves" package [36]. Comparison subjects should include curves for full treatment (assuming everyone is treated) and no treatment (assuming no one is treated). The closer the decision curve is to the top, the higher the clinical value of the model.
The nomogram is constructed by converting gene expression levels from a predictive model into scores based on the model’s coefficients by using "nomogramFormula" package [37]. For individual patients, doctors can determine corresponding scores based on the values of each variable in the nomogram and sum the scores of all variables to obtain a total score. This total score is then used to predict the prognosis or the likelihood of an event occurring for the patient. The nomogram visually displays individual patient risk, aiding doctors in making personalized treatment decisions based on specific patient characteristics.
ROC analysis and correlation analysis
The oxidative stress prediction model was applied to three different types of carotid atherosclerosis data, and ROC analysis was adopted to verify its accuracy based on "pROC" [38]. The correlation between genes and immune cells was evaluated by calculating the Pearson correlation coefficient based on R (version 4.3.0) programming environment.
Analysis of single-cell data from carotid plaques
"Seurat" package [39] was used for processing single-cell data. We implemented a quality-control criterion that required cells to have gene expression levels ranging between 200 and 5,000, and a mitochondrial content exceeding 20%. A total of 48292 cells from GSE159677 [28] were included in the analysis. Samples were integrated using the top 2,000 hypervariable genes. Principal component analysis (PCA) was carried out to map high-dimensional single-cell data into two-dimensional space. The resolution parameter (set to 0.6 in this study) determines the granularity of clustering in single-cell analysis. Based on data characteristics and preliminary testing, a resolution of 0.6 provided an optimal balance between cluster granularity and biological relevance, as verified through visualization techniques (cluster trees). We used t-Distributed Stochastic Neighbor Embedding (tSNE) for dimensionality reduction and visualization of high-dimensional data, with parameters set to perplexity = 30, learning rate = 200, and 1000 iterations. t-SNE was also employed to visualize cell clusters. Cell clusters were manually annotated with reference to the cellmark database and published studies [4–7]. Afterward, the expression of model genes was evaluated in each cell cluster.
Quantification and statistical analysis
Statistical analysis was performed using R (version 4.3.0). Data are expressed as mean ± standard deviation (SD) or as medians with interquartile ranges. Statistical significance between two groups was determined using the Student’s unpaired t-test. A p-value < 0.05 was considered significant. One asterisk (*) indicates a p-value < 0.05, two asterisks (**) indicate a p-value < 0.01, three asterisks (***) indicate a p-value < 0.001, and four asterisks (****) indicate a p-value < 0.0001.
Results
Oxidative stress profile and screening of key oxidative stress-related genes in carotid atherosclerosis
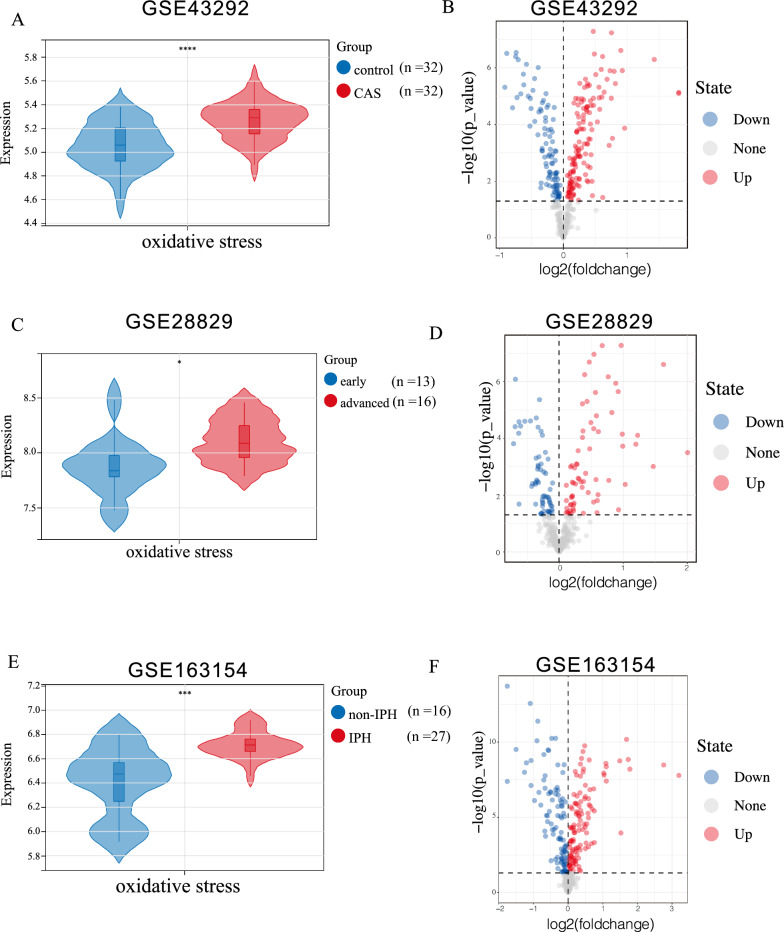
The analysis revealed significant differences in oxidative stress scores between healthy carotid samples and carotid atherosclerotic plaque samples (CAS), early CAS and advanced CAS, and non-intraplaque hemorrhage (non-IPH) and intraplaque hemorrhage (IPH) samples (Fig. 1A, C, E). In addition, the oxidative stress scores were significantly higher in the CAS group compared with the control group, and higher in the advanced CAS and intraplaque hemorrhage groups compared with the early CAS and non-IPH groups, respectively. These results collectively indicated that the occurrence, progression, and plaque rupture of CAS are closely correlated with oxidative stress. We performed a differential analysis of oxidative stress-related genes across three datasets and identify those with consistent expression differences in all three as key oxidative stress-related genes: 141 up-regulated oxidative stress-related genes and 96 down-regulated oxidative stress-related genes were identified In the control and CAS groups (Fig. 1B); 69 up-regulated oxidative stress-related genes and 61 down-regulated oxidative stress-related genes were identified in the early CAS group and advanced CAS group (Fig. 1D); 117 up-regulated oxidative stress-related genes and 101 down-regulated oxidative stress-related genes were identified in the non-IPH and IPH groups (Fig. 1F). Total of 83 key oxidative stress-related genes were identified as the hub oxidative stress-related genes (Fig. 2A).
Fig. 1.
A: Oxidative stress scores in the GSE43292 dataset (CAS and control samples). B: Volcano plot illustrating oxidative stress-related genes in the GSE43292 dataset (CAS and control samples). C: Oxidative stress scores in the GSE28829 dataset (Early plaque and advanced plaque groups). D: Volcano plot displaying oxidative stress-related genes in the GSE28829 dataset (Early plaque and advanced plaque groups). E: Oxidative stress scores in the GSE163154 dataset (non-IPH and IPH). F: Volcano plot depicting oxidative stress-related genes in the GSE163154 dataset (non-IPH and IPH). Analyzed with Student’s unpaired t-test. * p-value < 0.05; ** p-value < 0.01; *** p-value < 0.001; **** p-value < 0.0001
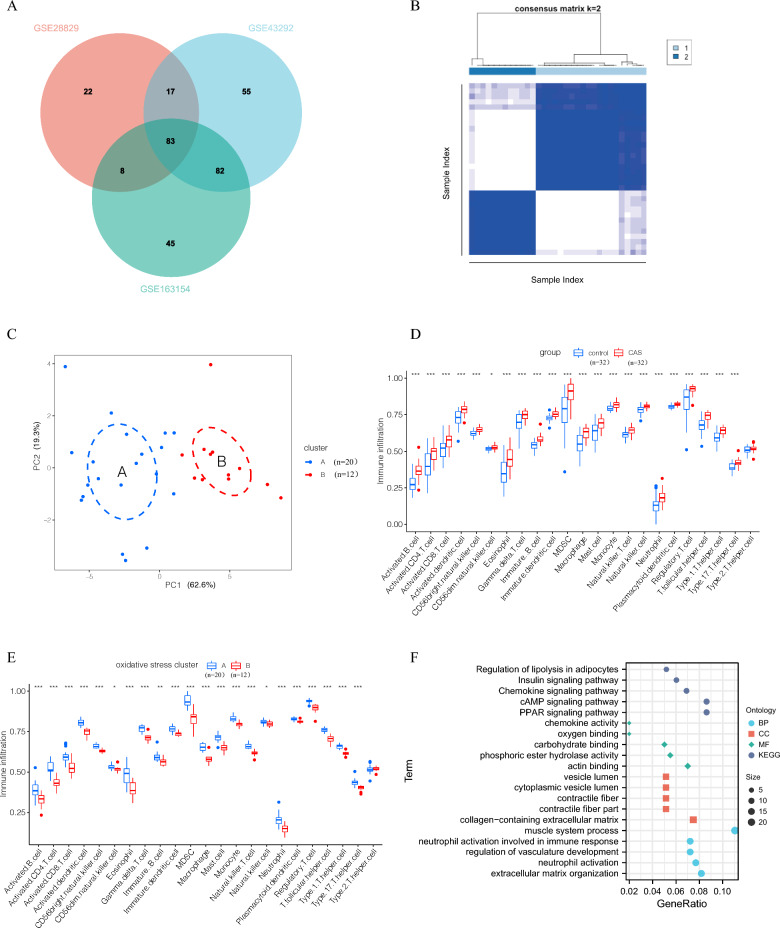
Fig. 2.
A: Venn diagram presenting differentially expressed oxidative stress genes across the three datasets. B: Consistency matrix clustering analysis. C: PCA analysis of oxidative stress clusters (PCA plot showing the distribution of samples in the first two principal components. Dashed circles represent clusters of samples identified based on their proximity in the PCA space, illustrating groupings with shared characteristics). D: Infiltration level of 23 immune-related cells in the CAS and control groups. E: Infiltration of 23 immune-related cells in the oxidative stress cluster. F: GO and KEGG analysis of differentially expressed genes in the oxidative stress cluster (BP: biological process CC: cellular component MF: molecular function). * p-value < 0.05; ** p-value < 0.01; *** p-value < 0.001; **** p-value < 0.0001
Analysis of consistency clusters
According to the 83 key oxidative stress-related genes and the results of the consistency cluster matrix analysis (Supplementary Fig. 1A, B), carotid atherosclerotic plaque samples were categorized into two clusters (Carotid atherosclerotic plaque samples are from GSE43292) (Fig. 2B). Also, PCA analysis determined that carotid atherosclerotic plaque samples in the two clusters could be well distinguished (Fig. 2C). This indicates that carotid atherosclerotic plaque samples can be effectively divided into two clusters based on oxidative stress-related genes. Given the association of oxidative stress with immune inflammation and its link to atherosclerosis, the level of immune cell infiltration was compared between the control and CAS groups, as well as between the two clusters. (cluster A and cluster B), was examined. We assessed immune cell infiltration in the samples based on the markers of various immune cell types. The percentage of infiltration of 23 immune-related cells was significantly higher in the CAS group compared to the control group, indicating that carotid atherosclerosis is associated with immuno-inflammatory processes (Fig. 2D). However, in the two clusters (A and B), with the exception of the higher abundance of type 2 helper T cells in cluster B, the remaining 22 immune-related cells were highly expressed in cluster A, suggesting that oxidative stress is significantly correlated with immune cell infiltration, with cluster A displaying a higher immune-inflammatory profile compared with cluster B (Fig. 2E). Following this, differential analysis was performed between cluster A and cluster B samples, and 231 differentially expressed genes were identified. GO and KEGG enrichment analyses of these differentially expressed genes demonstrated that these genes were largely involved in the regulation of adipocyte lipolysis, chemokine signaling pathway, peroxisome proliferators-activated receptors (PPAR) signaling pathway, oxygen binding, neutrophil activation, extracellular matrix tissue synthesis, and other processes (Fig. 2F, Supplementary Fig. 1C).
Construction of the oxidative stress prediction model
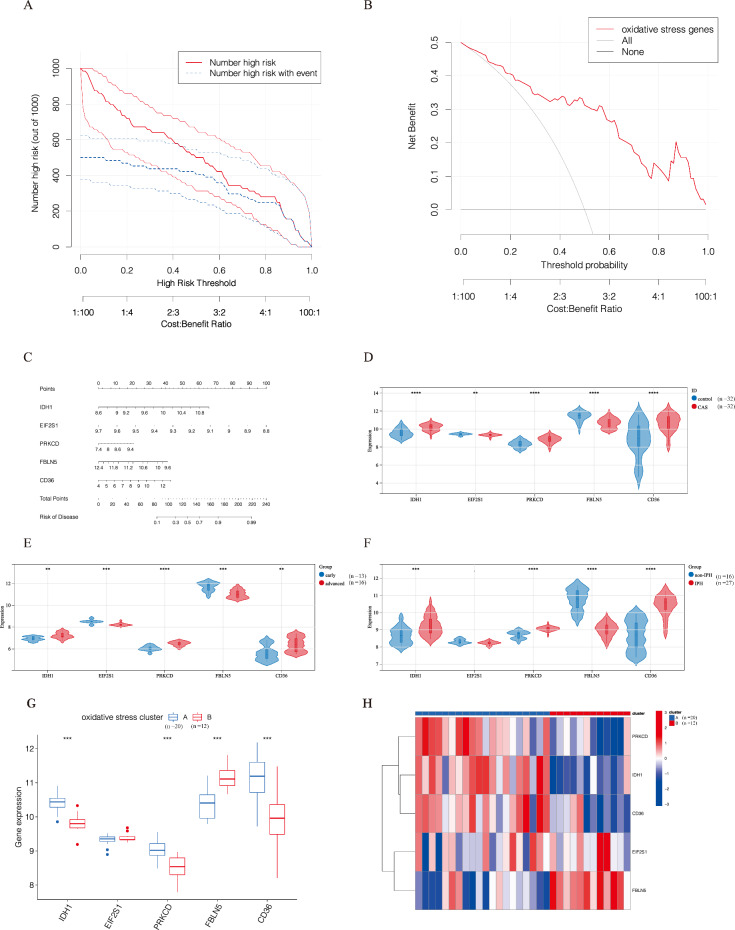
We used 83 key oxidative stress-related genes to construct predictive models using random forest and SVM methods. The model construction was based on the dataset GSE43292. The predictive models constructed using the random forest method had lower residual values (i.e., higher confidence) (Supplementary Fig. 1D, Supplementary Fig. 2A, B), whilst ROC analysis validated that the predictive model constructed using this method had higher area under the curve (AUC) values (Supplementary Fig. 2C). The importance scores of predictive model’s genes were determined from Supplementary Fig. 2D (calculated according to the random forest method), and the top 5 genes (IDH1, EIF2S1, PRKCD, FBLN5, and CD36) were selected to construct the predictive model. The accuracy curve of the predictive model was consistent with the ideal curve and the deviation correction curve (Supplementary Fig. 2E), highlighting the high accuracy of the predictive model. Similarly, DCA analysis confirmed that, compared to ‘treat-all’ or ‘treat-none’ strategies, the oxidative stress predictive model provides a higher net benefit, accurately predicts the prognosis of CAS patients, and helps doctors assess whether further interventions are necessary for these patients (Fig. 3A, B). Following this, the nomogram was established based on the predictive model composed of IDH1, EIF2S1, PRKCD, FBLN5, and CD36. As anticipated, the results demonstrated that the nomogram could accurately predict the risk of CAS (Fig. 3C).
Fig. 3.
A: Clinical impact curves of oxidative stress prediction models. B: DCA curve analysis of oxidative stress models. C: Nomogram of the oxidative stress prediction model. D: Relative expression levels of oxidative stress model genes in the CAS and control groups. E: Relative expression levels of oxidative stress model genes in the early plaque group and the advanced plaque group. F: Relative expression levels of oxidative stress model genes in the non-IPH and IPH groups. G: Relative expression of oxidative stress model genes in the oxidative stress clusters. H: Heatmap of expression of oxidative stress model genes in oxidative stress clusters. * p-value < 0.05; ** p-value < 0.01; *** p-value < 0.001; **** p-value < 0.0001
The predictive model of oxidative stress for analysis of gene expression and immune characteristics
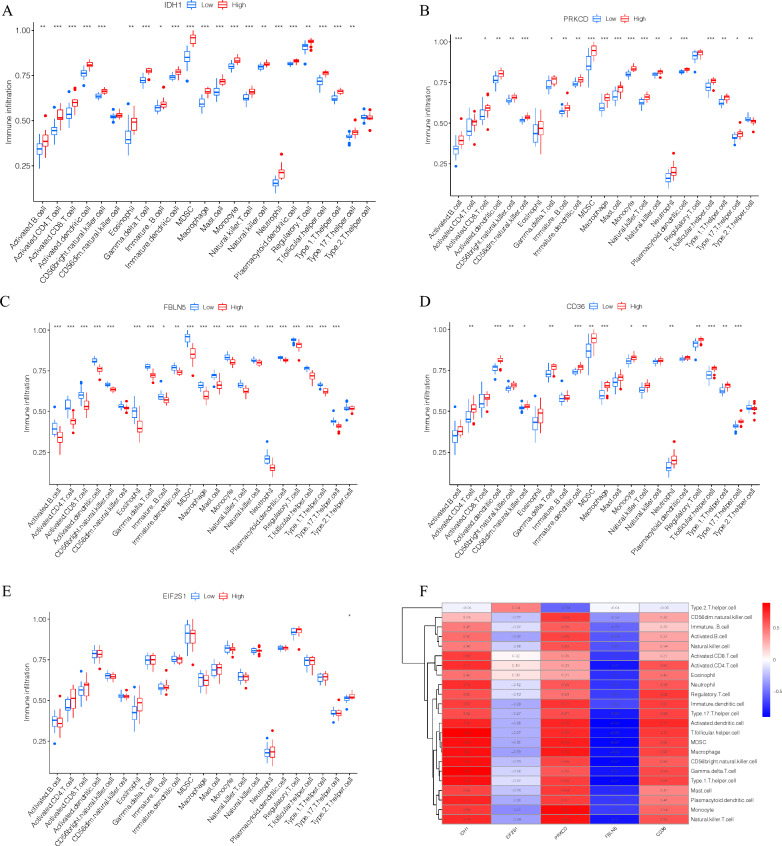
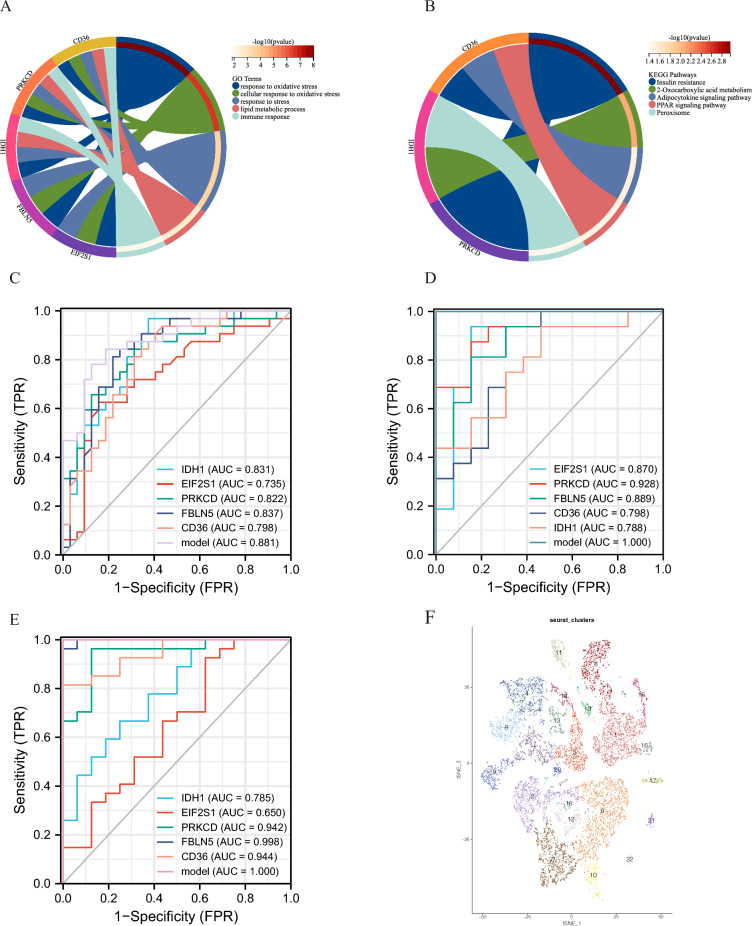
The expression of IDH1, EIF2S1, PRKCD, FBLN5, and CD36 in three different datasets of carotid atherosclerosis samples was analyzed. As delineated in Fig. 3D-F, the expression level of IDH1, PRKCD, and CD36 was increased in the CAS, advanced, and IPH groups, whereas that of EIF2S1 and FBLN5 was lower. Similarly, the expression of these genes was assessed in clusters A and B, and the results showed that, with the exception of EIF2S1, the expression of the remaining four genes varied across clusters. Among them, the expression level of IDH1, PRKCD, and CD36 was up-regulated in cluster A, whereas that of FBLN5 was up-regulated in cluster B (Fig. 3G, H). Our previous analysis established that cluster A was associated with high levels of immune cell infiltration (Fig. 2D). Therefore, the immune cell characteristics of these five genes were investigated. Differences in the expression level of IDH1, PRKCD, FBLN5, and CD36 led to changes in the proportion of most immune cell infiltration. Meanwhile, EIF2S1 was not associated with immune cell infiltration (Fig. 4A–E). Further analysis revealed that the expression levels of IDH1, PRKCD, and CD36 were positively correlated with immune infiltration. According to the expression heatmap which shows correlation between IDH1, PRKCD, FBLN5, CD36, EIF2S1 and 23 immune cells, IDH1 had the strongest positive correlation with T follicular helper cells, while the expression levels of FBLN5 was negatively correlated with immune infiltration. Besides, FBLN5 had the strongest negative correlation with activated dendritic cells (Fig. 4F). GO analysis was performed for IDH1, EIF2S1, PRKCD, FBLN5, and CD36 and determined that these genes were mainly involved in response to oxidative stress, cellular response to oxidative stress, response to stress, lipid metabolic process, and immune response (Fig. 5A). At the same time, KEGG analysis showed that IDH1, PRKCD, and CD36 were involved in insulin resistance, 2-Oxocarboxylic acid metabolism, adipocytokine signaling pathway PPAR signaling pathway, and peroxisome (Fig. 5B).
Fig. 4.
A: Infiltration characteristics of IDH1 and 23 immune cells. B: Infiltration characteristics of PRKCD and 23 immune cells. C: Infiltration characteristics of FBLN5 and 23 immune cells. D: Infiltration characteristics of CD36 and 23 immune cells. E: Infiltration characteristics of EIF2S1 and 23 immune cells. F: Expression heatmap which shows correlation between IDH1, PRKCD, FBLN5, CD36, EIF2S1 and 23 immune cells. * p-value < 0.05; ** p-value < 0.01; *** p-value < 0.001; **** p-value < 0.0001
Fig. 5.
A: GO analysis of oxidative stress prediction model genes. B: KEGG analysis of oxidative stress prediction model genes. C: ROC analysis of oxidative stress prediction model in the GSE43292 dataset (including 32 carotid plaques and 32 control samples). D: ROC analysis of the oxidative stress prediction model in the GSE28829 dataset (16 advanced and 13 early carotid plaques). E: ROC analysis of oxidative stress prediction model in the GSE16315 dataset (including 27 IPH samples and 16 non-IPH samples); F: tSNE plot of the carotid plaque group
Validation of oxidative stress prediction models
Based on GSE28829, GSE163154 and GSE43292, ROC analysis delineated the high discriminatory power of the model in identifying carotid atherosclerosis (Fig. 5C), plaque stability (Fig. 5D), and carotid plaque progression (Fig. 5E), with its predictive accuracy being substantially higher than that of individual genes. This finding signified that the model and the model genes were related to not only the occurrence of carotid atherosclerosis but also plaque stability and progression.
The relationship between the development and progression of carotid atherosclerosis and the abnormal expression of key genes in macrophages
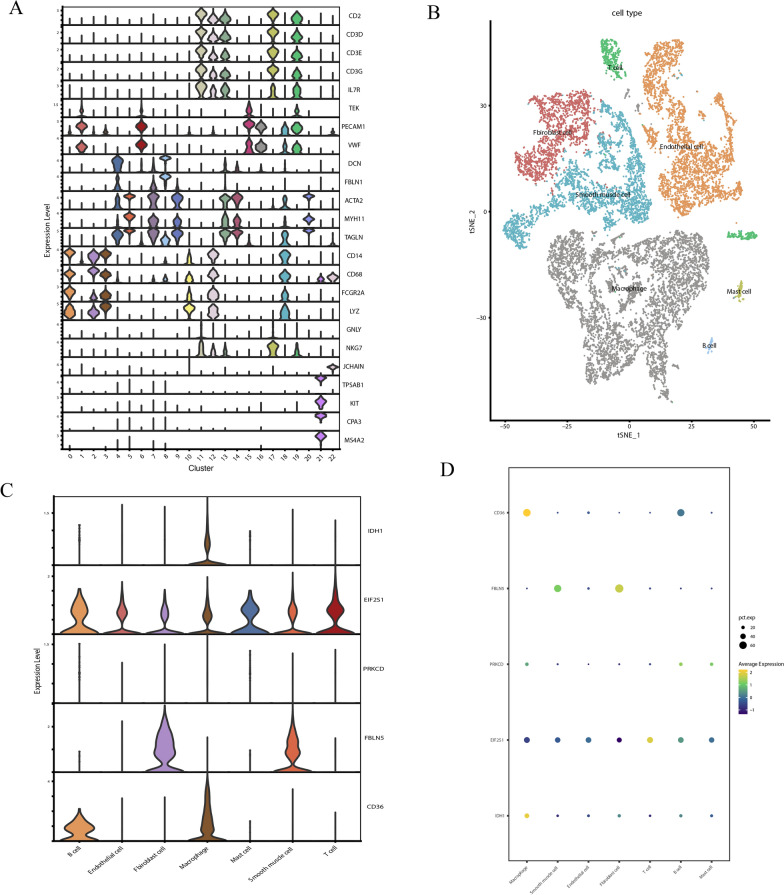
Based on GSE159677, cluster analysis performed on single-cell data identified 23 clusters at a resolution of 0.6 (Fig. 5F, Supplementary Fig. 3A, B). According to the relevant markers, the cells were divided into T cells (CD2, CD3D, CD3E, CD3G, TEK, and IL7R), NK cells (GNLY and NKG7), endothelial cells (PECAM1 and VWF), fibroblasts (DCN and FBLN1), vascular smooth muscle cells (ACTA2, MTH11, and TAGLN), macrophages (CD14, CD68, FCGR2A, and LYZ), B cells (JCHAIN), and mast cells (TPSAB1, KIT, CPA3, and MS4A2) (Fig. 6A, B). Supplementary Fig. 3C presents the top genes in the 23 clusters. Then, the expression of five oxidative stress genes (IDH1, EIF2S1, PRKCD, FBLN5, and CD36) was detected, revealing that CD36 and IDH1 were highly expressed in macrophages, FBLN5 was abundantly expressed in fibroblasts and smooth muscle cells, EIF2S1 was mainly expressed in T cells, and PRKCD expression was not expressed in all cell types (Fig. 6C, D).
Fig. 6.
A: Visualization of marker gene expression in each cell type. B: Cluster analysis of carotid plaque groups. C: Graph of IDH1, PRKCD, FBLN5, CD36, and EIF2S1 expression in each cell population. D: Dot plot of IDH1, PRKCD, FBLN5, CD36, EIF2S1 expression in each cell population
The comparison between calcified atherosclerotic core (AC) plaques and the patient-matched proximal adjacent portions of carotid artery (PA) showed the most pronounced difference was in the macrophage cluster, with macrophage counts being significantly higher in the plaque group (Supplementary Fig. 3B). Importantly, when comparing the expression of IDH1, EIF2S1, PRKCD, FBLN5, and CD36 between the two groups, it was found that IDH1, PRKCD, CD36, and EIF2S1 showed an upward trend in the plaque group, while FBLN5 exhibited a downward trend. (Supplementary Fig. 3D). Given the association between IDH1, PRKCD, and CD36 with high levels of immune cell infiltration, they may potentially be related to macrophage clusters in the plaque group. Moreover, the expression of IDH1 and CD36 was assessed in the plaque group, demonstrating that they were highly expressed in macrophage clusters, which was in line with the results of previous studies (Supplementary Fig. 3E).
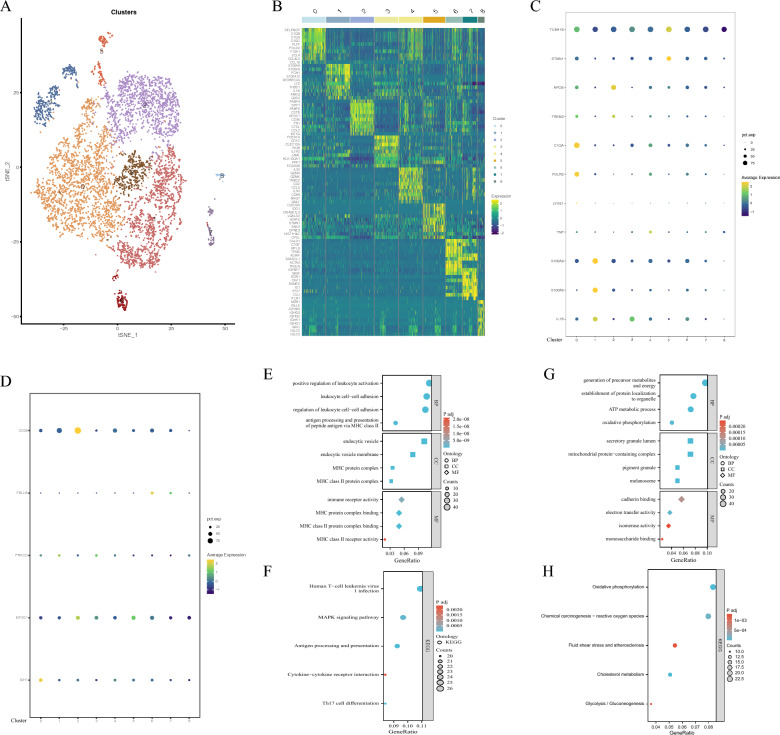
Our previous results demonstrated highly expressed IDH1 and CD36 may be related to macrophage infiltration in carotid atherosclerosis. To further investigate the relationship between them, macrophages were further analyzed. We re-clustered macrophages to further explore the roles of IDH1 and CD36 in macrophage subpopulations (Fig. 7A). Figure 7B presents the top genes in each cluster. Subgroup analysis of macrophages according to relevant marks demonstrated that they can roughly be divided into inflammatory macrophages (clusters 1, 3, 4, 6, and 7), resident-like macrophages (cluster 0), foam macrophages (clusters 1, 2, 3, 4, 6, and 7), and proliferating macrophages (cluster 5) (Fig. 7C). CD36 and IDH1 were significantly expressed in clusters 2 and 0, respectively (Fig. 7D). Cluster 2 comprised foam macrophages, and CD36 has been shown to be related to lipid phagocytosis by macrophages, consistent with our results. In addition, cluster 0 represented resident-like macrophages, and the apparent expression of IDH1 in this cluster drove the generation of foam cells by promoting macrophage ferroptosis, a crucial step in the pathogenesis of atherosclerosis. In addition, GO and KEGG analyses were performed on cluster 0 (Fig. 7E, F), demonstrating that cluster 0 was involved in positive regulation of leukocyte activation, leukocyte cell–cell adhesion, gene expression, regulation of leukocyte cell–cell adhesion, human T-cell, leukemia virus 1 infection, and other immune response processes. GO and KEGG analyses were performed on cluster 2, unveiling that it participated in the generation of precursor metabolites and energy, the establishment of protein localization to organelle, oxidative phosphorylation, chemical carcinogenesis- reactive oxygen species, and other metabolism processes (Fig. 7G, H).
Fig. 7.
A: tSNE plots of macrophage subsets. B: Heat map of differentially expressed genes in each macrophage cluster. C: Dot plot of marker genes in each cluster of macrophages. D: Dot plot of IDH1, PRKCD, FBLN5, CD36, and EIF2S1 expression in each cluster of macrophages. E: GO analysis of cluster 0. F: KEGG analysis of cluster 0. G: GO analysis of cluster 2. H: KEGG analysis of cluster 2
Discussion
Increased oxidative stress and systemic inflammation are risk factors for carotid atherosclerosis. Recent studies have evinced that hyperlipidemia, smoking, and lack of physical activity, which are risk factors for atherosclerosis, drive oxidative stress and inflammation [39, 40]. Increased oxidative stress and inflammation lead to endothelial cell dysfunction, shifting macrophages to the pro-inflammatory phenotype, and vascular smooth muscle cell proliferation, which further accelerate the progression of carotid atherosclerosis and plaque formation.
Our study comprehensively analyzed the expression of oxidative stress-related genes and immune-inflammatory profiles in carotid atherosclerotic samples in silico. In agreement with the results of existing studies, the expression of oxidative stress-related genes was significantly higher in carotid atherosclerosis samples. Additionally, the samples were clustered according to the differentially expressed oxidative stress genes. The results showed that the samples could be well distinguished based on the expression of oxidative stress genes, and the infiltration level of the 23 immune inflammation-related cells varied across oxidative stress clusters. Among them, the difference in macrophages was more pronounced. Macrophages play a central role in atherosclerosis as regulators of inflammation. Activated macrophages and foam cells are implicated in the formation of atherosclerotic plaques, with the former promoting plaque necrosis and the thinning of protective collagen scars (fibrous caps) [40]. Ginhoux et al. demonstrated [41] that macrophages in plaques can adopt highly inflammatory characteristics, leading to tissue destruction, while M0 macrophages are non-activated macrophages that can differentiate into pro-inflammatory M1 or anti-inflammatory M2 macrophages following exposure to inflammatory factors [42, 43]. Therefore, oxidative stress may play a key role in the development of atherosclerosis by inducing macrophage differentiation. It is worth noting that activated CD4 T cells and activated CD8 T cells also showed strong heterogeneity. Previous studies have concluded that a large proportion of T cells in atherosclerotic plaques exhibit a memory phenotype. After stimulation by antigens, stimuli, and cytokines, these CD4 T cells differentiate into various T cell subsets and participate in immune and inflammatory processes [44, 45]. CD8 T cells are not as abundant as CD4 T cells in atherosclerotic plaque, but the content of the former markedly increases in severe atherosclerotic plaque lesions, signaling that CD8 T cells are closely related to inflammation and plaque progression. Earlier studies have pointed out that CD8 T cells contribute to inflammation and necrotic cores within plaques, which can lead to plaque instability and rupture, thereby promoting the development of severe cardiovascular and cerebrovascular diseases [46]. Taken together, these findings suggest that T cells may be involved in the formation and progression of atherosclerotic plaques caused by oxidative stress, which is consistent with the results of the present study.
Herein, the random forest method was used to construct an oxidative stress model incorporating five oxidative stress-related genes (IDH1, EIF2S1, PRKCD, FBLN5, and CD36) to predict the risk of carotid atherosclerosis. Of note, a retrospective study showed that polymorphisms in IDH1 were negatively associated with the development of ischemic brain damage in patients undergoing carotid endarterectomy, while another study determined that IDH1 was associated with oscillatory shear stress on endothelial progenitor cells [47, 48]. Hyperlipidemia and diabetes can increase the expression of PRKCD in monocytes, which has been shown to alter macrophage activity and foam cell formation by regulating PI3K (phosphatidylinositol 3-kinases) / PKB (protein kinase B) and ERK (extracellular signal-regulated kinase) expression. Interestingly, mechanical stress can up-regulate PRKCD expression and enhance the proliferative and migratory abilities of vascular smooth muscle cells, suggesting that PRKCD may be a potential target for the treatment of atherosclerosis [49–51]. Smoking is an important risk factor for atherosclerosis. Zhou et al. demonstrated that nicotine up-regulates the expression of CD36 and peroxisome proliferator-activated receptor-γ (PPARγ) in macrophages to accelerate the development of atherosclerosis. Similarly, Lin et al. evinced that PRKCD can regulate the expression of CD36 in macrophages and exert a coordinated effect [50, 52]. Extracellular superoxide dismutase (ecSOD) plays an instrumental role in atherosclerosis and endothelial function by governing the level of superoxide anion (O2−) in the extracellular space. Nguyen et al. [53] identified fibulin-5 as the major binding protein of ecSOD. Its interaction with fibulin-5 is a prerequisite for the binding of ecSOD to vascular tissue, which regulates vascular O2− levels. Further studies have found that ecSOD-fibulin-5 interactions can regulate vascular redox status in extracellular space, which is a novel mechanism that mediates the development of various cardiovascular disorders, including atherosclerosis [53]. Although EIF2S1 was not differentially expressed in the oxidative stress cluster, it was identified as a differential gene in carotid atherosclerosis. However, studies on EIF2S1 and atherosclerosis are scarce. Nevertheless, previous studies have reported that EIF2S1 is an endoplasmic reticulum stress gene that can predict the prognosis of hepatocellular carcinoma [54]. Moreover, EIF2S1-mediated chronic ER stress promotes placental malformation, which is associated with adverse pregnancy outcomes [55]. Importantly, Bai et al. found that the dephosphorylation of EIF2S1 contributes to ROS accumulation and insulin resistance sensitivity in triple-negative breast cancer cells by disrupting activating transcription factor 4 (ATF4) -mediated glutathione biosynthesis [56]. These studies highlight the role of EIF2S1 in oxidative stress and cell damage [54–56]. We constructed an oxidative stress model, which was initially applied to CAS samples. The results demonstrated high discriminatory power, and DCA analysis was used to evaluate the necessity and benefits of treatment, providing a theoretical reference for clinical decision-making. In addition, the oxidative stress model was also validated across two types of samples. As expected, the oxidative stress model was highly accurate for predicting the risk of carotid atherosclerosis and plaque progression and stability in the two types of samples. The above-mentioned results also corroborated that oxidative stress genes play a crucial role in carotid atherosclerosis.
Next, the single-cell data derived from calcified atherosclerotic core (AC) plaques and patient-matched proximal adjacent (PA) portions of carotid artery were investigated. Following normalization and quality control of samples, 23 clusters were identified. According to the relevant markers, these clusters were divided into 8 groups of cells (T cells, NK cells, endothelial cells, fibroblasts, vascular smooth muscle cells, macrophages, B cells, and mast cells). The macrophage and smooth muscle cell clusters were significantly enriched in the carotid plaque group. Macrophages are known to play a central role in atherosclerosis, and IDH1 and CD36 were observed to be enriched in macrophages. Therefore, subgroup analyses were performed on the macrophage clusters, and the expression of five oxidative stress genes (IDH1, EIF2S1, PRKCD, FBLN5, and CD36) was analyzed in each subgroup. The results showed that IDH1 and CD36 were highly expressed in resident macrophages and foam cells, respectively. Moreover, they were also highly expressed in macrophage clusters in the carotid plaque group. CD36 has been associated with phagocytosis of lipids by macrophages, which is consistent with our results. Macrophage foams are known to play a vital role in atherosclerosis progression owing to their strong affinity and uptake of oxidized LDL. However, following IDH mutation, the binding of CD36 with NRF-2 resulted in decreased GPX4 expression, a key enzyme participating in the clearance of lipid ROS [57]. This disruption also accelerated glutathione depletion, promoted ROS accumulation, increased oxidized LDL content, and promoted macrophage foam cell formation [58]. At the same time, the levels of ferrous ions were also increased, which further promoted the ferroptosis of macrophages and aggravated the formation of foam cells [59]. After foam cell death, lipids accumulate on the arterial wall, thereby exacerbating atherosclerotic lesions [60]
By the best of our knowledge, this study is the first to integrate transcriptomic data, single-cell sequencing, and machine learning algorithms to identify key oxidative stress-related genes and immune cell infiltration factors involved in the formation, progression, and stability of carotid plaques, thus enhancing our understanding of the pathophysiology of carotid atherosclerosis. Through single-cell resolution data analysis, we further examined expression patterns of key genes across various cell types, revealing that the high expression of IDH1 and CD36 in resident macrophages and foam cells may play a crucial role in influencing plaque stability [57, 61]. These findings not only deepen our understanding of the mechanisms by which oxidative stress and immune cells contribute to plaque instability but also provide a theoretical basis for potential therapeutic targets (e.g., IDH1 and CD36) for carotid atherosclerosis, laying the groundwork for future therapeutic strategies.
In addition, our predictive model, comprising five oxidative stress-related genes (IDH1, EIF2S1, PRKCD, FBLN5, and CD36), demonstrated significant predictive value for identifying carotid plaque stability and progression. Unlike single-gene biomarkers, this multi-gene model enhances prediction accuracy, aiding early screening of high-risk populations, supporting personalized treatment strategies, and showing promise for clinical application in early intervention and risk assessment of plaques. However, as a bioinformatics-based study, these results require further validation through prospective studies and clinical data to confirm the functionality of the key genes. Future studies should also gather more clinical data to improve the robustness and accuracy of the model developed.
Compared with existing studies, such as the model by Wang et al. [62], which used machine learning to predict carotid plaque progression without incorporating single-cell sequencing data, our study integrated single-cell data, allowing a more refined analysis of cell–cell interactions and specifically identifying the contributions of foam cells and resident macrophages to plaque instability. Additionally, our findings align with those of Wang et al. [62], who identified M1 macrophages as a primary factor in plaque instability. Our study not only confirmed the role of these genes in oxidative stress but also revealed their distinct expression patterns across various immune cell types, offering a comprehensive understanding of the regulatory mechanisms underlying carotid plaque stability.
This study may facilitate clinical translation. High expression levels of IDH1 and CD36 may serve as therapeutic targets for carotid atherosclerosis by mitigating oxidative stress and lipid accumulation, thereby slowing plaque progression and enhancing stability. The oxidative stress model demonstrated high discriminatory power in detecting and predicting carotid plaque progression, facilitating early identification of high-risk patients and enabling personalized treatment strategies. This study advances fundamental knowledge and informs clinical practice.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- CVDs
Cardiovascular diseases
- ROS
Reactive oxygen species
- TNF
Tumor necrosis factor
- IKK
Inhibitory kappa B kinase
- NF-κB
Nuclear factor-κB
- NEMO
NF-κB essential modulator
- Nox
NADPH oxidase
- iNOS
Inducible nitric oxide synthase
- nNOS
Neuronal nitric oxide synthase
- ssGSEA
Single sample gene set enrichment analysis
- PCA
Principal component analysis
- CAS
Carotid atherosclerotic plaque samples
- AC
Calcified atherosclerotic core plaques
- PA
Proximal adjacent portions of carotid artery
- IPH
Intraplaque hemorrhage
- non-IPH
Non-intraplaque hemorrhage
- GO
Gene ontology
- KEGG
Kyoto encyclopedia of genes and genomes
- SVM
Support vector machine
- ROC
Receiver operating characteristic
- DCA
Decision curve analysis
- tSNE
T-distributed stochastic neighbor embedding
- UMAP
Uniform manifold approximation and projection
- PPAR
Peroxisome proliferator-activated receptors
- AUC
Area under the curve
- PI3K
Phosphatidylinositol 3-kinases
- PKB
Protein kinase B
- ERK
Extracellular signal-regulated kinase
- ecSOD
Extracellular superoxide dismutase
- ATF4
Activating transcription factor 4
Author contributions
D.M and Y.Y performed the data analysis and wrote the manuscript.
Funding
Not applicable.
Availability of data and materials
The data are available in Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) and AmiGO 2 website (https://amigo.soybase.org/amigo/amigo/landing).
Declarations
Ethics approval and consent to participate.
Not applicable.
Consent for publication.
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.World Health Organization. Health topics, Cardiovascular diseases (CVDs). Available online at: https://www.who.int/health-topics/cardiovascular-diseases. Accessed Mar 22, 2023.
- 2.Libby P. Inflammation in atherosclerosis. Nature. 2002;420(6917):868–74. 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 3.Kanter JE, Kramer F, Barnhart S, et al. Diabetes promotes an inflammatory macrophage phenotype and atherosclerosis through acyl-CoA synthetase 1. Proc Natl Acad Sci USA. 2012;109(12):E715–24. 10.1073/pnas.1111600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golledge J, Greenhalgh RM, Davies AH. The symptomatic carotid plaque. Stroke. 2000;31(3):774–81. 10.1161/01.str.31.3.774. [DOI] [PubMed] [Google Scholar]
- 5.GBD 2015 DALYs and HALE Collaborators. Global, regional, and national disability-adjusted life-years (DALYs) for 315 diseases and injuries and healthy life expectancy (HALE), 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388(10053):1603–58. 10.1016/S0140-6736(16)31460-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrington W, Lacey B, Sherliker P, et al. Epidemiology of atherosclerosis and the potential to reduce the global burden of atherothrombotic disease. Circ Res. 2016;118(4):535–46. 10.1161/CIRCRESAHA.115.307611. [DOI] [PubMed] [Google Scholar]
- 7.Tabas I, Williams KJ, Borén J. Subendothelial lipoprotein retention as the initiating process in atherosclerosis: update and therapeutic implications. Circulation. 2007;116(16):1832–44. 10.1161/CIRCULATIONAHA.106.676890. [DOI] [PubMed] [Google Scholar]
- 8.Camejo G, Lalaguna F, López F, Starosta R. Characterization and properties of a lipoprotein-complexing proteoglycan from human aorta. Atherosclerosis. 1980;35(3):307–20. 10.1016/0021-9150(80)90129-x. [DOI] [PubMed] [Google Scholar]
- 9.Ross R. Atherosclerosis—an inflammatory disease. N Engl J Med. 1999;340(2):115–26. 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 10.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473(7347):317–25. 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 11.Tabas I, García-Cardeña G, Owens GK. Recent insights into the cellular biology of atherosclerosis. J Cell Biol. 2015;209(1):13–22. 10.1083/jcb.201412052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montezano AC, Touyz RM. Reactive oxygen species and endothelial function—role of nitric oxide synthase uncoupling and Nox family nicotinamide adenine dinucleotide phosphate oxidases. Basic Clin Pharmacol Toxicol. 2012;110:87–94. [DOI] [PubMed] [Google Scholar]
- 13.Sies H, Berndt C, Jones DP. Oxidative stress. Annu Rev Biochem. 2017;86:715–48. 10.1146/annurev-biochem-061516-045037. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Horke S, Förstermann U. Oxidative stress in vascular disease and its pharmacological prevention. Trends Pharmacol Sci. 2013;34(6):313–9. 10.1016/j.tips.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Libby P. Current concepts of the pathogenesis of the acute coronary syndromes. Circulation. 2001;104(3):365–72. 10.1161/01.cir.104.3.365. [DOI] [PubMed] [Google Scholar]
- 16.Kaartinen M, Penttilä A, Kovanen PT. Mast cells of two types differing in neutral protease composition in the human aortic intima. Arterioscler Thromb. 1994;14(6):966–72. 10.1161/01.atv.14.6.966. [DOI] [PubMed] [Google Scholar]
- 17.Döring Y, Manthey HD, Drechsler M, et al. Auto-antigenic protein-DNA complexes stimulate plasmacytoid dendritic cells to promote atherosclerosis. Circulation. 2012;125(13):1673–83. 10.1161/CIRCULATIONAHA.111.046755. [DOI] [PubMed] [Google Scholar]
- 18.Liu F, Xia Y, Parker AS, Verma IM. IKK biology. Immunol Rev. 2012;246:239–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IκB-NF-κB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–5. [DOI] [PubMed] [Google Scholar]
- 20.Morgan MJ, Liu Z. Crosstalk of reactive oxygen species and NF-κB signaling. Cell Res. 2011;21(1):103–15. 10.1038/cr.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelisek J, Eckstein HH, Zernecke A. Pathophysiological mechanisms of carotid plaque vulnerability: impact on ischemic stroke. Arch Immunol Ther Exp. 2012;60(6):431–42. 10.1007/s00005-012-0192-z. [DOI] [PubMed] [Google Scholar]
- 22.Dunmore BJ, McCarthy MJ, Naylor AR, Brindle NP. Carotid plaque instability and ischemic symptoms are linked to immaturity of microvessels within plaques. J Vasc Surg. 2007;45(1):155–9. 10.1016/j.jvs.2006.08.072. [DOI] [PubMed] [Google Scholar]
- 23.Sluimer JC, Gasc JM, van Wanroij JL, et al. Hypoxia, hypoxia-inducible transcription factor, and macrophages in human atherosclerotic plaques are correlated with intraplaque angiogenesis. J Am Coll Cardiol. 2008;51(13):1258–65. 10.1016/j.jacc.2007.12.025. [DOI] [PubMed] [Google Scholar]
- 24.Chistiakov DA, Orekhov AN, Bobryshev YV. Contribution of neovascularization and intraplaque haemorrhage to atherosclerotic plaque progression and instability. Acta Physiol. 2015;213(3):539–53. 10.1111/apha.12438. [DOI] [PubMed] [Google Scholar]
- 25.Döring Y, Manthey HD, Drechsler M, Lievens D, et al. Auto-antigenic protein-DNA complexes stimulate plasmacytoid dendritic cells to promote atherosclerosis. Circulation. 2012;125(13):1673–83. 10.1161/CIRCULATIONAHA.111.046755. [DOI] [PubMed] [Google Scholar]
- 26.Jin H, Goossens P, Juhasz P, et al. Integrative multiomics analysis of human atherosclerosis reveals a serum response factor-driven network associated with intraplaque hemorrhage. Clin Transl Med. 2021;11(6): e458. 10.1002/ctm2.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayari H, Bricca G. Identification of two genes potentially associated in iron-heme homeostasis in human carotid plaque using microarray analysis. J Biosci. 2013;38(2):311–5. 10.1007/s12038-013-9310-2. [DOI] [PubMed] [Google Scholar]
- 28.Alsaigh T, Evans D, Frankel D, Torkamani A. Decoding the transcriptome of calcified atherosclerotic plaque at single-cell resolution. Commun Biol. 2022;5(1):1084. 10.1038/s42003-022-04056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hänzelmann S, Castelo R, Guinney A. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7): e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–3. 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, Carter SL, Getz G, Stemke-Hale K, Mills GB, Verhaak RG. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2(3):100141. 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liaw A, Wiener M. Classification and regression by random forest. R News. 2002;2(3):18–22. [Google Scholar]
- 35.Karatzoglou A, Smola A, Hornik K, Zeileis A. kernlab - An S4 Package for Kernel Methods in R. J Stat Softw. 2004;11(9):1–20. [Google Scholar]
- 36.Pfeiffer RM, Gail MH. Estimating the decision curve and its precision from three study designs. Biometrical J Biometrische Zeitschrift. 2020;62(3):764–76. 10.1002/bimj.201800240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong H, Hong S. simpleNomo: a python package of making nomograms for visualizable calculation of logistic regression models. Health Data Sci. 2023;3:0023. 10.34133/hds.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hao Y, Stuart T, Kowalski MH, Choudhary S, Hoffman P, Hartman A, Srivastava A, Molla G, Madad S, Fernandez-Granda C, Satija R. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat Biotechnol. 2024;42(2):293–304. 10.1038/s41587-023-01767-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. 2013;13(10):709–21. 10.1038/nri3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol. 2016;17(1):34–40. 10.1038/ni.3324. [DOI] [PubMed] [Google Scholar]
- 42.Frostegård J, Ulfgren AK, Nyberg P, Hedin U, Swedenborg J, Andersson U, Hansson GK. Cytokine expression in advanced human atherosclerotic plaques: dominance of pro-inflammatory (Th1) and macrophage-stimulating cytokines. Atherosclerosis. 1999;145(1):33–43. 10.1016/S0021-9150(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 43.Huang SC, Everts B, Ivanova Y, O’Sullivan D, Nascimento M, Smith AM, Beatty W, Love-Gregory L, Lam WY, O’Neill CM, Yan C, Du H, Abumrad NA, Urban JF Jr, Artyomov MN, Pearce EL, Pearce EJ. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol. 2014;15(9):846–55. 10.1038/ni.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tabas I, Lichtman AH. Monocyte-macrophages and T cells in atherosclerosis. Immunity. 2017;47(4):621–34. 10.1016/j.immuni.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kyaw T, Winship A, Tay C, Kanellakis P, Hosseini H, Cao A, Li P, Tipping P, Bobik A, Toh BH. Cytotoxic and proinflammatory CD8+ T lymphocytes promote development of vulnerable atherosclerotic plaques in apoE-deficient mice. Circulation. 2013;127(9):1028–39. 10.1161/CIRCULATIONAHA.112.001347. [DOI] [PubMed] [Google Scholar]
- 46.Kolbus D, Ramos OH, Berg KE, Persson J, Wigren M, Björkbacka H, Fredrikson GN, Nilsson J. CD8+ T cell activation predominate early immune responses to hypercholesterolemia in Apoe-/- mice. BMC Immunol. 2010;11:58. 10.1186/1471-2172-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vasuri F, de Biase D, Vacirca A, Acquaviva G, Sanza V, Gargiulo M, Pasquinelli G. Gene polymorphism in tissue epidermal growth factor receptor (EGFR) influences clinical and histological vulnerability of carotid plaques. Pathol Res Pract. 2022;229: 153721. 10.1016/j.prp.2021.153721. [DOI] [PubMed] [Google Scholar]
- 48.Yu J, Fu J, Zhang X, Cui X, Cheng M. The integration of metabolomic and proteomic analyses revealed alterations in inflammatory-related protein metabolites in endothelial progenitor cells subjected to oscillatory shear stress. Front Physiol. 2022;13: 825966. 10.3389/fphys.2022.825966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Q, Park K, Xia Y, Matsumoto M, Qi W, Fu J, Yokomizo H, Khamaisi M, Wang X, Rask-Madsen C, King GL. Regulation of macrophage apoptosis and atherosclerosis by lipid-induced PKCδ isoform activation. Circ Res. 2017;121(10):1153–67. 10.1161/CIRCRESAHA.117.311606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin CS, Lin FY, Ho LJ, Tsai CS, Cheng SM, Wu WL, Huang CY, Lian CH, Yang SP, Lai JH. PKCδ signalling regulates SR-A and CD36 expression and foam cell formation. Cardiovasc Res. 2012;95(3):346–55. 10.1093/cvr/cvs189. [DOI] [PubMed] [Google Scholar]
- 51.Li C, Wernig F, Leitges M, Hu Y, Xu Q. Mechanical stress-activated PKCdelta regulates smooth muscle cell migration. FASEB J. 2003;17(14):2106–8. 10.1096/fj.03-0150fje. [DOI] [PubMed] [Google Scholar]
- 52.Zhou MS, Chadipiralla K, Mendez AJ, Jaimes EA, Silverstein RL, Webster K, Raij L. Nicotine potentiates proatherogenic effects of oxLDL by stimulating and upregulating macrophage CD36 signaling. Am J Physiol Heart Circ Physiol. 2013;305(4):H563–74. 10.1152/ajpheart.00042.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen AD, Itoh S, Jeney V, Yanagisawa H, Fujimoto M, Ushio-Fukai M, Fukai T. Fibulin-5 is a novel binding protein for extracellular superoxide dismutase. Circ Res. 2004;95(11):1067–74. 10.1161/01.RES.0000149568.85071.FB. [DOI] [PubMed] [Google Scholar]
- 54.Liu P, Wei J, Mao F, Xin Z, Duan H, Du Y, Wang X, Li Z, Qian J, Yao J. Establishment of a prognostic model for hepatocellular carcinoma based on endoplasmic reticulum stress-related gene analysis. Front Oncol. 2021;11: 641487. 10.3389/fonc.2021.641487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Capatina N, Hemberger M, Burton GJ, Watson ED, Yung HW. Excessive endoplasmic reticulum stress drives aberrant mouse trophoblast differentiation and placental development leading to pregnancy loss. J Physiol. 2021;599(17):4153–81. 10.1113/JP281994. [DOI] [PubMed] [Google Scholar]
- 56.Bai X, Ni J, Beretov J, Wasinger VC, Wang S, Zhu Y, Graham P, Li Y. Activation of the eIF2α/ATF4 axis drives triple-negative breast cancer radioresistance by promoting glutathione biosynthesis. Redox Biol. 2021;43: 101993. 10.1016/j.redox.2021.101993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang TX, Liang JY, Zhang C, Xiong Y, Guan KL, Yuan HX. The oncometabolite 2-hydroxyglutarate produced by mutant IDH1 sensitizes cells to ferroptosis. Cell Death Dis. 2019;10:755. 10.1038/s41419-019-1995-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li B, Wang C, Lu P, Ji Y, Wang X, Liu C, Lu X, Xu X, Wang X. IDH1 promotes foam cell formation by aggravating macrophage ferroptosis. Biology (Basel). 2022;11(10):1392. 10.3390/biology11101392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo Y, Duan H, Qian Y, Feng L, Wu Z, Wang F, Feng J, Yang D, Qin Z, Yan X. Macrophagic CD146 promotes foam cell formation and retention during atherosclerosis. Cell Res. 2017;27:352–72. 10.1038/cr.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J, Zu Y, Dhanasekara CS, Li J, Wu D, Fan Z, Wang S. Detection and treatment of atherosclerosis using nanoparticles. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2017;9: e1412. 10.1002/wnan.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ben-Aicha S, Anwar M, Vilahur G, Martino F, Kyriazis PG, de Winter N, Punjabi PP, Angelini GD, Sattler S, Emanueli C. Small extracellular vesicles in the pericardium modulate macrophage immunophenotype in coronary artery disease. JACC Basic Transl Sci. 2024;9(9):1057–72. 10.1016/j.jacbts.2024.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, Kang Z, Liu Y, Li Z, Liu Y, Liu J. Identification of immune cell infiltration and diagnostic biomarkers in unstable atherosclerotic plaques by integrated bioinformatics analysis and machine learning. Front Immunol. 2022;13: 956078. 10.3389/fimmu.2022.956078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are available in Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) and AmiGO 2 website (https://amigo.soybase.org/amigo/amigo/landing).