INTRODUCTION
Pathologic response (PR) to preoperative chemotherapy has been shown to be associated with improved overall survival (OS) in patients who undergo hepatectomy for colorectal liver metastasis (CLM).1, 2 The role of biological factors, including RAS, TP53, APC, SMAD4, BRAF, and FBXW7, on the prognosis is well established, but the effect of these gene mutations on pathologic response has been scarcely studied.3 Only RAS alteration was previously reported to decrease the rate of PR.4 In this study, we assessed the effect of known gene alterations on PR and the correlation between gene mutations and PR on OS in patients with CLM.
PATIENTS AND METHODS
The Institutional Review Board at MD Anderson Cancer Center approved this study protocol (#2023-0050). From a prospectively maintained database, we collected data on patients who underwent initial R0 or R1 hepatectomy for CLM after receiving a maximum of 12 cycles of first-line preoperative chemotherapy between January 2004 and December 2020. Patients with missing data were excluded. Next-generation sequencing using tumor DNA from primary or CLM specimens was performed with an AmpliSeq cancer-related multigene panel including at least 46 genes using the Ion Torrent Personal Genome Machine (Life Technologies, Carlsbad, CA) in a Clinical Laboratory Improvement Amendment–certified molecular diagnostic laboratory. Major pathologic response (majorPR) was defined as tumor viability of less than 50%. Minor pathologic response (minorPR) was defined as tumor viability above 50%.2 Clinicopathologic and biologic factors associated with majorPR and OS were evaluated by multivariate analyses, using backward elimination with P < 0.05 to select variables. All statistical tests were 2-sided, and P < 0.05 was considered statistically significant.
RESULTS
Patient Characteristics
Median [interquartile range (IQR)] follow-up and OS of the whole cohort (N = 458) was 3.8 [2.3-5.4] and 6.9 [3.4–not reached] years, respectively. During the follow-up period, 167 patients (36.5%) died. MajorPR was achieved in 252 (55.0%) patients. Median [IQR] percentage of pathological tumor viability was significantly lower in patients with TP53 wild-type than those with TP53 alteration (30 [10–50] % vs 47 [24–70] %, P < 0.001) (Figure 1). Multivariate analysis revealed that oxaliplatin-containing regimen (risk ratio (RR): 2.54, 95% confidence intervals (CI): 1.58–4.07, P < 0.001), bevacizumab-containing regimen (RR: 2.15, 95%CI: 1.36–3.39, P = 0.001) and TP53 alteration (RR: 0.42, 95%CI: 0.27–0.66, P < 0.001) were independently associated with majorPR (Table 1).
Figure 1.
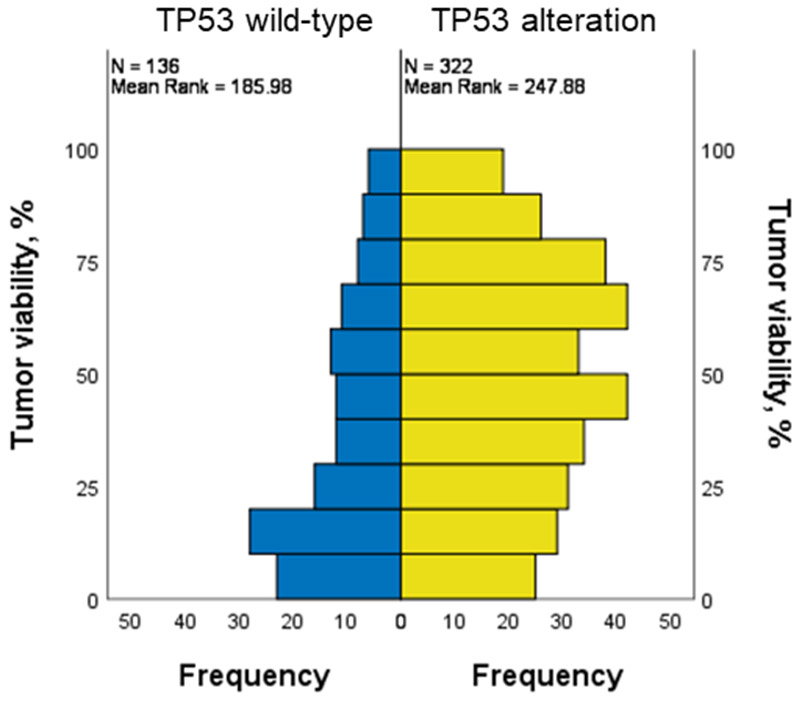
Histogram of pathological tumor viability categorized by TP53 alteration.
Table 1.
Factors associated with major pathologic response
| Factors | No. of Patients |
No. of Events |
Multivariable RR† |
95% CI | P value |
|---|---|---|---|---|---|
| Oxaliplatin-containing | |||||
| Yes | 353 | 213 | 2.54 | 1.58–4.07 | < 0.001 |
| No | 105 | 39 | 1 (referent) | ||
| Bevacizumab-containing | |||||
| Yes | 343 | 205 | 2.15 | 1.36–3.39 | 0.001 |
| No | 115 | 47 | 1 (referent) | ||
| TP53 alteration | |||||
| Yes | 322 | 161 | 0.42 | 0.27–0.66 | < 0.001 |
| No | 136 | 91 | 1 (referent) | ||
Abbreviations: RR, risk ratio; CI, confidence interval
The Binary logistic model analysis initially included age (continuous), sex, primary T stage (> T3 vs. ≤ T3), oxaliplatin-containing regimen, bevacizumab-containing regimen, RAS or BRAF, APC, TP53, PIK3CA, SMAD4 and FBXW7 alterations.
The cohort was divided into 3 groups: TP53 wild-type with majorPR (N = 91, 19.9%), TP53 alteration with majorPR (N = 161, 35.2%), and minorPR (N = 206, 45.0%). Five-year OS of the different groups is shown in Figure 2. Multivariate analysis revealed that patients in the TP53 wild-type with majorPR group (hazard ratio (HR): 0.49, 95%CI: 0.31–0.77, P = 0.002) and those in the TP53 alteration with majorPR group (HR: 0.70, 95%CI: 0.49–1.00, P = 0.048) had significantly better OS compared to those in the minor PR group (Table 2).
Figure 2.
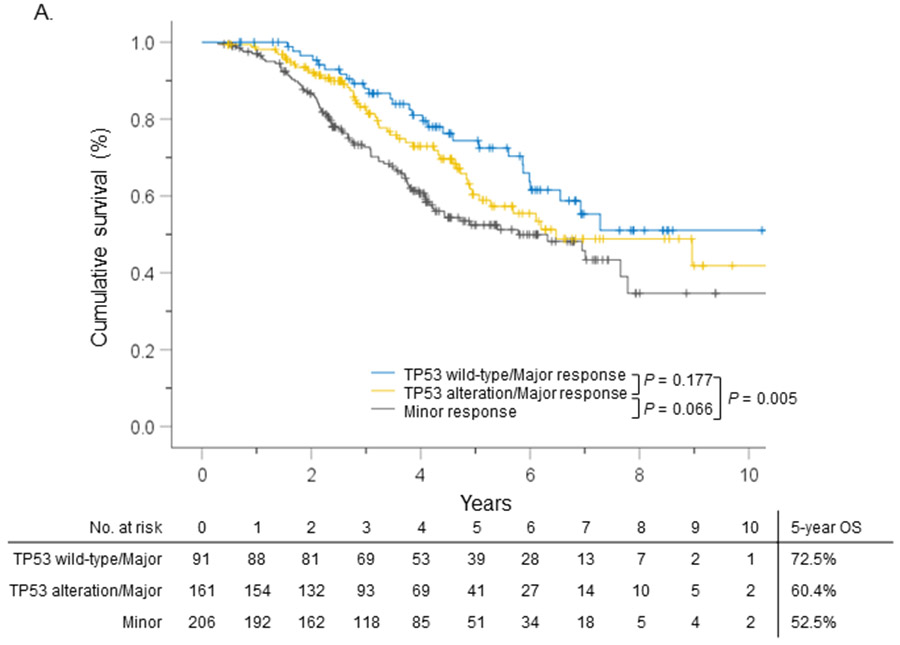
Overall survival (OS) stratified by TP53 alteration and pathologic response.
Table 2.
Factors associated with overall survival
| Factors | No. of Patients |
No. of Events |
Multivariable HR† |
95% CI | P value |
|---|---|---|---|---|---|
| Extrahepatic metastasis | |||||
| Yes | 101 | 48 | 1.63 | 1.15–2.33 | 0.007 |
| No | 357 | 119 | 1 (referent) | ||
| RAS or BRAF alteration | |||||
| Yes | 264 | 102 | 1.43 | 1.03–1.99 | 0.035 |
| No | 194 | 65 | 1 (referent) | ||
| SMAD4 alteration | |||||
| Yes | 50 | 26 | 1.96 | 1.26–3.03 | 0.003 |
| No | 408 | 141 | 1 (referent) | ||
| APC alteration | |||||
| Yes | 248 | 82 | 0.62 | 0.45–0.86 | 0.004 |
| No | 210 | 85 | 1 (referent) | ||
| Combination of TP53 alteration and PR | |||||
| TP53 wild-type/Major PR | 91 | 28 | 0.49 | 0.31–0.77 | 0.002 |
| TP53 alteration/Major PR | 161 | 52 | 0.70 | 0.49–1.00 | 0.048 |
| Minor PR | 206 | 87 | 1 (referent) |
Abbreviations: HR, hazard ratio; CI, confidence interval; PR, pathologic response;
The Cox proportional hazard model analysis initially included age (continuous), sex, primary T stage (> T3 vs. ≤ T3), primary lymph node metastasis, synchronous liver metastasis, extrahepatic metastasis, tumor size (continuous), tumor number (continuous), oxaliplatin-containing regimen, bevacizumab-containing regimen, surgical margin (R0 vs R1), posthepatectomy chemotherapy, RAS or BRAF, APC, PIK3CA, SMAD4 and FBXW7 alteration and combination of TP53 alteration and PR.
DISCUSSION
This is the first study showing TP53 alteration is associated with worse PR. This effect can be due to TP53-mediated resistance to chemotherapy as this has been shown in colorectal cancer cell lines.5 On the other hand, the statistical difference between the Kaplan-Meier curve (Figure 2) and multivariate analysis (Table 2) can be explained by the factors in the multivariate analysis such as chemotherapy regimen, co-existing mutations, or extrahepatic disease. Further studies targeting the combined association of TP53 and these factors with pathologic response and survival can help clarify the role of TP53 in CLM.6
ACKNOWLEDGMENTS
The authors thank Ms. Ruth Haynes for administrative support in the preparation of this manuscript.
FUNDING SUPPORT
Supported by the National Cancer Institute under award number P30CA016672, which supports the MD Anderson Cancer Center Clinical Trials Support Resource.
Footnotes
CONFLICT OF INTEREST DISCLOSURES
Nothing to disclose.
REFERENCES
- 1.Blazer DG 3rd, Kishi Y, Maru DM, Kopetz S, Chun YS, Overman MJ, Fogelman D, Eng C, Chang DZ, Wang H, Zorzi D, Ribero D, Ellis LM, Glover KY, Wolff RA, Curley SA, Abdalla EK, Vauthey JN. Pathologic response to preoperative chemotherapy: a new outcome end point after resection of hepatic colorectal metastases. J Clin Oncol 2008;26(33): 5344–5351. [DOI] [PubMed] [Google Scholar]
- 2.Rubbia-Brandt L, Giostra E, Brezault C, Roth AD, Andres A, Audard V, Sartoretti P, Dousset B, Majno PE, Soubrane O, Chaussade S, Mentha G, Terris B. Importance of histological tumor response assessment in predicting the outcome in patients with colorectal liver metastases treated with neo-adjuvant chemotherapy followed by liver surgery. Ann Oncol 2007;18(2): 299–304. [DOI] [PubMed] [Google Scholar]
- 3.Kawaguchi Y, Kopetz S, Kwong L, Xiao L, Morris JS, Tran Cao HS, Tzeng CD, Chun YS, Lee JE, Vauthey JN. Genomic Sequencing and Insight into Clinical Heterogeneity and Prognostic Pathway Genes in Patients with Metastatic Colorectal Cancer. J Am Coll Surg 2021;233(2): 272–284 e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmitti G, Shindoh J, Mise Y, Kopetz S, Loyer EM, Andreou A, Cooper AB, Kaur H, Aloia TA, Maru DM, Vauthey JN. RAS mutations predict radiologic and pathologic response in patients treated with chemotherapy before resection of colorectal liver metastases. Ann Surg Oncol 2015;22(3): 834–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer J, McLean EG, Aroori S, Wilson P, McCulla A, Carey PD, Longley DB, Johnston PG. Characterization of p53 wild-type and null isogenic colorectal cancer cell lines resistant to 5-fluorouracil, oxaliplatin, and irinotecan. Clin Cancer Res 2004;10(6): 2158–2167. [DOI] [PubMed] [Google Scholar]
- 6.Michel M, Kaps L, Maderer A, Galle PR, Moehler M. The Role of p53 Dysfunction in Colorectal Cancer and Its Implication for Therapy. Cancers (Basel) 2021;13(10). [DOI] [PMC free article] [PubMed] [Google Scholar]


