Molecular markers provide an important method for detecting tuberculosis (TB) transmission that has been followed by rapid progression to active disease.1 Restriction fragment length polymorphism (RFLP) typing of Mycobacterium tuberculosis has been used to confirm outbreaks, through the demonstration of shared isolates.2 In population-based studies, it has therefore been inferred that patients with TB whose infecting organisms are characterized by shared RFLP “patterns” (“fingerprints”) have likely been involved in recent transmission.3,4 We used public health reporting data and RFLP-based DNA fingerprinting to determine the extent to which ongoing transmission contributes to TB incidence in Montreal and to evaluate factors associated with such TB transmission.
RFLP analysis using the insertion sequence (IS) 61105 was used to classify the 303 culture-positive cases of TB reported in Montreal during 1997 and 1998. We restricted the analysis to those isolates with 5 or more bands, because isolates with few or no copies of the IS6110 element cannot be reliably classified by this method.5 Reactivation of remotely acquired tuberculous infection was considered likely if the RFLP pattern was unique within the database. Recent transmission was considered likely if an isolate matched at least one other by identical, or near-identical, criteria. “Identical” isolates were characterized by identical numbers of bands on gel electrophoresis, following digestion by the restriction endonuclease; all such bands had to have matching molecular weights. “Near-identical” isolates were characterized by a single band difference (a single band addition, band loss or band shift). This study was approved by the research ethics committee of the Montreal General Hospital, which is responsible for the infectious diseases unit of the Montreal public health department.
We obtained RFLP results for 288 of the culture- positive cases. We found that 243 isolates had at least 5 bands: 183 unique isolates, 43 near-identical isolates and 17 identical isolates. The 17 identical isolates belonged to 7 “clusters” of 2 or 3 people each. The 60 identical and near-identical isolates belonged to 16 clusters of 2 to 13 people each. Individuals with unique and matched isolates did not differ with respect to age, sex, proportion who were foreign-born and a number of clinical characteristics (Table 1).
Table 1
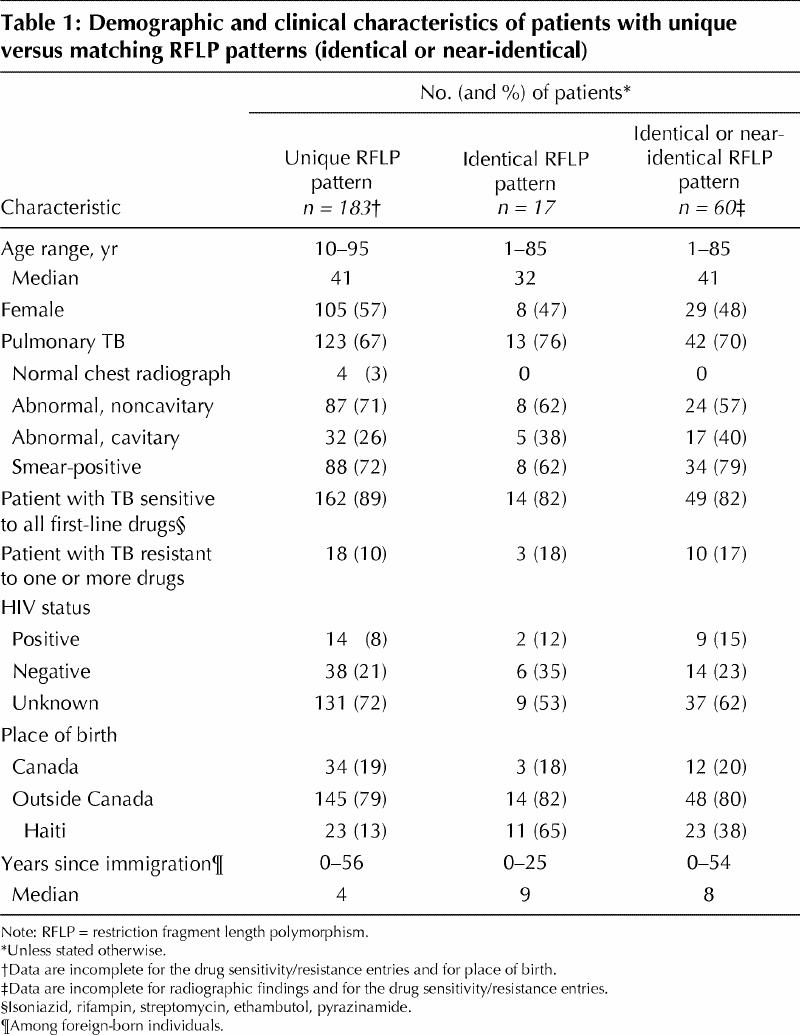
The only characteristic associated with matching RFLP patterns was Haitian birth; Haitian-born individuals accounted for 19% of isolates with adequate fingerprints, but 65% of the identical isolates and 37% of the identical and near-identical isolates together. The crude odds ratio estimates for matching isolates, comparing Haitian-born with non-Haitian-born individuals, were 9.7 (95% confidence interval [CI] 3.0–34.0) when identical RFLP pattern was used as the matching criterion, and 3.9 (95% CI 1.9–8.2) when both identical and near-identical RFLP patterns were considered. Potential explanations for this finding include the following: (1) limited genetic diversity among M. tuberculosis organisms originally acquired in Haiti, (2) involvement in new transmission in Montreal or (3) accelerated progression to active disease. However, only 5 of 23 Haitian-born individuals with identical or near-identical isolates were HIV-seropositive, making the last explanation unlikely.
We assumed that one person per “cluster” had reactivated latent TB and then transmitted it to the others. The proportion of disease due to recent transmission was therefore estimated to be 4%, that is, (17 – 7)/243 (95% CI 2%–7%) or 18%, that is, (60 – 16)/243 (95% CI 13%–24%), depending on the matching criterion used: identical RFLP patterns or identical and near-identical patterns respectively.
Our 2-year study in Montreal suggested that 82%–96% of incident active TB cases were the result of reactivation. Secondary active cases of TB appear uncommon in Montreal, which is likely the result of a well-functioning program of contact investigation and management.6 These results are consistent with recent US7 and Canadian8 recommendations: where there is little ongoing spread of disease, further reductions in TB incidence will require increased emphasis on the treatment of latent infection.
β See related articles pages 349 and 355
Acknowledgments
We wish to thank Ms. Elizabeth Hamer, Ms. Naomi Koerner and Dr. May Zaoudé for their assistance with data collection and processing. We also wish to thank Michael Purdy for his patient assistance in preparing the fingerprint files prior to analysis.
Footnotes
This article has been peer reviewed.
Contributors: Ms. Kulaga participated in all phases of the study design and data collection, conducted the statistical analyses reported here and served as primary author of the manuscript. Dr. Behr played a key role in the design and conduct of this study, supervised all molecular typing and contributed extensively to the manuscript. He also secured funding for the molecular typing. Dr. Musana aided in study design, collected the initial epidemiological data, performed a number of preliminary analyses and contributed revisions to the manuscript. Ms. Brinkman conducted all DNA extraction, molecular typing and formatting of the molecular data. She also critically reviewed the manuscript. Dr. Menzies first developed this research project as well as the necessary collaborations, secured initial funding, handled much of the data and extensively revised the manuscript. Dr. Brassard extracted the necessary data from the public health database, suggested analyses and contributed revisions to the manuscript. Dr. Kunimoto provided initial molecular typing data and contributed revisions to the manuscript. Dr. Tannenbaum was responsible for tuberculosis reporting and control at the public health department at the time of the study. She contributed extensively to the conception and conduct of the study and provided revisions to the manuscript. Ms. Thibert was responsible for initial laboratory management of all tuberculosis isolates reported in the manuscript. This includes identification, drug susceptibility testing, as well as preparation of relevant isolates for DNA extraction. She also critically reviewed the manuscript. Dr. Joseph aided in study protocol development and supervision of statistical analyses. He also contributed revisions to the manuscript. Dr. Boivin aided in the study protocol development and contributed revisions to the manuscript. Dr. Kevin Schwartzman developed the initial study protocol, supervised all phases of the epidemiologic data collection and analysis, and supervised preparation of the manuscript, including extensive revision. He also secured funding for the epidemiologic data collection.
This project was funded by the Association pulmonaire du Québec and by the Canadian Institutes of Health Research. Ms. Kulaga was supported by a McGill Faculty of Medicine Studentship Award. Dr. Behr is the recipient of a New Investigator career award from the Canadian Institutes of Health Research. Dr. Musana was supported by a Commonwealth Foundation Scholarship. Dr. Menzies is the recipient of an Investigator career award from the Canadian Institutes of Health Research. Dr. Kunimoto was an Alberta Heritage Foundation for Medical Research Scholar. Dr. Joseph is the recipient of a Senior Investigator career award from the Canadian Institutes of Health Research. Dr. Schwartzman is the recipient of a Chercheur-boursier clinicien career award from the Fonds de la recherche en santé du Québec.
Competing interests: None declared.
Correspondence to: Dr. Kevin Schwartzman, Respiratory Epidemiology Unit ,1110 Pine Ave. W, Montreal QC H3A 1A3; fax 514 398-8981; kevin.schwartzman@mcgill.ca
References
- 1.Kulaga S, Behr MA, Schwartzman K. Genetic fingerprinting in the study of tuberculosis transmission. CMAJ 1999;161(9):1165-9. [PMC free article] [PubMed]
- 2.Daley CL, Small PM, Schecter GF, Schoolnik GK, McAdam RA, Jacobs WR Jr, et al. An outbreak of tuberculosis with accelerated progression among persons infected with the human immunodeficiency virus. An analysis using restriction-fragment-length polymorphisms. N Engl J Med 1992;326(4):231-3. [DOI] [PubMed]
- 3.Small PM, Hopewell PC, Singh SP, Paz A, Parsonnet J, Ruston DC, et al. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N Engl J Med 1994; 330 (24): 1703-9. [DOI] [PubMed]
- 4.Alland D, Kalkut GE, Moss AR, McAdam RA, Hahn JA, Bosworth W, et al. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N Engl J Med 1994; 330(24):1710-6. [DOI] [PubMed]
- 5. van Soolingen D, de Haas PEW. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology. J Clin Microbiol 1993;31:1897-95. [DOI] [PMC free article] [PubMed]
- 6.Dasgupta K, Schwartzman K, Marchand R, Tannenbaum T, Brassard P, Menzies D. Comparison of cost-effectiveness of tuberculosis screening of close contacts and foreign-born populations. Am J Respir Crit Care Med 2000;162:2079-86. [DOI] [PubMed]
- 7.American Thoracic Society. Targeted tuberculin testing and treatment of latent tuberculosis infection. Am J Respir Crit Care Med 2000;161:S221-47. [DOI] [PubMed]
- 8.Long R, editor. Canadian tuberculosis standards. 5th ed. Canadian Lung Association, Canadian Thoracic Society, Health Canada; 2000. Available: www.hc-sc.gc.ca/hpb/lcdc/publicat/cts-ncla00 (accessed 2002 June 13).


