Abstract
This research delves into the therapeutic implications of utilizing small interfering RNA (siRNA) to target the ribosomal protein S19 (RPS19) gene in chronic myeloid leukemia (CML) using the K562 cell line model. The primary objective was to investigate how gene silencing affects apoptosis promotion and cell cycle arrest. The study employed bioinformatics tools and databases to explore the interactions involving RPS19 and neighboring proteins. Subsequently, siRNA-mediated gene silencing was utilized to suppress RPS19 expression in K-562 cells, with assessments conducted on cell cycle progression and apoptosis through flow cytometry analysis. Furthermore, real-time PCR was employed to evaluate the expression levels of RPS19, along with the closely associated RPS16 and RPS18 genes. Silencing the RPS19 gene in siRNA-transfected K-562 cells led to an increase in apoptotic cells by over 20%, with a significant accumulation in the sub-G1 and G1 phases. Additionally, the knockdown of RPS19 resulted in a 75% decrease in RPS16 expression and a 50% decrease in RPS18 expression. These results demonstrate the therapeutic potential of targeting RPS19 in CML cells, suggesting a promising approach for precise treatment strategies in leukemia and potentially other types of cancer.
Key Words: RPS19, siRNA, chronic myeloid leukemia, gene silencing, cell cycle, apoptosis
Introduction
Chronic myeloid leukemia (CML) is a form of blood cancer characterized by the presence of the Philadelphia chromosome, a genetic anomaly resulting from a chromosomal exchange between chromosomes 9 and 22 (1). This genetic rearrangement leads to the formation of the breakpoint cluster region-Abelson murine leukemia viral oncogene homolog (BCR-ABL) fusion gene, which generates an overactive tyrosine kinase that stimulates the uncontrolled growth of myeloid cells (2). Targeted therapies have significantly improved the prognosis and quality of life for individuals with CML (3).
siRNA therapy shows promise in treating various cancers by selectively targeting and silencing genes involved in cancer progression. This precise gene silencing method offers a potential strategy for inhibiting the expression of oncogenes and key genes driving tumor development (4). By delivering siRNA molecules precisely to cancer cells, crucial pathways essential for cancer cell survival, proliferation, and metastasis can be disrupted, potentially enhancing the effectiveness of traditional cancer treatments and overcoming drug resistance mechanisms (5).
The RPS19 gene, an essential element of ribosomal protein S19, holds a key position in protein production and cellular operations. Changes or irregularities in the RPS19 gene have been associated with a range of conditions, such as Diamond-Blackfan anemia and specific cancer types (6). Research has revealed that modifications in RPS19 gene activity can influence cellular development, replication, and specialization, underscoring its importance in regular bodily functions and the onset of diseases (7). Investigating the complex mechanisms involved in the RPS19 gene is crucial to discover its potential therapeutic applications and progress toward precise interventions for related disorders. The increased expression of RPS19 has been associated with various types of cancer, playing a significant role in the onset and progression of the disease (8). RPS19, an essential protein in the ribosome responsible for protein production, has been found to be elevated in multiple forms of cancer, including breast (9), colorectal (10), and prostate (11). These heightened RPS19 levels have been linked to enhanced cell growth, resistance to cell death, and promotion of cancer cell metastasis. Exploring the mechanisms behind the upregulation of RPS19 in cancer could provide valuable insights for developing targeted therapies that aim to control RPS19 expression and disrupt the pathways that support cancer cell proliferation and survival.
Through our research, we aimed to elucidate the impact of RPS19 gene silencing on promoting cell apoptosis, inducing cell cycle arrest, and altering the expression levels of key RPS19-related genes, including RPS16 and RPS18. By understanding the underlying mechanisms of RPS19 gene knockdown in CML cells using the K562 (12) cell line model, we aimed to make significant contributions to the advancement of innovative and precise treatment approaches for addressing leukemia and potentially other forms of cancer.
Materials and methods
Bioinformatics Analysis
In this research, we utilized PyMol software to examine the amino acids involved in the interactions between the RPS19 protein and other proteins within the 40S complex, along with LigPlot. Additionally, we utilized the STRING database to identify potential proteins interacting with RPS19. To identify key proteins directly interacting with RPS19 in the ribosomal 40S complex, we carefully analyzed Protein Data Bank (PDB) files related to the structure of the 40S subunit in various modes. These files can be accessed in the PDB database under the access numbers 6G4W, 6G4S, 6G18, 6G53, 6G5I, 6G5H, 6G51, and 6ZVH (http://www.uniprot.org).
Cell Culture
To assess the impact of RPS19 gene silencing on K-562 lymphoblast cells, the cell line was procured from the Pasteur Institute of Iran in Tehran. The cells were initially grown in RPMI culture medium supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 units/mL streptomycin. They were then incubated for 24 hours at 37°C in a humidified atmosphere with 5% CO2. Upon reaching 80% confluency, the cells were either sub cultured or utilized for transfection procedures.
Transfection Process
The siRNA oligonucleotide designed to target the RPS19 gene (Forward: GCUCCCUACGAUGA GAAC; Reverse: AGUUCUCAUCGUAGGGAG) was acquired from BIONEER Company in Korea (catalog No. QF-210204-0008). For cell preparation prior to transfection, 80,000 K-562 cells were plated in each well of a 24-well plate with 500 µl of culture medium devoid of serum and antibiotics. Subsequently, 100 µl of the transfection suspension was introduced into each well, followed by a 3-hour incubation period to ensure effective transfection. After incubation, 10% FBS (fetal bovine serum) was added to the cells without any antibiotics to prevent potential interference with the transfection process mediated by lipofectamine. To allow sufficient time for lipofectamine to aid in siRNA entry into the cells, an additional 48-hour incubation period was provided, after which 1 µl of penicillin/streptomycin antibiotic was supplemented into the medium to prevent any possible bacterial or fungal infections.
Gene Expression Analysis through Real-time PCR
To assess the impact of RPS19 gene silencing on the expression levels of potentially affected genes, real-time PCR was conducted. Initially, total RNA was extracted from the transfected cells using Trizol reagent from Invitrogen (USA). Briefly, cells were centrifuged, treated with RNX solution, and mixed with chloroform. After phase separation, RNA was precipitated with isopropanol and washed with ethanol. The final RNA pellet was dissolved in DEPC-treated water, and its concentration and purity were measured using a Nanodrop device. The RNA concentration and purity were assessed using a Nanodrop device, measuring absorbance at 260 and 280 nm after blanking with distilled water. Two microliters of RNA were diluted in 48 microliters of distilled water. Additionally, RNA quality was evaluated through agarose gel electrophoresis following extraction. Twenty-five μL of UltraPure DNAse/RNAse-Free distilled water was added, and the quality and quantity of the extracted RNAs were assessed using spectrophotometry. Following this, 1 μg of RNA was utilized for cDNA synthesis with the aid of universal primers Oligo-dT and Random Hexamer from ParToos in Iran. The relative expression levels of RPS16, RPS18, and RPS19 were then determined through real-time PCR employing the Applied Biosystems StepOne Real-Time PCR system according to the manufacturer's instructions. The melting temperature of the primers was 60 °C. Data analysis was conducted using the 2−∆∆Ct method (13). Additionally, the primers used in this procedure are detailed in Table 1.
Table 1.
The list of primers utilized in this study.
| Name | Sequence (5'-3') | product length | Accession No. |
|---|---|---|---|
| RPS16 | F: ACGTTGTCTAGTCCACGCTC R: CACAGCTGTCGCTGTCTTC |
92 | NM_001321111.2 |
| RPS18 | F: ATGCTCATGTGGTGTTGAGG R: CAGTCTGGGATCTTGTACTGG |
127 | NM_022551.3 |
| RPS19 | F: CTGTAAAAGACGTGAACCAGCA R: GAAGCAGCTCGCGTGTAGAA |
163 | NM_001321483.2 |
| GAPDH | F: ACACCCACTCCTCCACCTTTG R: TCCACCACCCTGTTGCTGTAG |
112 | NM_001357943.2 |
Apoptosis Evaluation in SiRNA-Transfected Cells
In the present study, apoptosis was evaluated 48 hours after transfection. To assess apoptosis in siRNA-transfected cells, the Annexin V-FITC/PI staining kit was utilized according to the manufacturer's instructions (Roche, Germany). Initially, the cells were collected, washed with PBS, centrifuged, and suspended in a binding buffer (comprising 25 mM CaCl2, 100 mM HEPES, 100 mM NaCl, pH 7.4). Subsequently, 10 µl of V-FITC Annexin was added to the cells, followed by a 15-minute incubation in darkness at room temperature. After incubation, the cells were washed and treated with 5 µl of Propidium Iodide (PI) solution. The cells were then examined using a flow cytometry device from Becton Dickinson. In this analysis, living cells were identified as negative for Annexin V and PI, cells in early apoptosis were identified as positive for Annexin V and negative for PI, cells in late apoptosis were identified as positive for both Annexin V and PI, and necrotic cells were identified as negative for Annexin V and positive for PI.
Cell Cycle Analysis
To evaluate the impact of transfection on the cell cycle using flow cytometry, the cells were initially incubated for 48 hours post-transfection. Subsequently, the transfected cells were rinsed with PBS, followed by the addition of 150 μl of cold PBS and gentle mixing. Next, 1 mL of cold ethanol (70%) was introduced to the samples. Following a 2-hour gentle agitation period, the cells were maintained at 4 ℃ for an additional 2 hours. The cells were then centrifuged, ethanol was discarded, and a further wash with PBS was conducted. Subsequently, PBS was carefully aspirated, and 1 mL of PI MASTER MIX, comprising 40 μl of Propidium Iodide (PI), 950 μl of PBS, and 10 μl of RNase, were added to the cells. The cells were then incubated in the dark at 37˚C for 30 minutes. Finally, the samples were analyzed using a flow cytometer from BD in San Diego, CA, USA, and the data were processed using FlowJo™ software.
Statistical Data Analysis
The data underwent statistical analysis utilizing the t-test within GraphPad Prism 6. Results were expressed as mean ± standard deviation. Statistical significance was determined for p-values less than 0.05.
Results
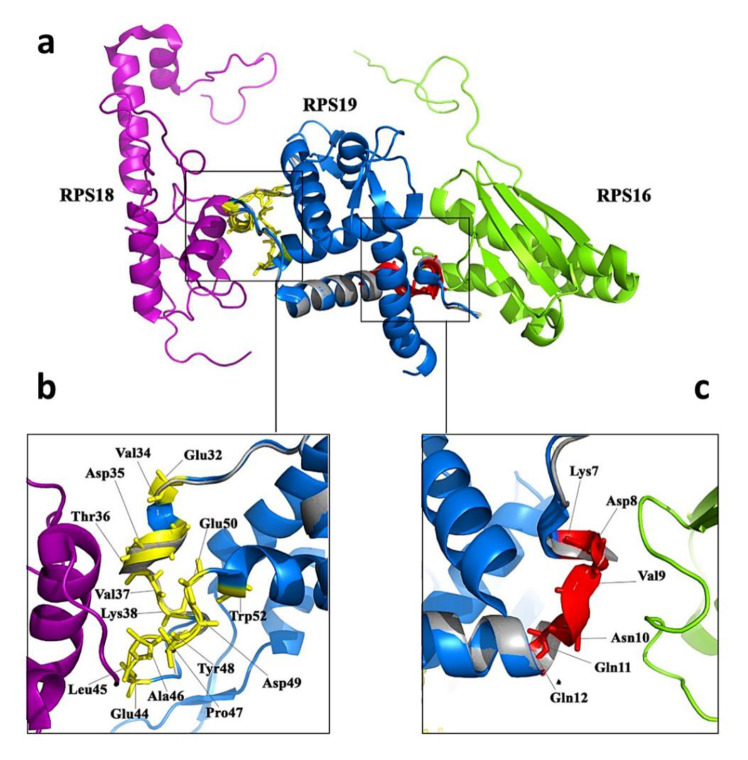
The utilization of PyMol software for detecting PDB files revealed the spatial configuration of RPS19 within various modes of the 40S subunit, enabling the close proximity of at least four proteins, namely RPS16, RPS18, RPS25, and RRP12. Notably, RPS16 and RPS18 emerged as key interactors with RPS19 in the 40S subunit (source: http://www.uniprot.org). Furthermore, an in-depth examination of the amino acids involved in these interactions across all files was conducted using PDBsum and LIGPLOT software. This analysis elucidated that RPS19 engages with RPS16 through amino acids Lys7, Asp8, Val9, Asn10, Gln11, and Gln12. Additionally, the residues Glu32, Val34, Asp35, Thr36, Val37, Lys38, Glu44, Leu45, Ala46, Pro47, Tyr48, Asp49, Glu50, and Trp52 were identified as mediating the interactions between RPS19 and RPS18 (Figure 1).
Fig. 1.
The details of the interaction of RPS19 protein with RPS16 and RPS18 proteins. a) Spatial arrangement of RPS18 (purple) and RPS16 (green) proteins on both sides of RPS19 protein. Normal RPS19 protein is marked in blue and mutant protein is marked in gray. b) Residues involved in the RPS19 and RPS18 interaction are shown in yellow. c) Residues involved in the RPS19 and RPS16 interaction are shown in red. (source: http://www.uniprot.org).
Expression of RPS19, RPS18, and RPS16 Genes
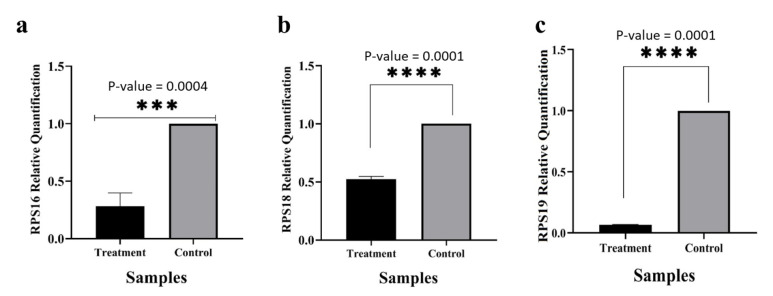
To validate the efficacy of RPS19 gene silencing and its impact on the expression of RPS16 and RPS18 genes in the K-562 cell line, reverse transcription quantitative polymerase chain reaction (RT-qPCR) was conducted. The results of the analysis (Figure 2) demonstrated that the introduction of siRNA successfully suppressed RPS19 gene expression compared to the control group, affirming the effectiveness of the gene silencing. Moreover, the downregulation of RPS19 gene expression significantly reduced the levels of RPS16 and RPS18 gene expression by 75% and 50%, respectively, in the transfected cells.
Fig. 2.
The expression level of RPS16 (a), RPS18 (b), and PRS19 (c) genes in K-562 cells transfected with RPS19-specific siRNA after 48 hours of incubation compared to normal cells. (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001).
Apoptosis Assay Results
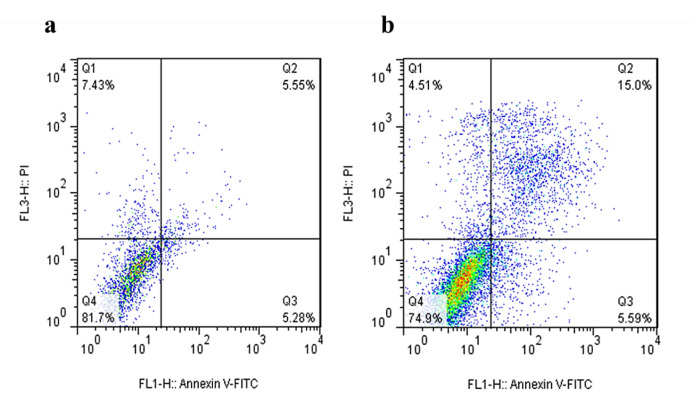
To explore the impact of RPS19 silencing on apoptosis induction in K-562 leukemia cells, flow cytometry analysis was employed. The findings revealed that the suppression of RPS19 gene expression led to a notable elevation in the apoptosis rate within the manipulated cells as opposed to the control group. The percentage of cells undergoing early and late apoptosis in the treated group surpassed 20%. The number of surviving cells decreased at 48 hours post-transfection with 10 pmol of RPS19 siRNA compared to the control group (Figure 3).
Fig. 3.
The effect of siRNA targeting RPS19 on the apoptosis rate of K-562 leukemia cells after 48 hours of incubation (b) compared to the nontreated cells (a). Q1: Necrosis, Q2: Late Apoptosis, Q3: Early Apoptosis, Q4: Live Cells.
Cell Cycle Results
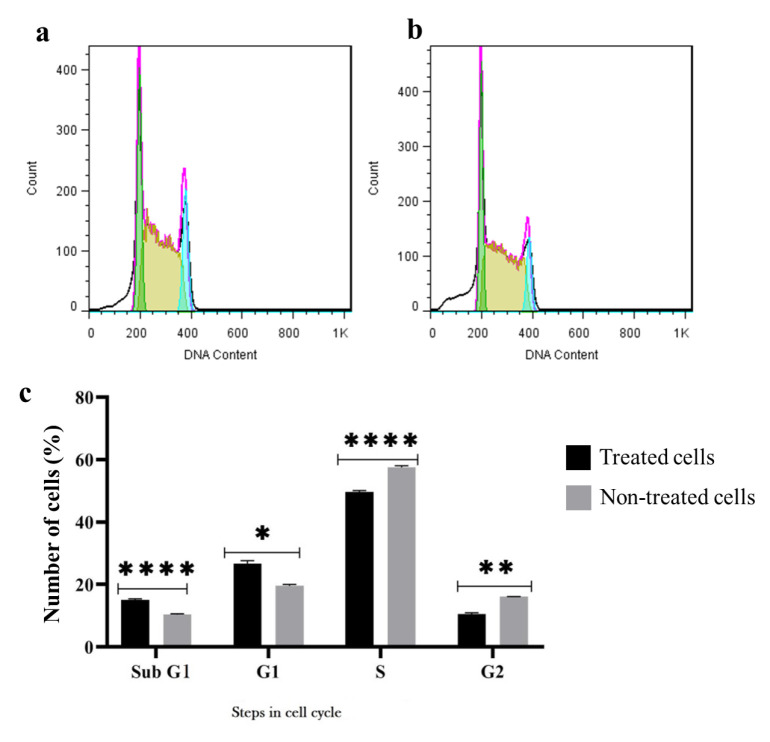
The impact of RPS19 gene silencing on the cell cycle progression after 48 hours was assessed through flow cytometry. The findings indicated that the suppression of this gene resulted in cell cycle arrest in K-562 leukemia cells. Specifically, the knockdown of RPS19 gene substantially increased the population of cells in the sub-G1 and G1 phases while decreasing the number of cells in the S and G2 phases compared to untreated cells (Figure 4).
Fig. 4.
The effect of RPS19 knockdown on the cell phase of K-562 leukemia cells after 48 hours. a) Histogram of the control group, b) siRNA-treated K-562 cells group, and c) bar plot analysis of the cell phase of K-562 cells treated with siRNA against RPS19 compared to control cells. The analysis was calculated by the use of the student t-test. P-value < 0.05 was displayed as statistical significance. (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001).
Discussion
The human RPS19 protein plays a crucial role in the maturation of 40S ribosomal subunits, impacting cellular functions and disease development (7). Knocking down RPS19 expression using siRNAs has been shown to disrupt ribosome biogenesis, decrease 18S rRNA synthesis, and trigger apoptosis in cell types like HeLa cells (14).
Ribosome biogenesis is a complex and energy-demanding process crucial for protein synthesis and overall cellular function. Disruptions in this process can have significant cellular consequences, including cell cycle arrest, senescence, or apoptosis, primarily through the activation of the tumor suppressor protein p53. Insufficient levels of a ribosomal protein disrupt ribosome biogenesis and result in the accumulation of free ribosomal proteins. These free proteins can bind to the MDM2 gene, which acts as a repressor of p53. The release of p53 leads to an increased expression of the P21 and P27 genes. This heightened expression ultimately contributes to the destruction of blood progenitor cells, which can lead to disease. While enhanced ribosome production is typically linked to the rapid proliferation of cancer cells, some aggressive tumors do not display this characteristic. This observation suggests a nuanced relationship between ribosome biogenesis and cancer progression (15).
Our study delved into the interactions between RPS19 and other proteins within the 40S complex, examining the amino acids involved. Using a specific siRNA, we silenced RPS19 to explore its impact on protein interactions in K-562 cells. We evaluated the expression levels of key RPS19-associated proteins (RPS18 and RPS16), apoptosis rates, and cell cycle progression in transfected cells. The successful use of siRNA targeting the RPS19 gene to enhance apoptosis and induce cell cycle arrest in a chronic myeloid leukemia cell line marks a significant step forward in cancer therapy research.
Badhai et al. conducted a study examining the impact of RPS19 deficiency on ribosomal protein levels and 18S rRNA expression, particularly in the context of Diamond-Blackfan anemia (DBA). Utilizing TF1 human erythroleukemia cell lines, they demonstrated that the depletion of RPS19 results in alterations to ribosomal function. This deficiency can activate both p53-dependent and p53-independent pathways, ultimately influencing cell proliferation and survival (16).
In addition, Vysochinskaya et al. in their study, reviewed advancements and future prospects in molecularly targeted therapies and siRNA applications for CML. They highlighted the potential of siRNA as a novel therapeutic strategy. The authors examined siRNA technology, emphasizing its ability to specifically inhibit the expression of the BCR-ABL1 oncogene transcript. Additionally, they analyzed recent findings from both in vitro and in vivo studies that focus on siRNA targeting critical genes associated with CML progression. Their research provides the promising role of molecularly targeted therapies and siRNA in enhancing treatment options for CML (17).
The findings from our study revealed a substantial increase of 20% in both early and late apoptosis in cells treated with the siRNA. While RPS19 plays essential roles in cellular processes (18), its direct involvement in apoptosis pathways is not explicitly mentioned in the search results provided. Further investigation may be necessary to establish a direct link between RPS19 and cellular apoptosis pathways. Our findings align with previous research emphasizing the critical involvement of ribosomal proteins in cancer development (19).
Furthermore, our study identified significant cell cycle arrest in treated cells, characterized by a higher percentage of cells in the G1 and sub-G1 phases. This suggests that manipulating RPS19 gene expression can impact cell cycle progression, potentially offering a promising approach to disrupt cancer cell growth.
Miyake et al. conducted a study focused on developing cell models for DBA by inducing RPS19 deficiency through siRNA. They established stable cell lines with inducible expression of siRNA targeting RPS19, and confirmed knockdown efficiency using quantitative PCR and western blot analysis. The RPS19-deficient cells exhibited impaired erythroid differentiation. When subjected to differentiation protocols, these cells demonstrated reduced hemoglobin synthesis and a decrease in colony-forming units (CFUs) compared to control cells. In terms of cell proliferation, RPS19 knockout cells displayed a diminished growth rate, accompanied by increased apoptosis. This suggests that the loss of RPS19 may activate apoptotic pathways. Additionally, the study highlighted defects in ribosome biogenesis in RPS19-deficient cells (20).
In several types of cancer cells, manipulating ribosomal proteins (RPs) has demonstrated significant impacts on cell proliferation, apoptosis, and cell cycle progression (8). Knockdown of RPS15A in HepG2 cells and lung cancer cells led to cell cycle arrest at the G0/G1 phase, suggesting a potential therapeutic target and biomarker in liver and lung cancers, respectively (21, 22). In breast cancer cell lines, the knockdown of RPS15A suppressed proliferation and induced apoptosis (23, 24). Additionally, the inhibition of RPL26 and RPL29 in pancreatic cancer cells correlated with reduced proliferation, increased apoptosis, and cell cycle blockage, indicating the therapeutic potential of targeting these ribosomal proteins in pancreatic cancer treatment (25). Moreover, targeting RPS6 induced G0/G1 phase arrest in non-small cell lung cancer cells (26). Furthermore, the knockdown of RPS3 in colon and breast cancer cells inhibited proliferation, invasion, and migration while increasing apoptosis rates, suggesting its significance as a therapeutic target (27, 28). Additionally, the knockdown of RPL9 and RPL34 in colon and pancreatic cancer cells, respectively, led to apoptosis induction both in vitro and in vivo (29, 30). The Knockdown of RPL34 in NSCLC cells significantly decreased proliferation and increased apoptosis and cell cycle arrest (31). Lastly, knockdown of RPL39 by siRNA in pancreatic cancer resulted in cancer cell regression and enhanced apoptosis both in vivo and in vitro (32). These findings collectively emphasize the crucial roles of various ribosomal proteins in different types of cancers and suggest their potential as targets for therapeutic interventions.
In the study conducted by Markiewski et al., the immunosuppressive role of RPS19 in cancer was thoroughly investigated. The authors discovered that RPS19 levels are elevated in human breast and ovarian cancer cells. This protein interacts with the complement C5a receptor 1 (C5aR1) on myeloid-derived suppressor cells (MDSC), promoting their recruitment to tumors and facilitating tumor growth. The findings demonstrate that RPS19 not only enhances the production of immunosuppressive cytokines, but also shifts T cell responses towards a T helper 2 phenotype. Importantly, reducing RPS19 levels or blocking its interaction with C5aR1 disrupts immunosuppression and slows tumor progression in a breast cancer model (33).
Furthermore, Bee et al. conducted a study examining the impact of disrupting the RPL19 gene on the invasive phenotype of human prostate cancer, specifically utilizing the PC-3M cell line. They achieved a significant reduction in RPL19 expression through siRNA knockdown. Invasion assays revealed a substantial decrease in the invasive potential of cells with RPL19 knockdown. The number of invasive cells dropped from over 300 in the control group to just 60 in the RPL19 knockdown group, indicating a five-fold reduction in invasiveness. Importantly, despite these alterations, the proliferation rate of si-RPL19 cells remained comparable to that of control cells. In xenograft models, tumors derived from si-RPL19-PC-3M cells exhibited significant growth compared to control tumors, further supporting the hypothesis that RPL19 is crucial for prostate cancer aggressiveness. The findings of this study underscore the important role of RPL19 not only as a ribosomal protein but also as a regulator of gene expression associated with prostate cancer malignancy. This research suggests that targeting RPL19 could be an effective therapeutic strategy for mitigating aggressive phenotypes in prostate cancer (34).
Expression analysis in our study demonstrated a significant 100% decrease in RPS19 gene expression, accompanied by 75% and 50% reductions in RPS16 and RPS18 gene expression, respectively. The downregulation of the RPS19, RPS16, and RPS18 genes following siRNA treatment provides valuable insights into the regulatory networks governing cell survival and proliferation in CML.
The enhanced apoptosis and cell cycle arrest observed in our study upon silencing the RPS19 gene emphasize the potential benefits of targeting ribosomal proteins in cancer therapy. Overall, our research outcomes illuminate the promising implications of RPS19 gene silencing through siRNA in CML treatment.
The results of our study represent a significant advancement in the potential treatment of CML through the silencing of the RPS19 gene using siRNA in the K562 cell line. Our findings suggest that RPS19 may play a crucial role in the progression of CML, positioning it as a promising therapeutic target for inducing apoptosis in affected cells. This research paves the way for further investigations in cancer therapy and underscores the potential of targeted gene silencing as an effective strategy for addressing CML and possibly other cancers.
Conflicts of Interests:
All authors declare that they have no conflict of interest.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Ethical considerations
The study protocol adheres to the guidelines of the Ethics committee of Islamic Azad University (IR.IAU.DAMGHAN.REC.1401.023).
References
- 1.Sampaio MM, Santos MLC, Marques HS, et al. Chronic myeloid leukemia-from the Philadelphia chromosome to specific target drugs: A literature review. World J Clin Oncol. 2021;12:69. doi: 10.5306/wjco.v12.i2.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rana A, Hussain Shah S, Rehman N, et al. Chronic myeloid leukemia: Attributes of break point cluster region-abelson (BCR-ABL) J Cancer Res Exp Oncol. 2011;3:62–6. [Google Scholar]
- 3.Daskalakis M, Feller A, Noetzli J, et al. Potential to improve therapy of chronic myeloid leukemia (CML), especially for patients with older age: incidence, mortality, and survival rates of patients with CML in Switzerland from 1995 to 2017. Cancers. 2021;13:6269. doi: 10.3390/cancers13246269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo W, Chen W, Yu W, et al. Small interfering RNA-based molecular therapy of cancers. Chin J Cancer. 2013;32:488. doi: 10.5732/cjc.012.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charbe NB, Amnerkar ND, Ramesh B, et al. Small interfering RNA for cancer treatment: overcoming hurdles in delivery. Acta Pharm Sin B. 2020;10:2075–109. doi: 10.1016/j.apsb.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campagnoli MF, Ramenghi U, Armiraglio M, et al. RPS19 mutations in patients with Diamond‐Blackfan anemia. Hum Mutat. 2008;29:911–20. doi: 10.1002/humu.20752. [DOI] [PubMed] [Google Scholar]
- 7.Kang J, Brajanovski N, Chan KT, et al. Ribosomal proteins and human diseases: molecular mechanisms and targeted therapy. Signal Transduct Target Ther. 2021;6:323. doi: 10.1038/s41392-021-00728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Khoury W, Nasr Z. Deregulation of ribosomal proteins in human cancers. Biosci Rep. 2021;41:BSR20211577. doi: 10.1042/BSR20211577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markiewski MM, Vadrevu SK, Sharma SK, et al. The ribosomal protein S19 suppresses antitumor immune responses via the complement C5a receptor 1. J Immunol. 2017;198:2989–99. doi: 10.4049/jimmunol.1602057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondoh N, Schweinfest CW, Henderson KW, et al. Differential expression of S19 ribosomal protein, laminin-binding protein, and human lymphocyte antigen class I messenger RNAs associated with colon carcinoma progression and differentiation. Cancer Res. 1992;52:791–6. [PubMed] [Google Scholar]
- 11.Arthurs C, Murtaza BN, Thomson C, et al. Expression of ribosomal proteins in normal and cancerous human prostate tissue. PLoS One. 2017;12:e0186047. doi: 10.1371/journal.pone.0186047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein E, Vánky F, Ben‐Bassat H, et al. Properties of the K562 cell line, derived from a patient with chronic myeloid leukemia. Int J Cancer. 1976;18:421–31. doi: 10.1002/ijc.2910180405. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Choesmel V, Bacqueville D, Rouquette J, et al. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109:1275–83. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turi Z, Lacey M, Mistrik M, et al. Impaired ribosome biogenesis: mechanisms and relevance to cancer and aging. Aging. 2019;11:2512. doi: 10.18632/aging.101922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Badhai J, Fröjmark AS, Razzaghian HR, et al. Posttranscriptional down-regulation of small ribosomal subunit proteins correlates with reduction of 18S rRNA in RPS19 deficiency. FEBS Lett. 2009;583:2049–53. doi: 10.1016/j.febslet.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vysochinskaya V, Dovbysh O, Gorshkov A, et al. Advancements and Future Prospects in Molecular Targeted and siRNA Therapies for Chronic Myeloid Leukemia. Biomolecules. 2024;14:644. doi: 10.3390/biom14060644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Nag S, Zhang X, et al. Ribosomal proteins and human diseases: pathogenesis, molecular mechanisms, and therapeutic implications. Med Res Rev. 2015;35:225–85. doi: 10.1002/med.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goudarzi KM, LINDSTRöM MS. Role of ribosomal protein mutations in tumor development. Int J Oncol. 2016;48:1313–24. doi: 10.3892/ijo.2016.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyake K, Flygare J, Kiefer T, et al. Development of cellular models for ribosomal protein S19 (RPS19)-deficient Diamond–Blackfan anemia using inducible expression of siRNA against RPS19. Mol Ther. 2005;11:627–37. doi: 10.1016/j.ymthe.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Xu M, Wang Y, Chen L, et al. Down-regulation of ribosomal protein S15A mRNA with a short hairpin RNA inhibits human hepatic cancer cell growth in vitro. Gene. 2014;536:84–9. doi: 10.1016/j.gene.2013.11.075. [DOI] [PubMed] [Google Scholar]
- 22.Zhao X, Shen L, Feng Y, et al. Decreased expression of RPS15A suppresses proliferation of lung cancer cells. Tumor Biol. 2015;36:6733–40. doi: 10.1007/s13277-015-3371-9. [DOI] [PubMed] [Google Scholar]
- 23.Kong L, Wei Q, Hu X, et al. Ribosomal protein small subunit 15A (RPS15A) inhibits the apoptosis of breast cancer MDA‐MB‐231 cells via upregulating phosphorylated ERK1/2, Bad, and Chk1. J Cell Biochem. 2020;121:587–95. doi: 10.1002/jcb.29304. [DOI] [PubMed] [Google Scholar]
- 24.Feng W, Liang C, Wang C, et al. Knockdown of ribosomal protein S15A inhibits proliferation of breast cancer cells through induction of apoptosis in vitro. Cytotechnology. 2018;70:1315–23. doi: 10.1007/s10616-018-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muro S, Miyake Y, Kato H, et al. Serum anti-60S ribosomal protein L29 antibody as a novel prognostic marker for unresectable pancreatic cancer. Digestion. 2015;91:164–73. doi: 10.1159/000371545. [DOI] [PubMed] [Google Scholar]
- 26.Chen B, Tan Z, Gao J, et al. Hyperphosphorylation of ribosomal protein S6 predicts unfavorable clinical survival in non-small cell lung cancer. J Exp Clin Cancer Res. 2015;34:1–16. doi: 10.1186/s13046-015-0239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alam E, Maaliki L, Nasr Z. Ribosomal protein S3 selectively affects colon cancer growth by modulating the levels of p53 and lactate dehydrogenase. Mol Biol Rep. 2020;47:6083–90. doi: 10.1007/s11033-020-05683-1. [DOI] [PubMed] [Google Scholar]
- 28.Ono H, Iizumi Y, Goi W, et al. Ribosomal protein S3 regulates XIAP expression independently of the NF-κB pathway in breast cancer cells. Oncol Rep. 2017;38:3205–10. doi: 10.3892/or.2017.6008. [DOI] [PubMed] [Google Scholar]
- 29.Baik IH, Jo G-H, Seo D, et al. Knockdown of RPL9 expression inhibits colorectal carcinoma growth via the inactivation of Id-1/NF-κB signaling axis. Int J Oncol. 2016;49:1953–62. doi: 10.3892/ijo.2016.3688. [DOI] [PubMed] [Google Scholar]
- 30.Wei F, Ding L, Wei Z, et al. Ribosomal protein L34 promotes the proliferation, invasion and metastasis of pancreatic cancer cells. Oncotarget. 2016;7:85259. doi: 10.18632/oncotarget.13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang S, Cui J, Yang Y, et al. Over-expressed RPL34 promotes malignant proliferation of non-small cell lung cancer cells. Gene. 2016;576:421–8. doi: 10.1016/j.gene.2015.10.053. [DOI] [PubMed] [Google Scholar]
- 32.Li Co, Chen D, Luo M. Knockdown of ribosomal protein L39 by RNA interference inhibits the growth of human pancreatic cancer cells in vitro and in vivo. Biotechnol J. 2014;9:652–63. doi: 10.1002/biot.201300321. [DOI] [PubMed] [Google Scholar]
- 33.Markiewski MM, Vadrevu SK, Sharma SK, et al. The ribosomal protein S19 suppresses antitumor immune responses via the complement C5a receptor 1. J Immunol. 2017;198:2989–99. doi: 10.4049/jimmunol.1602057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bee A, Brewer D, Beesley C, et al. siRNA knockdown of ribosomal protein gene RPL19 abrogates the aggressive phenotype of human prostate cancer. PLoS One. 2011;6:e22672. doi: 10.1371/journal.pone.0022672. [DOI] [PMC free article] [PubMed] [Google Scholar]