Abstract
The export of mRNA from the nucleus to the cytoplasm involves interactions of proteins with mRNA and the nuclear pore complex. We isolated Crp79p, a novel mRNA export factor from the same synthetic lethal screen that led to the identification of spMex67p in Schizosaccharomyces pombe. Crp79p is a 710-amino-acid-long protein that contains three RNA recognition motif domains in tandem and a distinct C-terminus. Fused to green fluorescent protein (GFP), Crp79p localizes to the cytoplasm. Like Mex67p, Crp79-GFP binds poly(A)+ RNA in vivo, shuttles between the nucleus and the cytoplasm, and contains a nuclear export activity at the C-terminus that is Crm1p-independent. All of these properties are essential for Crp79p to promote mRNA export. Crp79p import into the nucleus depends on the Ran system. A domain of spMex67p previously identified as having a nuclear export activity can functionally substitute for the nuclear export activity at the C-terminus of Crp79p. Although both Crp79p and spMex67p function to export mRNA, Crp79p does not substitute for all of spMex67p functions and probably is not a functional homologue of spMex67p. We propose that Crp79p is a nonessential mRNA export carrier in S. pombe.
INTRODUCTION
A large number of proteins associate with the unspliced and mature mRNA in the nucleus to perform a variety of functions (Izaurralde and Adam, 1998; Stutz and Rosbash, 1998; Nakielny and Dreyfuss, 1999; Conti and Izaurralde, 2001; Zenklusen and Stutz, 2001). The mature mRNA, bound to some of these proteins as ribonucleoprotein (mRNP) complexes, is exported through the nuclear pore. Genetic and biochemical studies in yeast, mammalian, and viral systems have identified a large number of proteins that are thought to be involved in the export of mRNA (Stutz and Rosbash, 1998; Nakielny and Dreyfuss, 1999). These export factors include heterogeneous RNA–associated nuclear proteins (e.g., human hnRNP A1, or Saccharomyces cerevisiae Npl3) (Michael et al., 1995; Singleton et al., 1995; Lee et al., 1996), RNA export factor (REF) class of proteins (e.g., Yra1/ALY/REF) (Lee et al., 1996; Strasser and Hurt, 2000; Stutz et al., 2000; Rodrigues et al., 2001), splicing-related factors (e.g., UAP56/Sub2/HEL, U2AF) (Gatfield et al., 2001; Strasser and Hurt, 2001; Zolotukhin et al., 2002), exon-junction proteins (e.g., Y14) (Le Hir et al., 2000; Kim et al., 2001), putative carriers of mRNA (e.g., Rev, Tap/Mex67) (Segref et al., 1997; Kang and Cullen, 1999; Katahira et al., 1999; Yoon et al., 2000), and finally, components of the nuclear pore complex (NPC) (Yang et al., 1998; Strambio-de-Castillia et al., 1999). In addition, protein import-export pathways are probably indirectly involved in mRNA export processes (Dasso and Pu, 1998). A few proteins, like Mex67 or Tap, that function as export carriers of mRNA characteristically shuttle, associate with the substrate mRNA directly or via adapter proteins like REFs, and connect the mRNP complexes to the nuclear pore proteins to promote nuclear mRNA export (Conti and Izaurralde, 2001; Zenklusen and Stutz, 2001). The carrier-mediated export of mRNA functionally separates the carriers from noncarrier export factors that may be passively exported along with mRNA.
Several potential mRNA carriers have been identified and their mechanism of function studied (Stutz and Rosbash, 1998; Nakielny and Dreyfuss, 1999). One prototypic carrier is the Rev protein of HIV-1. It exports viral transcripts containing an RNA sequence called the Rev-responsive element (Stutz and Rosbash, 1998). Rev binds to the Rev-responsive element and uses a nuclear export signal (NES) to export the mRNA cargo (Malim et al., 1989; Fischer et al., 1995; Pollard and Malim, 1998). The Rev NES is a short leucine-rich peptide that interacts with its export receptor Crm1p (Xpo1 in S. cerevisiae), a member of the importin-β class of import/export receptors (Fornerod et al., 1997; Stade et al., 1997). Crm1p directs the export of NES-containing proteins along with their cargo mRNA by interacting with the NPC (Stutz and Rosbash, 1998).
scMex67p of S. cerevisiae, spMex67p of S. pombe, and their human homologue Tap are non–importin-β type mRNA carriers. scMEX67 is an essential gene, and at nonpermissive temperatures, its temperature-sensitive (ts) mutants accumulate poly(A)+RNA in the nucleus (Segref et al., 1997). scMex67p binds to poly(A)+ RNA, to REFs, and to the GLFG regions of Nup116p and Nup100p (Segref et al., 1997; Strasser and Hurt, 2000; Strasser et al., 2000; Strawn et al., 2001). In contrast, spmex67 is not essential, and its involvement in poly(A)+ RNA export is revealed only by its synthetic lethality with a mutation in rae1, an essential gene required for mRNA export in S. pombe (Brown et al., 1995; Yoon et al., 2000). spMex67p binds poly(A)+ RNA, shuttles between the nucleus and cytoplasm, and functions to export mRNA that is dependent on a nuclear export activity present within its middle domain (Yoon et al., 2000). Recent in vitro binding experiments suggest that the splicing factor UAP56/Sub2p interacts with the REF protein Aly/Yra1 and that Tap/Mex67p can displace UAP56/Sub2p from Aly/Yra1 (Luo et al., 2001; Strasser and Hurt, 2001). Thus, Tap/Mex67 may interact with the mRNP via Aly/Yra1. Conversely, Tap/Mex67p independently or with cofactors p15/Mtr2p can interact with the NPC to target the mRNP complexes to the nuclear pores (Bachi et al., 2000; Strasser et al., 2000; Strawn et al., 2001).
In S. pombe, Rae1p is an essential protein that is required for export of mRNA and cell cycle progression (Brown et al., 1995). At the restrictive temperature, the ts mutant rae1–167 rapidly accumulates poly(A)+ RNA in the nucleus (Brown et al., 1995). The rae1–167 allele is also synthetically lethal with nup184–1 mutation. Nup184p is a nonessential nucleoporin in S. pombe that does not contain degenerate FG repeats. Its orthologue in S. cerevisiae, Nup188p, has been implicated in nuclear membrane biogenesis (Nehrbass et al., 1996; Zabel et al., 1996)
In this work, we describe the isolation and functional characterization of a novel cytoplasmic protein, Crp79p, which, like spMex67p, complements the synthetic lethality of a rae1–16 nup184–1 double mutation by preventing accumulation of poly(A)+ RNA in the nucleus. We show that like spMex67p, Crp79p shuttles between the nucleus and the cytoplasm, can be UV cross-linked to poly(A)+ RNA in vivo, and mediates export of mRNA via a nuclear export activity present at the C-terminus. We argue that Crp79p can function as a carrier in transporting mRNA from the nucleus to the cytoplasm.
MATERIALS AND METHODS
Strains and Culture
The basic genetic and cell culture techniques used have been described (Moreno et al., 1991; Alfa et al., 1993). The strains used are wild-type 217: h− leu1–32 ura4-d18; SL27: h− leu1–32 ura4-d18 rae1–167 nup184–1/pREP81X-rae1+ (Whalen et al., 1999); h− rae1–167 (Brown et al., 1995); h− rae1–167 pim1-d1 (this work); pim1-d1 (Demeter et al., 1995); h− Δcrp::ura4 leu1–32 ura4-d18; h− Δcrp::ura4 Δmex::kan leu1–32 ura4-d18 (this work); and h− rae1–167 npp106–1/pREP81X-rae1+ leu1–32 ura4-d18 (Yoon et al., 1997). Appropriately supplemented–essential minimal media EMM medium was used to express genes from the pREP plasmids containing the nmt promoter (Maundrell, 1993). The nmt promoter was repressed by the addition of 10 μM thiamine in EMM medium (Forsburg, 1993).
Isolation of crp79
SL27 (rae1–167 nup184–1/pREP81X-rae1+) cells were transformed with a library of partially digested genomic DNA constructed at the SalI site of pUR18 (Berbet et al., 1992), and Ura+ colonies were isolated that could grow in the presence of thiamine at 28°C. After elimination of the rae1+ clones by virtue of their ability to confer growth of SL27 cells at 36°C in the presence of thiamine, plasmids were rescued and retransformed into the SL27 strain. Analysis of the plasmids showed that eight different genes had been isolated. One clone, p27–9, encoded several open reading frames with significant similarity to the RNA recognition motif (RRM) found in RNA binding proteins. Using reverse transcription and PCR, we isolated a cDNA whose sequences revealed the presence of three introns within the gene of crp79.
Construction of Crp79p-GFP Fusion Proteins
The full-length Crp79p-GFP fusion in pAB3 was constructed by introduction of a KpnI site at the termination codon of the crp79 gene in p27–9, followed by insertion of a KpnI-SacI fragment bearing the gene for the green fluorescent protein (GFP) (S65T). Except for pAB58, the truncated derivatives of pAB3 shown in Figure 2 and pAG116 in Figure 5 were constructed by introduction of a BglII site at the end points of the deletion. The C-terminal deletion (pAB58) was made by introduction of a KpnI site at the deletion endpoint, and the crp79 open reading frame was fused in frame to GFP as in pAB3. The point mutant Crp79p(1–122, F74V) (pAB106F74V) was made by PCR mutagenesis of two overlapping fragments containing the F74V mutation. These fragments were used as templates in a crossover PCR to produce the full-length construct that was cloned into pAB86.
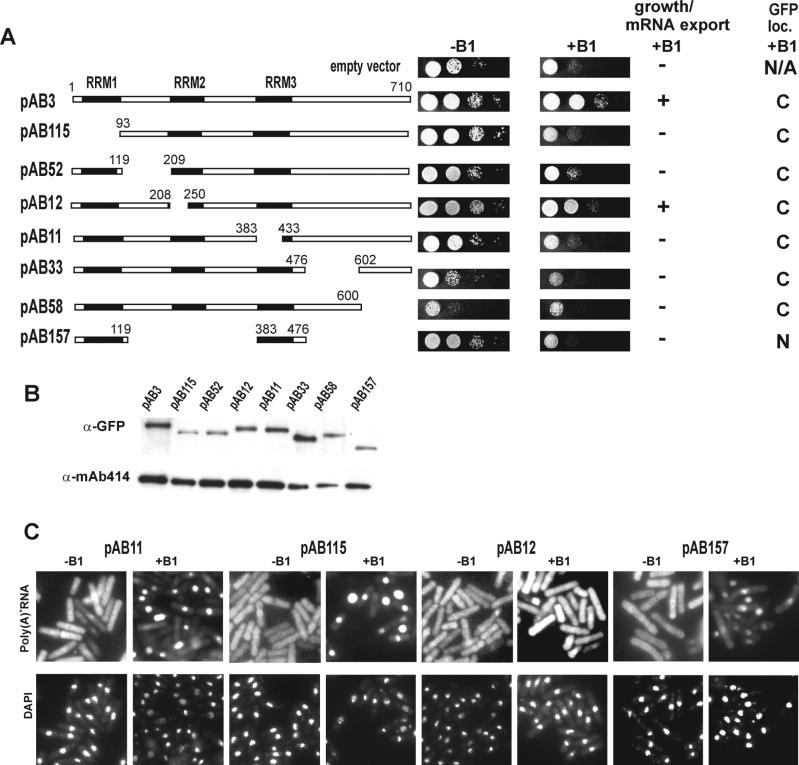
Figure 2.
Growth and mRNA export of SL27 cells. (A) Complementation of synthetic lethality of rae1–167 nup184–1/pREP81X-rae1 (SL27) by Crp79p-GFP and its deletions. A schematic diagram of the regions surrounding the deletions along with deletion endpoints is shown (for details of plasmid construction, see MATERIALS AND METHODS). Growth of SL27 cells was monitored for 4 d with (−B1) and without (+B1) expression of rae1. Ability and inability to restore growth and mRNA export to SL27 cells under conditions of synthetic lethality is indicated and summarized by (+) and (−). The GFP loc. indicates cellular localization of relevant fusion protein; C, cytoplasm; N, nucleus; N/A, not applicable. (B) Determination of steady-state protein levels of Crp79p-GFP and deletion derivatives in SL27 cells grown when expression of rae1 was repressed (in presence of B1). Top, Western blot analysis using a monoclonal anti-GFP antibody. Bottom, Control for protein loading by Western blot for a protein that reacts with NPC-specific mAb414 mAb. (C) Poly(A)+ RNA was detected in SL27 cells carrying pAB11, pAB12, pAB115, and pAG157 (see Figure 2A for deletion endpoints) grown in EMM to midlog phase in absence (−B1) or 18 h in presence of thiamine (10 μM). Coincident DAPI staining is shown at bottom.
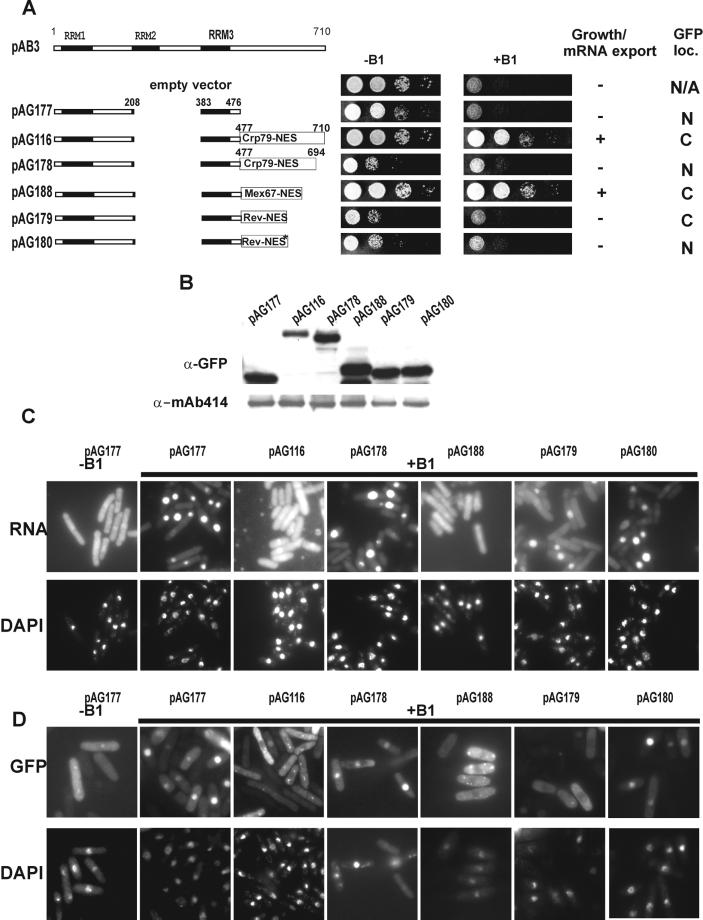
Figure 5.
Nuclear export activity–dependent suppression of growth and mRNA export defects of SL27 cells. (A) Schematic diagram of various GFP fusions. The coordinates for the spMex67 are aa 439–505 (Yoon et al., 2000). The Rev-NES sequence is 23 aa long, containing LPPLERLTL (wild-type) or LPPAERATL (mutant) peptide sequence (Love et al., 1998). Growth of rae1–167 nup184–1 cells was monitored for 4 d, expressing different GFP fusion constructs with or without rae1 expression (−B1 and +B1, respectively). + indicates ability to complement, and − indicates inability to complement under synthetic lethal conditions. The GFP loc. indicates cellular localization of relevant Crp79p-GFP fusion; C, cytoplasm; N, nucleus; and N/A, not applicable. (B) Steady-state levels of GFP fusion proteins used in A following a procedure similar to that used in Figure 2B. (C) Poly(A)+ RNA localization of various GFP fusions in A as indicated in SL27 cells after 18 h of growth in presence of B1 (10 μM) are shown. (D) Localization of the GFP fusions in A as indicated in the absence (−B1) or 18 h after addition of B1 to the media. Coincident DAPI staining is shown at bottom.
Plasmids for the HeLa export were constructed with pXRGG as the source plasmid (Love et al., 1998), into which a polylinker was introduced at the XhoI-BglII sites. The new plasmid, pAG112, was used to clone various PCR fragments as XhoI-BglII fragments.
Plasmids pAG177-pAG188 were made from pAG116 by replacing its BglII-KpnI fragment, followed by insertion of a polylinker at the unique KpnI site to generate pAG177. pAG179 and pAG180 were made by placing wild-type or mutant Rev-NES sequence at the polylinker between the XhoI and BglII sites of pAG177. These sequences were generated by PCR using pXRGG as the template and wild-type or mutant oligonucleotide primers. pAG188 was constructed similarly by placing a DNA fragment encoding for the nuclear export activity (435–509 aa) of spMex67p. pAG178 was constructed by replacing the BglII–KpnI fragment of pAG116 with a BglII-KpnI fragment containing 477–694 aa of Crp79p. Plasmids for UV cross-linking experiments were constructed by fusing the RRM domains to the C-terminus of GST in a pREP3X-vector. For the full-length Crp79p, a GFP fusion was used.
Construction of crp79 Null and the Genomic Integration of crp79-gfp
The full-length crp79 insert from p27–9 was cloned into pSK+, and a deletion between amino acids (aa) 125 and 651 with a HindIII site placed at the deletion junction was constructed. A HindIII fragment bearing ura4+ was placed in the deletion junction. This plasmid was cut with PstI and XhoI, and the crp79::ura4 fragment was gel-purified and transformed into the SP286 diploid strain. ura+ transformants were selected and screened by Southern blotting for the disruption of one of the crp79 loci. Analysis of 12 tetrads showed a 2:2 segregation of ura4+, and the crp79::ura4 colonies grew as well as crp79+ colonies. For chromosomal integration of the full-length crp79p-gfp fusion, the ars1 sequence was removed from pAB3 by digestion with AatII and NheI. The resulting plasmid was linearized by digestion with AgeI and transformed into JBP217. Stable ura4+ transformants were selected and screened for integration at the crp79 locus by Southern blotting.
Spot Assay for Growth
Single transformants were first patched on EMM-agar plates in the absence of thiamine (−B1) with appropriate auxotrophic selection and then grown into 10 ml liquid medium until the stationary phase (3 d). Tenfold dilutions were plated in the presence or absence of thiamine (final concentration of B1 was 10 μM) on EMM plates without leucine and/or uracil as appropriate.
In Situ Hybridization
In situ hybridization was performed as described previously (Amberg et al., 1992) with the following modifications. Oligo dT50 carrying an α-digoxygenin at the 3′ end was used as the hybridization probe. FITC-antidigoxygenin or rhodamine-antidigoxygenin, as appropriate, was used for detecting the hybridization probe by fluorescence microscopy. 4′,6′-Diamidino-2-phenylindole (DAPI) was used for observing the DNA.
UV Cross-Linking
Plasmids expressing full-length Crp79p-GFP (pAB3) or C-terminal GST fusions of RRM1, RRM (F74V) mutant, RRM2, and RRM3 were introduced into wild-type strains. The methods for UV cross-linking, mRNA isolation, and Western blotting were as described previously (Yoon et al., 2000). Monoclonal anti-GST, anti-GFP antibodies were used for detection of the fusion proteins using the Renaissance kit (NEN, Boston, MA).
Heterologous Nuclear Export Assay in HeLa Cells
Export activity of various regions of Crp79p were measured with a heterologous assay described previously (Love et al., 1998). Briefly, a vector containing glucocorticoid receptor (Gr)–GFP fusions with specific protein segments of interest was used to transfect HeLa cells. In the presence of 2 μM corticosterone, the GFP fusion protein translocated from the cytoplasm into the nucleus. One hour after incubation with hormone, cells were washed extensively, and the export of proteins was assayed in presence of cycloheximide (10 μg/ml) by monitoring depletion of the fusion protein in the nucleus and its redistribution into the cytoplasm for 1 h at 37°C. As a positive control for export, a fusion of spMex67p-NES-Gr-GFP was used.
RESULTS
Isolation of crp79 as a Multicopy Suppressor of rae1–167 nup184–1 Synthetic Lethality
The S. pombe SL27 (rae1–167 nup184–1/pREP81X-rae1) is a synthetic lethal strain that contains the ts rae1–167 mutation in Rae1p and a mutation nup184–1 in Nup184p (Whalen et al., 1999). In this article, rae1–167 nup184–1/pREP81X-rae1 cells are referred to as SL27 cells for the purpose of brevity. The SL27 strain is kept viable by expression of rae1 gene from a pREP81X plasmid under the control of a weak, thiamine-repressible, nmt1 promoter. In the absence of thiamine (−B1), Rae1p is synthesized, and the SL27 cells grow normally. In the presence of thiamine (+B1), the expression of Rae1p is repressed, resulting in inhibition of growth. The SL27 cells accumulate poly(A)+ RNA in the nucleus under synthetic lethal conditions 12–18 h after addition of thiamine to the medium.
Previously, by screening an S. pombe genomic library, we isolated spmex67 as a multicopy suppressor of growth and mRNA export defects of the SL27 strain (Yoon et al., 2000). Using the same strategy, we isolated another multicopy suppressor plasmid, p27–9, that suppressed both growth (data not shown) and the mRNA export defects of SL27 under synthetic lethal conditions (+B1, 18 h) (compare +B1 panels for empty vector vs. p27–9 in Figure 1A).
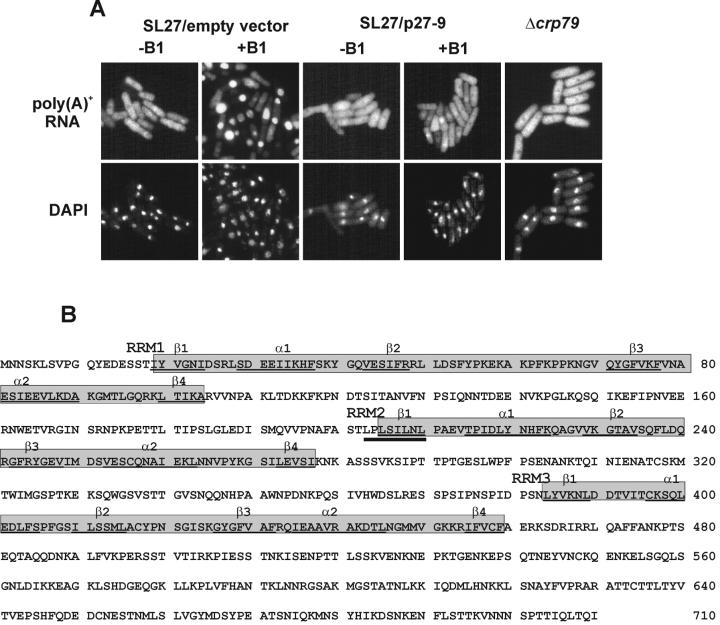
Figure 1.
Suppression of the mRNA export defect in SL27 (rae1–167 nup184–1/pRXE81X-rae1) cells. Poly(A)+ RNA localization in SL27 cells and in cells with the Δcrp79 mutation. (A) Poly(A)+ RNA was detected in Δcrp79 cells grown in EMM medium. The SL27 cells carrying an empty vector or a plasmid (p27–9) expressing Crp79p were grown in EMM to midlog phase expressing rae1 (absence, −B1) or without expression of rae1 (18 h in presence of 10 μM B1). Coincident DAPI staining is shown at bottom. (B) aa sequence and structural organization of Crp79p of S. pombe. The sequence of Crp79p has been deposited in the GenBank data base with accession No. AF432874. Shaded boxes indicate the three RRM domains. The secondary structure predictions of α-helix and β-sheets are indicated and underlined (Burd and Dreyfuss, 1994a). The dark underline indicates the position of the leucine-rich NES.
Sequence analyses of p27–9 and its inserts of cDNA revealed a single gene with three introns encoding a 710-aa protein of predicted molecular weight ∼79 kDa (Figure 1B) (GenBank accession No. AF432874). We named this protein Crp79p (cytoplasmic RRM containing protein of 79 kDa) to indicate its cytoplasmic localization in the wild-type strain (see below). A strain with a deletion of crp79 (Δcrp79::ura4) grew normally at temperatures between 18 and 37°C (data not shown) without any detectable mRNA export defects (Figure 1A). Thus, crp79 is a nonessential gene, and loss of the protein has no significant impact on mRNA export in S. pombe.
The HMMER program in the Pfam database identified three RRMs in Crp79p located sequentially from the N-terminus, with designations of RRM1, RRM2, and RRM3 (shaded in Figure 1B). All three RRMs carry the conserved hexapeptide RNP2 (L/I F/Y V/I G/K N/L L) and the octapeptide RNP1 (K/R G F/Y G/A FVX F/Y) RNA binding consensus within the predicted βαββαβ fold (underlined in Figure 1B) (Burd and Dreyfuss, 1994a, 1994b). RRM2 also contains a leucine-rich, Rev-NES–like motif (LxLxxLxL) between residues 203 and 210. This is located within the RNP2 region (residues above thick underline in Figure 1B). In contrast, the C-terminus of Crp79p (from 460 to 710 aa) apparently lacked significant homology to any known protein or structural motif in the databases. BLASTP searches conducted with Crp79p showed similarities with other poly(A)+ RNA binding proteins; however, these were limited to the RRM1 and RRM3 domains only (Burd and Dreyfuss, 1994a). Recently, from a search for genes of S. pombe induced in meiosis, the same gene was isolated independently and named meu5 (Accession No. AB020594) (Watanabe et al., 2001).
Common Functional Determinants within Crp79p-GFP Required for Growth and mRNA Export
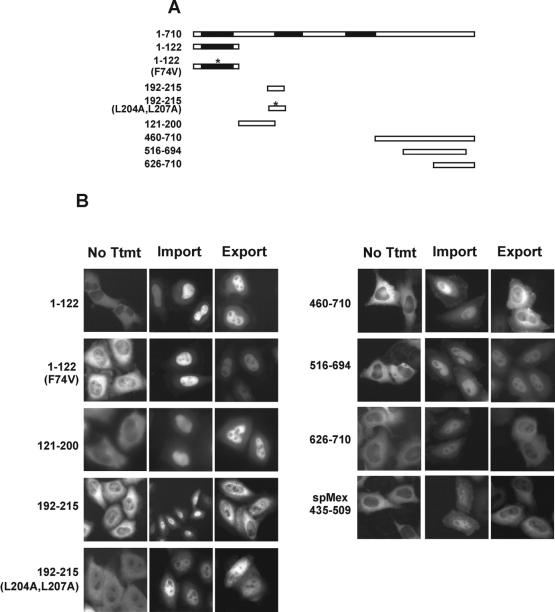
The genomic copy of crp79 was fused at the C-terminus to a gene fragment encoding the GFP. Crp79-GFP was fully functional and complemented growth (pAB3 in Figure 2A) and mRNA export (data not shown) phenotypes of rae1–167 nup184–1 (SL27) cells when rae1 expression was repressed (+B1). We undertook a systematic deletion analysis of the three RRM domains of Crp79-GFP, including the regions between the RRMs and the unique C-terminus, to determine which parts of the protein are necessary for its function. SL27 cells were transformed with plasmids carrying deletions of Crp79-GFP, and the ability of the transformants to grow in the presence (−B1) or absence (+B1) of rae1 expression was compared (Figure 2A). Complementation was achieved by the full-length version of Crp79-GFP (pAB3) or partially by a deletion in RRM2 (pAB12) (Figure 2A). However, deletions in RRM1 (pAB115), RRM3 (pAB11), or in the region between RRM1 and RRM2 (pAB52) failed to complement. A C-terminal deletion between 601 and 710 aa (pAB58) or between 477 and 601 aa (pAB33) also failed to rescue the growth of rae1–167 nup184–1 (SL27) cells. In the latter two cases, however, the transformants grew poorly in the absence of B1 also, indicating some dominant negative effects on cell growth. Another deletion construct (pAB157) that contained RRM 1 and RRM3 also failed to complement growth. The various deletion constructs or the full-length Crp79p-GFP expressed adequate amounts of intact protein (Figure 2B). Thus, the inability to complement growth by the truncated proteins was unlikely, because of an insufficient quantity of the functional proteins. Taken together, these results suggest that RRM2 is the only dispensable region in Crp79p for complementation of the synthetic lethality because of the combination of rae1–167 and nup184–1 mutations.
We tested whether Crp79-GFP and its various deletions could suppress mRNA export defects of SL27 cells when the expression of rae1 was repressed. For this, the total poly(A)+ RNA was detected by in situ hybridization in SL27 cells grown in the absence or in the presence of B1 for 18 h. Figure 2C shows in situ hybridizations in SL27 cells transformed with various deletion derivatives of Crp79-GFP. Like Crp79p (p27–9, shown in Figure 1A), deletion in RRM2 (pAB12) was able to suppress the nuclear accumulation of mRNA of SL27 cells depleted of Rae1p compared with cells containing an empty vector (compare Figures 2C and 1A). In contrast, cells expressing deletions that removed RRM1 (pAB115) or RRM3 (pAB11) exhibited extensive mRNA accumulation in the nucleus when rae1 expression was repressed (Figure 2C). These were fusions that were also unable to suppress the growth defect of SL27 cells under synthetic lethal conditions (Figure 2A). Similarly, the deletion that combined RRM1 and RRM3 (pAB157) showed mRNA accumulation in the nucleus (Figure 2C). Other deletions that were unable to rescue synthetic lethality (e.g., pAB52, pAB33, and pAB58) also did not rescue the mRNA export defects (not shown; data summarized in Figure 2A). Therefore, deletions of Crp79-GFP that complemented the growth defect also mitigated the mRNA export defect of SL27 cells.
A multicopy plasmid carrying spmex67 was shown previously to suppress the growth and mRNA export defects of rae1–167 nup184–1 or rae1–167 npp106–1 synthetic lethal mutants (Yoon et al., 1997, 2000). The same plasmid suppresses the ts phenotype of the rae1–167 mutation, albeit at the lower restrictive temperature of 32°C (Yoon et al., 2000). However, multicopy crp79 was unable to suppress either the synthetic lethality of rae1–167 npp106 mutant or the ts phenotype of rae1–167 at 32°C (data not shown). In addition, Crp79p expression was also unable to suppress the growth and mRNA export defect associated with rae1–167 Δmex67 synthetic lethality (data not shown). Thus, Crp79p does not substitute for and is not a functional homologue of spMex67p. We tested whether Δcrp79 and Δmex67 were synthetically lethal when combined. The double-null mutant cells grew normally and showed no detectable mRNA export defect (data not shown).
UV Cross-Linking of Crp79-GFP, RRM1, or RRM3 to poly(A)+ RNA In Vivo
The ability of Crp79-GFP to be cross-linked to RNA was tested by isolating poly(A)+ RNA from UV-treated or untreated SL27 cells transformed with the plasmid pAB3 expressing the full-length Crp79-GFP fusion protein. Figure 3A shows that Crp79-GFP was cross-linked to poly(A)+ RNA in vivo. We also tested the individual RRM domains for their ability to be UV cross-linked to poly(A)+ RNA. Figure 3A also shows the results of a UV cross-linking experiment with cells expressing wild-type RRM1, RRM2, and RRM3 and an RRM1 mutant, F74V, that carried a conserved phenylalanine to valine mutation known to abolish both specific and nonspecific binding to poly(A)+ RNA in RRM-containing proteins (Deardorff and Sachs, 1997). We found that individually RRM1 or RRM3 UV cross-linked to poly(A)+ RNA in vivo. Moreover, as expected, the RRM1 with the F74V mutation showed no detectable UV cross-linking to poly(A)+ RNA. However, the RRM2 domain failed to show any cross-linking to poly(A)+ RNA. Thus, although a deletion of either RRM in the context of the whole protein led to complete loss of mRNA export/growth complementation functions, individually the domains RRM1 and RRM3 cross-linked RNA. These data also suggest that proteins with the C-terminal deletions that retain the RRMs (pAB33 or pAB58) are expected to bind RNA, and as such, their inability to complement growth or mRNA export defects may reside in other aspects of Crp79p function.
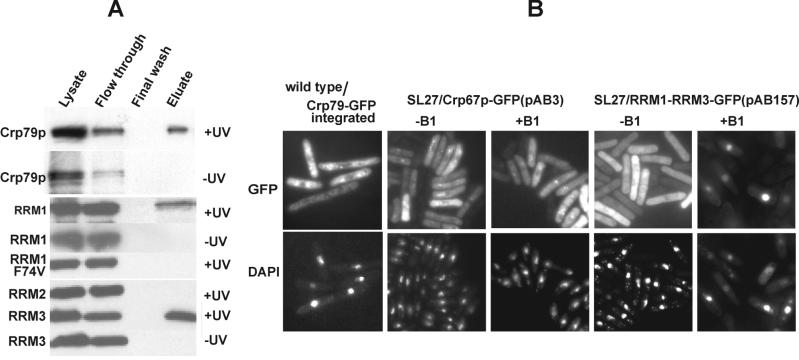
Figure 3.
UV cross-linking and localization of Crp79p-GFP. (A) Crp79-GFP and N-terminal GST-fusions of RRM1, RRM1 F74V, RRM2, and RRM3 were expressed from plasmids under the control of the genomic crp79 promoter or a heterologous (rae1) promoter, respectively. UV cross-linked (+UV) or uncross-linked (−UV) samples were prepared. The protein cross-linked to poly(A)+ RNA were detected by Western blot analysis using polyclonal anti-GST antibody. (B) Cells in which Crp79-GFP was integrated at its genomic locus were tested for GFP signal. For cellular localization of Crp79-GFP (pAB3) or a deletion carrying RRM1 and RRM2 fused to GFP (pAG157) in SL27 cells, transformants were grown with or without rae1 expression (−B1 or 18 h in presence of B1), and the proteins were visualized by GFP fluorescence. Coincident DAPI staining is shown at bottom.
Crp79-GFP Localizes in the Cytoplasm
In wild-type cells, Crp79p-GFP expressed from an integrated chromosomal locus (Figure 3B) or from multicopy plasmids (pAB3) (not shown) was found to be located predominantly in the cytoplasm. In most cells, some of the fusion protein was concentrated in the cytoplasm as dots or speckles. When the dots were photo-bleached, they reappeared (data not shown), suggesting that they are probably dynamic in nature. In SL27 cells, Crp79-GFP (pAB3) localized predominantly to the cytoplasm in the presence or absence of rae1 expression (Figure 3B). Moreover, with the exception of RRM1-RRM3 fused to GFP (pAG157), all other deletion derivatives of Crp79p-GFP were also located in the cytoplasm, irrespective of whether rae1 was expressed (see GFP localization +B1 data summary in Figure 2A). RRM1-RRM3 fusion to GFP (pAG157) in the presence of rae1 expression was located in the cytoplasm but accumulated in the nucleus under synthetic lethal conditions (Figure 3B). Thus, the RRM domains (pAB157) can be imported into the nucleus.
Identification of a Nuclear Export Activity at the C-Terminus of Crp79p
The above results raised the possibility that Crp79p may also be imported into the nucleus, but in the steady state, most of the protein is located in the cytoplasm. This led us to investigate whether there is an NES within Crp79p that could direct its export from the nucleus. Alternatively, there could also be a region within the protein that contains a nuclear export property by contributing to an interaction with an adapter protein(s) that contains the proper export signal. We used a heterologous nuclear export assay developed in HeLa cells to scan the entire protein sequence for nuclear export activities (Love et al., 1998; Yoon et al., 2000). Briefly, in this assay, DNA fragments corresponding to the protein segments of interest are fused to a Gr-GFP chimera. HeLa cells are transfected with plasmids expressing various Gr-GFP fusions. Untreated cells show the Gr-GFP fusion in the cytoplasm (no Ttmt samples in Figure 4). In the presence of micromolar concentrations of the steroid hormone, the fusion proteins are imported rapidly (∼30 min) into the nucleus (Import). When the hormone is washed off, the fusion protein can exit the nucleus (within 60 min) if the protein segment encodes an NES or an activity (Export). A previously characterized nuclear export activity from spMex67 (435–509 aa) was used as a positive control in the export assay (Yoon et al., 2000). Using this assay, we found that RRM1 (1–122 aa), RRM1 (F74V), and sequences between RRM1 and RRM2 (120–200 aa) were deficient in promoting export when fused to a version of Gr-GFP in the export assay (Figure 4A and 4B). A leucine-rich Rev-like signal sequence present within RRM2 (201–214 aa) (Figure 1B) showed an NES activity, and mutations of the two internal leucine residues to alanine abolished its export activity (Figure 4, A and B). However, when this activity was compared with the HIV-Rev NES, its relative nuclear export activity was weak (data not shown). A small amount of Gr-GFP fusion of RRM3 localized to the nucleus in the absence of the steroid (No Ttmt). Treatment with the steroid hormone resulted in further increase of the fusion protein in the nucleus. This nuclear accumulation was unaffected upon removal of the hormone (data not shown). The C-terminal region from 460–710 aa demonstrated a strong nuclear export activity (Figure 4B). This export activity was insensitive to leptomycin B, suggesting that the Crm1p pathway most likely was not involved in its export (data not shown). A smaller region from position 626 to 710 also showed a significant amount of export activity. When a large number of cells were compared, the activity of the 460–710 aa was slightly higher than the 626–710 aa region. However, the 662–710 aa and the 516–694 aa fragments failed to show any detectable export activity (data not shown and Figure 4B). We conclude that there is a minimal nuclear export activity contained within aa 626 and 710, whereas the region between residues 460 and 620 may have a stimulatory role. It is not known at this time whether the export activity is encoded directly by an NES sequence within this region or whether it is a result of an interaction with another NES-containing protein.
Figure 4.
Identification of nuclear export activities in Crp79p using a heterologous assay for protein export. (A) A schematic map of different deletions used in the nuclear export assay in B is indicated. (B) The regions of Crp79p as indicated were expressed as hormone-inducible Gr-GFP chimera proteins in HeLa cells. Import: treatment with 2 μM corticosteroid for 1 h at 37°C. Export: cells were washed three times with phosphate-buffered saline to remove hormone and incubated for an additional hour at 37°C in the presence of cycloheximide (10 μg/ml). spMex67(NES)-Gr-GFP was used as a positive control for the assay (Yoon et al., 2000).
mRNA Export by Crp79p Requires Its C-Terminal Nuclear Export Activity
Irrespective of the nature of the export activity, its presence provoked us to consider the possibility that the protein could be exported from the nucleus via its nuclear export activity and in the process carry mRNA out of the nucleus. Such a process may underlie the rescue of the synthetic lethal phenotype of rae1–167 nup184–1 (SL27) cells by ensuring nuclear export of essential mRNA. To test this hypothesis, we first made a GFP fusion that retained RNA binding (RRM1 and RRM3) but was devoid of any nuclear export activity by combining 1–208 aa with 383–460 aa of Crp79p (see Figures 1B and 4B). (This construct was different from the pAB157 used in Figure 2, which also contained a version of RRM1-RRM3-GFP fusion.) The ability of this fusion protein to support growth and mRNA export in rae1–167 nup184–1 cells under the synthetic lethal condition was examined. The resulting GFP fusion (pAG177) was unable to complement growth and the mRNA export defect of rae1–167 nup184–1 (SL27) synthetic lethal cells (Figure 5, A and C). The fusion protein was localized in the cytoplasm when rae1 was expressed (−B1) but accumulated in the nucleus when expression of rae1 was repressed (+B1) (Figure 5D; compare −B1 with +B1 panels). Next, we examined the effect of attaching the C-terminal region containing the nuclear export activity onto the RNA-binding segment in pAG177 on the complementation of growth and mRNA export defect of SL27 cells. For this, a region containing the full nuclear export activity (460–710 aa) or a truncated, export-deficient region (460–694 aa) was inserted into pAG177 to obtain RRM1-RRM3-NES (460–710)-GFP (pAG116) or RRM1-RRM3-NES (460–694)-GFP (pAG178) fusions (Figure 5A). As seen in Figure 5, A and C, cellular growth and significant rescue of mRNA export was achieved in cells carrying the pAG116 but not the pAG178 plasmid. These results suggest that the ability to complement the growth of rae1–167 nup184–1 (SL27) synthetic lethal strain and the rescue of the mRNA export defects probably depended on the presence of a functional nuclear export activity as well as an ability to bind RNA. Consistent with the complementation properties, the GFP fusion expressed from pAG116 accumulated in the cytoplasm, whereas that from pAG178 was seen to be located in the nucleus under the conditions of synthetic lethality (Figure 5D). The cytoplasmic localization of the NES-GFP fusion in the absence of rae1 could be the result of a steady-state localization of a dynamic protein that shuttles. These results raised the possibility that when the RRM domains are fused to a nuclear export activity, the resulting protein may be able to function as an mRNA export carrier by using the RRM domain to bind the cargo RNA and transporting them during its own export.
It is possible that the C-terminal region with nuclear export activity has additional functions that are necessary for mRNA export. We directly tested whether the export activity of Crp79p could be functionally substituted by a heterologous nuclear export function from spMex67p or the leucine-rich HIV-Rev NES (and its mutant) (Fischer et al., 1995; Yoon et al., 2000). By inserting the spMex67 export activity region (435–509 aa), the Rev-NES, or its mutant version into the RRM1-RRM3-GFP fusion (pAG177), we constructed the plasmids pAG188, pAG179, and pAG180, respectively (Figure 5A). RRM1-RRM3 fused to the spMex67 nuclear export activity could suppress the synthetic lethality as well as the mRNA export defects of rae1–167 nup184–1 (SL27) cells (Figure 5, A and C). In cells expressing the Rev-NES fusion (pAG179), slightly reduced accumulation of poly(A)+ RNA in the nucleus and increased levels in the cytoplasm were observed compared with rae1–167 nup184–1 (SL27) cells that expressed the mutant version of Rev-NES (pAG180) under similar conditions (Figure 5C). However, no clear growth complementation was achieved by either wild-type or the mutant Rev-NES GFP fusions. Fusion proteins containing either the spMex67p or Rev-NES both localized in the cytoplasm under the conditions of synthetic lethality (Figure 5D). In contrast, the fusion protein containing the mutant Rev-NES was located predominantly in the nucleus. The steady-state level of various proteins bearing nuclear export activity was comparable (Figure 5B). The ability of the spMex67-NES to functionally replace the nuclear export function of Crp79p suggests that mRNA export functions of the latter were indeed mediated by a nuclear export activity–dependent mechanism. Because the RRM1-RRM3-Rev-NES-GFP (pAG179) fusion was located in the cytoplasm in a Rev-NES–dependent manner, these results suggest that it also was capable of exporting mRNA, although the strong nuclear accumulation of poly(A)+ RNA may mean that the magnitude of mRNA export was not significant.
Crp79p-GFP Shuttles between the Nucleus and the Cytoplasm
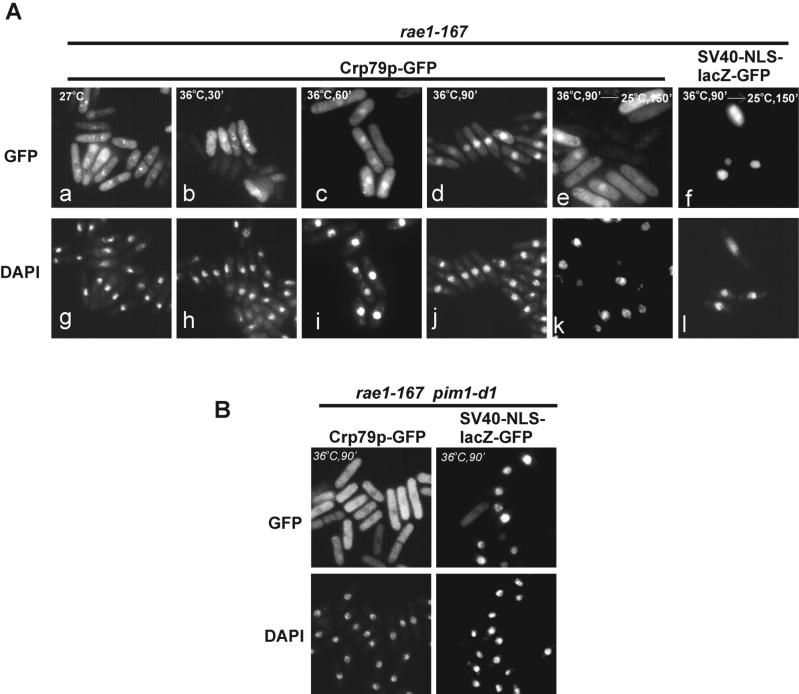
For Crp79p to function as an mRNA export carrier, it would be expected to shuttle between the nucleus and the cytoplasm. The nuclear localization of a GFP fusion consisting of RRM1 and RRM3 alone (pAG157) under the conditions of synthetic lethality already suggested this possibility (Figure 3B). To test directly whether shuttling occurred, we looked for genetic backgrounds in which Crp79p-GFP accumulated in the nucleus. After inactivation of Rae1p (36°C, rae1–167 mutation), Crp79-GFP translocated from the cytoplasm into the nucleus within 90 min (inactivation for 60 min resulted in only in a partial nuclear accumulation) (Figure 6, compare panels b, c, and d). It was previously shown that under similar conditions, a strong nuclear accumulation of poly(A)+ RNA took <30 min (Brown et al., 1995). Thus, Crp79p-GFP was capable of entering into the nucleus, but it took considerably more time to accumulate in the nucleus than poly(A)+ RNA.
Figure 6.
Shuttling of Crp79p-GFP and Ran-dependent import. (A) Shuttling of Crp79p-GFP fusion in rae1–167 cells. The nuclear accumulation of Crp79p-GFP in cells with rae1–167 mutation at 27°C (a) and after shift to 36°C for 30′, 60′, and 90′ (b–d) are indicated. Incubation of cells at 36°C for 90 min followed by additional incubation for 150 min at a permissive temperature of 25°C is shown in e. Localization of SV40-NLS-lacZ-GFP was used as a control; cells were incubated at 36°C followed by incubation at 25°C for 150 min (f). Coincident DAPI staining for all samples are shown at bottom (g–i). (B) Localization of Crp79p-GFP and SV40-NLS-lacZ-GFP in rae1–167 pim1-d1 strain after simultaneous inactivation of both proteins for 90 min at 36°C are shown. Coincident DAPI staining is shown below.
To determine whether Crp79p-GFP was capable of exiting the nucleus, the fusion protein was imported into the nucleus as above (Figure 6A, panel d). These cells were then returned to the permissive temperature of 25°C for 2.5 h in the presence of cycloheximide to prevent new protein synthesis. Results in Figure 6A (compare panels d and e) show that Crp79p-GFP was able to exit the nucleus after a shift back to the permissive temperature. As a control for nuclear envelope integrity, an SV40-(NLS)-GFP-lacZ reporter protein remained in the nucleus throughout the experiment (Figure 6A, panel f). The nuclear import and export of Crp79p together show that that the protein could shuttle between the nucleus and the cytoplasm. Crp79p-GFP fusions that possess the C-terminal nuclear export activity (Figure 2A) similarly could shuttle between the nucleus and cytoplasm (data not shown).
Nuclear Import of Crp79p Is Ran-Dependent
In our search for an import signal within Crp79p, no short sequences within the RRM domains was identified that could direct nuclear import of a lacZ-GFP fusion protein (data not shown). Most proteins that are imported into the nucleus either carry or bind to adapter protein(s) that contain a nuclear localization signal (NLS) for interaction with members of the β-importin–type import receptors. Typically, the import receptors require the functioning of the Ran system (Dasso and Pu, 1998). We examined whether the import of Crp79p-GFP into the nucleus depends on the Ran-GTPase switch system. For this, the Crp79-GFP fusion was expressed in a rae1–167 pim1-d1 strain, encoding ts mutations for both Rae1p and RCC1p proteins (Demeter et al., 1995). The latter is a guanine nucleotide exchange factor, and its ts mutation in S. cerevisiae confers defects in the nuclear import of proteins at restrictive temperatures (Demeter et al., 1995). After simultaneous inactivation of rae1–167 and pim1-dl at 36°C for 90 min, Crp79p-GFP was found to be located predominantly in the cytoplasm (Figure 6B). In some cells, a hollow nucleus could be detected. Because cells carrying the pim1 mutation may show disintegration of the nuclear envelope, we ensured the nuclear envelope integrity by using a reporter SV40-NLS-LacZ-GFP that remained nuclear during and after the inactivation treatment (Demeter et al., 1995). Only ∼10% of total cells showed a diffused localization of the reporter, suggesting that most cells maintained nuclear envelope integrity during the course of the experiment (Figure 6B). We conclude that the inability to import Crp79p-GFP is caused by a loss of RCC1 function, and thus, its import depends on the Ran system.
DISCUSSION
Crp79p Is an spMex67p-like mRNA Export Carrier
In this article, we describe the isolation and functional characterization of a novel poly(A)+ RNA binding protein from S. pombe, Crp79p, that can function in the nuclear export of mRNA. A schematic depicting the functional domains of Crp79p is shown in Figure 7. Like spMex67p, Crp79p was isolated as a multicopy suppressor of growth and mRNA export defects of the SL27 strain, suggesting that both support the export of similar mRNA cargo. spMex67p and Crp79p share some features. Both can UV cross-link poly(A)+ RNA in vivo and shuttle between the nucleus and the cytoplasm, and neither is essential for mRNA export in normal cells. Their simultaneous loss in Δcrp79 Δmex67 cells resulted in no apparent growth or mRNA export defects, suggesting functional redundancies of both proteins in S. pombe. Nonetheless, both Crp79p and spMex67p can function as auxiliary mRNA export carriers by using distinct, functionally essential nuclear export activities. Both properties of poly(A)+ RNA binding and nuclear export activity of Crp79p are required for exporting mRNA. Therefore, Crp79p may actually function directly as a carrier of mRNA.
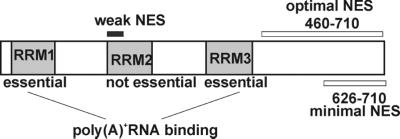
Figure 7.
Schematic summarizes the functional domains of Crp79p. A weak leucine-rich NES present in RRM2 is indicated, and the coordinates of the minimal and optimal nuclear export activity (NES) as identified in HeLa cells are indicated.
The Mode of Substrate Recognition of Crp79p May Be Different from That of spMex67p
Despite functional similarity, there was very little structural homology between Crp79p and spMex67p. spMex67p contains a single RNP-type RNA recognition motif and can bind mRNA, but by comparison with scMEX67p/Tap, it is likely that spMex67p uses adapter proteins, such as Yra1, to capture mRNA indirectly (Strasser et al., 2000; Zhou et al., 2000). In contrast, Crp79p, like the poly(A)-binding proteins, contains three RRM domains, two of which (RRM1 and RRM3) were essential for its function. The RRM-type motif is used directly for binding substrate RNA, as shown in the case of the U1A protein to U1 snRNA (Scherly et al., 1989). Although the role of Yra1-like proteins cannot be ruled out, it seems likely that Crp79p interacts directly with specific transcripts via its multiple RRM domains. Precedence for such a direct mode of substrate recognition is well known for Rev, a protein that binds an RNA sequence called Rev-responsive element present on HIV-1 transcripts (Pollard and Malim, 1998).
The mRNA Export Function of Crp79p Depends on Its C-Terminal Nuclear Export Activity
Several experimental observations suggest that nuclear export activity present at the C-terminus of Crp79p is necessary for mRNA export in SL27 cells under synthetic lethal conditions. First, the steady-state localization of Crp79p in cytoplasm in the presence of B1 correlates directly to the rescue of the mRNA export defect, whereas the nuclear localization of RRM1-RRM3-GFP fusion (pAG177) is contemporaneous with nuclear accumulation of mRNA. The simplest interpretation of these results is that nuclear export activity–dependent export of the protein directly exports bound mRNA. Second, nuclear export activity of spMex67p could substitute for that of Crp79p in shuttling as well as mRNA export functions of Crp79p. Third, after inactivation of Rae1p, RRM domains carrying the C-terminal nuclear export activity took 90 min, whereas the RRM domains without it took only 30 min, to accumulate in the nucleus. These results suggest that 1) Crp79p was still shuttling when mRNA accumulated in the nucleus, 2) Crp79p was probably exporting some mRNA during its exit from the nucleus, and 3) the nuclear exit of Crp79p is probably independent of Rae1p.
Interestingly, in some cases the presence of RRMs plus a nuclear export activity was not sufficient to rescue growth and mRNA export defects. Some deletion derivatives of Crp79p-GFP (Figure 2A) were unable to rescue the growth and mRNA export defects of rae1–167 nup184–1 (SL27) cells under synthetic lethal conditions, although they presumably are capable of binding RNA, contain an nuclear export activity, and shuttle between the nucleus and cytoplasm. Because all of these deletion derivatives were localized predominantly in the cytoplasm under the conditions of synthetic lethality, one possibility is that they exported some RNA but that the magnitude of mRNA export was too low and/or export of specific transcripts was affected by the internal deletions.
How Does Crp79p Interact with the NPC?
The current understanding of scMex67/Tap function in the export of mRNA suggests that they bridge mRNP complexes directly to the nuclear pores (Conti and Izaurralde, 2001; Zenklusen and Stutz, 2001). The C-subterminal regions of both scMex67 and Tap interact directly with the FG repeats of nuclear pore proteins directly or in the presence of Mtr2/p15 (Bachi et al., 2000; Strasser et al., 2000; Strawn et al., 2001). In the case of Rev, it has a distinct NES that is recognized by its receptor, Crm1p (Fornerod et al., 1997; Stade et al., 1997). The latter interacts with pore factors to navigate Rev-associated mRNA through the pores. It is not clear whether scMex67/Tap and Rev follow some common steps or whether they operate in completely separate pathways. At this point, it is not clear whether the C-terminus of Crp79p interacts directly with NPC proteins or binds to a karyopherin that, in turn, directs the Crp79p-mRNA complex out of the nucleus.
Does Crp79p Possess an NES or an Interaction Domain?
Two nuclear export activities were identified within Crp79p by use of HeLa cells in a heterologous assay of nuclear export (Figure 7). One activity was a leucine-rich NES (Figure 4B) within RRM2 that was not essential for the mRNA export function of Crp79p (see pAB12 in Figure 2A). The second nuclear export activity was present at the C-terminus (626–710 aa), was essential for Crp79p function, and was functionally conserved in human cells. However, the nuclear export activity at the C-terminus required an additional stimulatory region between aa 460 and 625 for full activity. Interestingly, in the context of S. pombe, the minimal nuclear export (626–710 aa) activity was sufficient for the export of the fusion protein (data not shown), but complementation of the mRNA export defect required a larger segment (460–710 aa) (see pAB33 in Figure 2, A and C). The nuclear export activity of the C-terminus may be complex, containing more than one activity or signal region. A small deletion could damage the export function by deleting one or more such noncontiguous elements. Alternatively, there could be one or more adapter proteins that interact with the C-terminal region, such that a larger section of the protein is needed for optimal function. We are currently investigating these possibilities.
The Nature of the RRM Domains
By analogy with other hnRNP-like proteins, the RRM domains of Crp79p may function by binding to RNA. Specifically, RRM1 and RMM3 each could independently bind poly(A)+ RNA, as determined by UV cross-linking experiments. Both RRM1 and RRM3 are essential for complementation of growth and mRNA export defect of rae1–167 nup184–1 cells. Most hnRNP proteins contain several RRM domains, and these may be required for cooperative binding, and/or they may have specificity for RNA sequences or structure (Burd and Dreyfuss, 1994b). Interestingly, the rae1–167 inactivation experiments suggest that RRM1 and RRM3, when fused to GFP, could shuttle between the nucleus and the cytoplasm (data not shown). Apparently, these domains do not possess a nuclear export activity, and their export from the nucleus is probably the result of binding RRM domains to poly (A)+ RNA that is exported by some other mechanism. And no discrete sequence with the RRMs was detected that could lead to import of lacZ-GFP fusions (data not shown). Nevertheless, the Ran-dependency of Crp79p import suggests the involvement of an importin-β type receptor.
Redundancy and Conservation of mRNA Export Pathways
Both spmex67 and crp79 are nonessential genes, so the mRNA export functions of spMex67p and Crp79p are redundant. In contrast, the orthologous S. cerevisiae MEX67 gene is essential for both growth and mRNA export. Moreover, both S. cerevisiae and humans lack a protein closely related to Crp79p. These observations suggest that each organism has a specific set of mRNA export carrier proteins that potentially use different export pathways. Despite this variation, the mechanism of export within a particular pathway is likely to be conserved. For example, both scMex67p and human Tap bind to FG-repeat nuclear pore proteins. Similarly, although an orthologue for Crp79p has not been found, its C-terminus can function efficiently to direct nuclear export in HeLa cells, suggesting that a similar export pathway may exist in human cells.
When different pathways are present, each export pathway may acquire some substrate specificity but not completely lose its ability to export other substrates. This redundancy would lead to the inability to detect an mRNA export defect when only one of the carrier proteins is defective. Substrate specificity would also play a role in the ability of a carrier protein gene to be a multicopy suppressor when different mRNA export pathways are defective. In this study, leucine-rich Rev NES or its likeness in RRM2 could not substitute for the C-terminal nuclear export functions of Crp79p, suggesting that mere export of the protein by any pathway would not constitute concurrent mRNA export. However, the Mex67p nuclear export activity could substitute for the C-terminal export functions of Crp79p, and the suppression pattern of the Crp79p-Mex67p fusion (pAG188) (ability to suppress rae1–167 nup184–1 but not rae1–167 at 32°C; A.G.T. and R.D., unpublished observations) was that of Crp79p and not of Mex67p. Presumably, the Mex67p export activity directs export of Crp79p along the same export pathway as it does when functioning to export Mex67p, or both export pathways have common steps. This suggests that the substrates that interact with Crp79p and Mex67p are different, with Mex67p able to export substrates when Rae1p is defective that Crp79p cannot. It is interesting to note in this regard that crp79 expression increases during meiosis (Watanabe et al., 2001). Given the redundancy in mRNA export pathways and the ability of multicopy level of expression of proteins to export less preferred substrates, defining the specificity of each individual carrier protein will require the identification of all of the carriers and examination of their specificities when expressed in a single copy.
ACKNOWLEDGMENTS
We thank Dr. Shelly Sazer for providing the S. pombe pim1-d1 strain. We thank Mike Radonovich and Dr. Cindy Pise-Masison for help with tissue culture and the members of our laboratory for helpful discussions. We are especially grateful to Dr. Henry Levin for a critical reading of the manuscript.
Abbreviations used:
- GFP
green fluorescent protein
- Gr
glucocorticoid receptor
- mRNP
ribonucleoprotein
- NES
nuclear export signal
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- REF
RNA export factor
- RRM
RNA recognition motif
- ts
temperature-sensitive
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E01–11–0133. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E01–11–0133.
REFERENCES
- Alfa C, Fantes P, Hyams J, Mcleod M, Warbrick E. Experiments with Fission Yeast. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1993. [Google Scholar]
- Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Bachi A, Braun IC, Rodrigues JP, Pante N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Gorlich D, Carmo-Fonseca M, et al. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbet N, Muriel WJ, Carr AM. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene. 1992;114:59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- Brown JA, Bharathi A, Ghosh A, Whalen W, Fitzgerald E, Dhar R. A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+ RNA export and in the cytoskeleton. J Biol Chem. 1995;270:7411–7419. doi: 10.1074/jbc.270.13.7411. [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994a;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994b;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E, Izaurralde E. Nucleocytoplasmic transport enters the atomic age. Curr Opin Cell Biol. 2001;13:310–319. doi: 10.1016/s0955-0674(00)00213-1. [DOI] [PubMed] [Google Scholar]
- Dasso M, Pu RT. Nuclear transport: run by Ran? Am J Hum Genet. 1998;63:311–316. doi: 10.1086/301990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff JA, Sachs AB. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J Mol Biol. 1997;269:67–81. doi: 10.1006/jmbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- Demeter J, Morphew M, Sazer S. A mutation in the RCC1-related protein pim1 results in nuclear envelope fragmentation in fission yeast. Proc Natl Acad Sci USA. 1995;92:1436–1440. doi: 10.1073/pnas.92.5.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Forsburg SL. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 1993;21:2955–2956. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Le Hir H, Schmitt C, Braun IC, Kocher T, Wilm M, Izaurralde I. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr Biol. 2001;11:1716–1721. doi: 10.1016/s0960-9822(01)00532-2. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Cullen BR. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung JU, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Yong J, Kataoka N, Abel L, Diem MD, Dreyfuss G. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J. 2001;20:2062–2068. doi: 10.1093/emboj/20.8.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- Love DC, Sweitzer TD, Hanover JA. Reconstitution of HIV-1 Rev nuclear export: independent requirements for nuclear import and export. Proc Natl Acad Sci USA. 1998;95:10608–10613. doi: 10.1073/pnas.95.18.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M-J, Zhou Z, Magni K, Christofrides C, Rappsilber J, Mann M, Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. The HIV-1 Rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- Maundrell K. Thiamine repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Transport of proteins and mRNA out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Nehrbass U, Rout MP, Maguire S, Blobel G, Wozniak RW. The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex. J Cell Biol. 1996;133:1153–1162. doi: 10.1083/jcb.133.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard VW, Malim MH. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- Rodrigues JP, Rode M, Gatfield D, Blencowe B, Carmo-Fonseca M, Izaurralde E. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc Natl Acad Sci USA. 2001;98:1030–1035. doi: 10.1073/pnas.031586198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherly D, Boelens W, van Venrooij WJ, Dathan NA, Hamm J, Mattaj IW. Identification of the RNA binding segment of human U1 A protein and definition of its binding site on U1 snRNA. EMBO J. 1989;8:4163–4170. doi: 10.1002/j.1460-2075.1989.tb08601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton DR, Chen S, Hitomi M, Kumagai C, Tartakoff AM. A yeast protein that bidirectionally affects nucleocytoplasmic transport. J Cell Sci. 1995;108:265–272. doi: 10.1242/jcs.108.1.265. [DOI] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Strambio-de-Castillia C, Blobel G, Rout MP. Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol. 1999;144:839–855. doi: 10.1083/jcb.144.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Bassler J, Hurt E. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J Cell Biol. 2000;150:695–706. doi: 10.1083/jcb.150.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413:648–652. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- Strawn LA, Shen T, Wente SR. The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. J Biol Chem. 2001;276:6445–6452. doi: 10.1074/jbc.M008311200. [DOI] [PubMed] [Google Scholar]
- Stutz F, Bachi A, Doerks T, Braun IC, Seraphin B, Wilm M, Bork P, Izaurralde E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Miyashita K, Saito TT, Yoneki T, Kakihara Y, Nabeshima K, Kishi YA, Shimoda C, Nojima H. Comprehensive isolation of meiosis specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res. 2001;29:2327–2337. doi: 10.1093/nar/29.11.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen WA, Yoon JH, Shen R, Dhar R. Regulation of mRNA export by nutritional status in fission yeast. Genetics. 1999;152:827–838. doi: 10.1093/genetics/152.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Love DC, Guhathakurta A, Hanover JA, Dhar R. Mex67p of Schizosaccharomyces pombe interacts with Rae1p in mediating mRNA export. Mol Cell Biol. 2000;20:8767–8782. doi: 10.1128/mcb.20.23.8767-8782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Whalen WA, Bharathi A, Shen R, Dhar R. Npp106p, a Schizosaccharomyces pombe nucleoporin similar to Saccharomyces cerevisiae Nic96p, functionally interacts with Rae1p in mRNA export. Mol Cell Biol. 1997;17:7047–7060. doi: 10.1128/mcb.17.12.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel U, Doye V, Tekotte H, Wepf R, Grandi P, Hurt EC. Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J Cell Biol. 1996;133:1141–1152. doi: 10.1083/jcb.133.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Stutz F. Nuclear export of mRNA. FEBS Lett. 2001;498:150–156. doi: 10.1016/s0014-5793(01)02482-6. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- Zolotukhin AS, Tan W, Bear J, Smulevitch S, Felber BK. U2AF participates in the binding of TAP (NXF1) to mRNA. J Biol Chem. 2002;277:3935–3942. doi: 10.1074/jbc.M107598200. [DOI] [PubMed] [Google Scholar]