Abstract
Regulation of the hTERT gene encoding the telomerase catalytic subunit plays an important role in human cell senescence, immortalization, and carcinogenesis. By examining the activity of various deleted or mutated hTERT promoter fragments, we show that an E-box element downstream of the transcription initiation site is critical to differential hTERT transcription between the telomerase/hTERT-positive renal cell carcinoma cell line (RCC23) and its telomerase/hTERT-negative counterpart containing a transferred, normal chromosome 3 (RCC23+3). This E-box element mediated repression of hTERT transcription in RCC23+3 but not in RCC23. A copy number–dependent enhancement of the repression suggested active repression, rather than loss of activation, in RCC23+3. Endogenous expression levels of c-Myc or Mad1, which could activate or repress hTERT transcription when overexpressed, did not account for the differential hTERT transcription. Gel mobility shift assays identified the upstream stimulatory factors (USFs) as a major E-box–binding protein complex in both RCC23 and RCC23+3 and, importantly, detected an RCC23+3-specific, E-box–binding factor that was distinct from the USF and Myc/Mad families. The E-box–mediated repression was also active in normal human fibroblasts and epithelial cells and inactive in some, but not all, telomerase/hTERT-positive cancer cells. These findings provide evidence for an endogenous, repressive mechanism that actively functions in telomerase/hTERT-negative normal cells and becomes defective during carcinogenic processes, e.g., by an inactivation of the telomerase repressor gene on chromosome 3.
INTRODUCTION
Telomeres are specialized structures at chromosome ends that consist of tandemly repeated DNA sequences and the associated proteins (König and Rhodes, 1997). A ribonucleoprotein enzyme, telomerase, catalyzes de novo synthesis of telomeric repeat DNA to maintain telomere length and structure (Bryan and Cech, 1999). Telomere shortening with each cell division in the absence of a telomere maintenance mechanism is suggested to function as an intrinsic clock that counts cell divisions and eventually causes permanent cell growth arrest (i.e., cellular or replicative senescence) in human cells (Chiu and Harley, 1997; Meyerson, 2000). Activation of telomerase is observed in ∼90% of human cancers but not in most normal somatic cells (Kim et al., 1994; Chiu and Harley, 1997; Meyerson, 2000). Forced expression of telomerase activity stabilizes telomeres in normal human cells and extends their replicative life span beyond cellular senescence (Bodnar et al., 1998). Conversely, inhibition of telomerase activity in cancer cells abolishes their telomere maintenance and immortal growth (Hahn et al., 1999). These findings establish an important role of telomerase-mediated telomere maintenance in human cell immortalization and carcinogenesis and suggest that telomerase repression may be a tumor-suppressive mechanism (Chiu and Harley, 1997; Meyerson, 2000). The expression level of the human telomerase reverse transcriptase (hTERT) gene encoding the telomerase catalytic subunit, which is primarily under transcriptional control, represents a major determinant of telomerase activity in human cells (Meyerson et al., 1997; Nakamura et al., 1997; Poole et al., 2001). Thus, investigation of transcriptional regulation of the hTERT gene should be essential for elucidating molecular mechanisms of telomerase regulation, cellular senescence, immortalization, and carcinogenesis in humans.
Several transcription factors have thus far been suggested as candidates for the transcriptional regulators of hTERT (Poole et al., 2001): the E-box–binding oncoprotein c-Myc (Greenberg et al., 1999; Wu et al., 1999; Kyo et al., 2000) and a ubiquitous transcription factor Sp1 (Kyo et al., 2000) are activators; the E-box–binding factor Mad1 (Günes et al., 2000; Oh et al., 2000), the tumor suppressor proteins p53 (Xu et al., 2000) and WT1 (Oh et al., 1999), and the zinc-finger factor MZF2 (Fujimoto et al., 2000) are possible repressors. Analyses of the activator or repressor function of these regulators, however, were based largely on the effect of overexpressed proteins. Among the consensus binding sequences of these regulators, two canonical E-box (CACGTG) elements located upstream and downstream of the transcription initiation site (−187 to −182 and +22 to +27, respectively), which are the potential binding sites for c-Myc and Mad1 (Sommer et al., 1998), have been most extensively analyzed. Although recombinant c-Myc and Mad1 proteins are able to bind these E-box elements (Wu et al., 1999; Kyo et al., 2000; Oh et al., 2000), there is no direct evidence that endogenously expressed Myc/Mad family of transcription factors contributes to transcriptional activation of the hTERT during transformation from normal to cancer cells. Only one report thus far provided direct evidence for a role of endogenous c-Myc and Mad1 in hTERT regulation during cell differentiation (Xu et al., 2001). Thus, the endogenous protein factors and their DNA binding sites that are critical to the regulation of hTERT transcription during human cell immortalization and carcinogenesis still remain to be identified.
Multiple mechanisms seem to play roles in activation and repression of the hTERT in cancer and normal cells, respectively (Devereux et al., 1999; Cong and Bacchetti, 2000; Poole et al., 2001); which of these mechanisms becomes functional, however, varies among individual tumors, different cell types, and cellular environments. To dissect each of these mechanisms, it is important to have an experimental system in which a specific regulatory mechanism can be analyzed. In this study, we used a pair of cell lines that have similar genetic backgrounds but differ in telomerase activity: a telomerase-positive, human renal cell carcinoma cell line (RCC23) and its telomerase-negative counterpart (RCC23+3) that was generated by microcell-mediated transfer of a normal human chromosome 3 into RCC23 cells (Table 1; Horikawa et al., 1998). Progressive shortening of telomeres as a function of cell population doublings and induction of cellular senescence at ∼40 population doublings after chromosome 3 transfer were observed in RCC23+3, in agreement with the repression of telomerase activity (Horikawa et al., 1998). The telomerase repression by chromosome 3 transfer was a result of the marked downregulation of hTERT mRNA expression (Table 1; Horikawa et al., 1998), suggesting the presence of a telomerase/hTERT-repressor gene on this chromosome. Thus, this experimental system consisting of two cell lines that are isogenic except for a transferred copy of chromosome 3 should be useful to dissect the specific regulation of hTERT that involves a putative repressor gene on chromosome 3. By systematic examination of the transcriptional activity of a series of truncated or mutated hTERT gene promoter fragments, we have identified a DNA element that is critical to the transcriptional control of hTERT in RCC23 and RCC23+3 cells. This analysis provides new insight into the endogenous regulation of hTERT expression in human cells. In addition, an artificial promoter generated during the course of this study may be a better tool for cancer gene therapy than the wild-type hTERT promoter.
Table 1.
Summary of characteristics of RCC23, RCC23+3, RCC23+3p, and REV cells
| Cell | Transferred chromosome 3a | Telomerase activityb | hTERT mRNAc | Telomere lengthd | Life span |
|---|---|---|---|---|---|
| RCC23 | none | + | + | maintained | immortal |
| RCC23+3 | intact 3 | − | −e | shortens | mortal |
| progressively | (41 PDs)f | ||||
| RCC23+3p | 3pter-q22 | − | −e | shortens | mortal |
| progressively | (28 PDs)f | ||||
| REV | 3pter-p23 | + | + | maintained | immortal |
| 3cen-q22 |
Transferred by means of microcell fusion.
TRAP (telomeric repeat amplification protocol) assay.
Conventional reverse transcription (RT)-PCR and quantitative real-time RT-PCR (Taqman) assays.
Terminal restriction fragment length by Southern blot.
At least 64-fold lower expression than RCC23 in Taqman assay.
Senesce at 41 or 28 population doublings after microcell fusion.
MATERIALS AND METHODS
Construction of Plasmids
A fragment of the hTERT promoter (−3915 to +40) was PCR-amplified from a bacterial artificial chromosome clone containing the hTERT genomic sequence (Horikawa et al., 1999) and inserted into SacI/SmaI sites of the luciferase reporter vector pGL3-Basic (Promega Corp., Madison, WI) to generate the pBT-3915. A series of unidirectional truncations from upstream (pBT-1125, pBT-949, pBT-385, pBT-304, pBT-255, pBT-88, and pBT-33) were generated by endonuclease digestion (SacI plus StuI, PstI, BstEII, BssHII, PvuII, SmaI, or SacII, respectively) of the pBT-3915 followed by end-polishing and self-circularization. The pBT-211 (previously named p2XEB), pBTdel-255, pBTdel-208, and pBTdel-130 were constructed as previously described (Horikawa et al., 1999). To make mutations in the pBT-255 construct, the QuikChange Site-Directed Mutagenesis kit (Stratagene Cloning Systems, La Jolla, CA) was used according to the supplier's protocol. For artificial promoters with additional E-box elements (pBT-255–2DEB and pBT-255–4DEB), one or three copies of the synthetic DNA (5′-CGCACGTGGG-3′; a canonical E-box italicized) were placed immediately downstream of the hTERT promoter (into XhoI/HindIII sites) in the pBT-255. For c-Myc and Mad1 expression constructs, human c-myc and mad1 cDNAs were amplified by RT–PCR and inserted into the mammalian expression vector pcDNA3.1(+) (Invitrogen Corp., San Diego, CA). All the plasmids were confirmed to have correct sequences by DNA sequencing.
Cells and Luciferase Assay
A renal cell carcinoma cell line, RCC23, and its derivative with a transferred copy of normal human chromosome 3 (RCC23+3) were previously described (Horikawa et al., 1998; clone 3-C was used as RCC23+3 in this study), and their properties are summarized in Table 1. RCC23+3p (clone 3-B in Horikawa et al., 1998) carries a transferred copy of partial human chromosome 3 (entire short arm plus cen-q22) and shows phenotypes similar to RCC23+3 (Table 1). REV is a revertant clone that emerged from senescent RCC23+3p culture with loss of the transferred 3p22-cen loci and reacquired the phenotypes of parental RCC23 cells (Horikawa et al., 1998, 2001; Table 1). For the luciferase assay, cells (8.0 × 104) were seeded on 24-well plates, cultured overnight and transfected with the hTERT promoter–luciferase plasmids (0.5 μg per well) by use of FuGENE6 transfection reagent (Roche Diagnostics, Indianapolis, IN). The ratio of DNA to FuGENE6 was 1:3, which resulted in similar transfection efficiencies in RCC23 and RCC23+3 cells. These conditions for transfection in this study made the comparison between these two cell lines more direct and reliable than in our previous study (Horikawa et al., 1999), in which the transfection efficiency in RCC23 was significantly higher than that in RCC23+3 (also see DISCUSSION). The pRL-SV40 (2 ng per well; Promega) driving Renilla reniformis luciferase was included in each transfection as a control to normalize the transcriptional activity of hTERT promoter fragments. The expression construct (c-Myc, Mad1, or vector alone; 1.0 μg per well) was included in cotransfection experiments. Preparation of cell lysates and measurement of luciferase activity were performed by use of the dual luciferase reporter assay system (Promega). All the data, expressed as the mean and SD, were from at least three independent experiments.
Normal human fibroblasts (NHFs) were derived from neonatal foreskin (Horikawa et al., 2001). Normal human prostate epithelial cells (PrECs) were obtained from BioWhittaker, Inc. (Walkersville, MD) and maintained according to the supplier's protocol. Rapidly proliferating NHFs and PrECs at early-passage culture were used in this study. The lack of telomerase activity and hTERT mRNA in NHFs and PrECs was confirmed as described previously (Horikawa et al., 1998; Devereux et al., 1999). Human cell lines used in this study that express telomerase activity and hTERT mRNA include CMV-Mj-HEL-1 (immortalized fibroblast cell line; a gift from Dr. Olivia Pereira-Smith, Baylor College of Medicine, Houston, TX); MCF-7 [breast cancer cell line; obtained from American Type Culture Collection (ATCC), Manassas, VA]; MDA-MB-435 (breast cancer cell line; obtained from ATCC); DU145 (prostate cancer cell line; obtained from ATCC); and TSU-Pr1 (“T24”) (a gift from Dr. Carrie Rinker-Schaeffer, University of Chicago, Chicago, IL), which was recently identified to be bladder cancer cells rather than of prostatic origin (van Bokhoven et al., 2001). Human mammary epithelial cells (strain 184; a gift from Dr. Martha Stampfer, Lawrence Berkeley National Laboratory, Berkeley, CA) and NHFs were infected with the LXIN retrovirus containing full-length hTERT cDNA (Nakayama et al., 1998; Mueller et al., 2000; Stampfer et al., 2001) to produce immortal 184-hTERT and NHF-hTERT cells, respectively. The luciferase assay using these cells as recipients (6.0 × 104 to 1.2 × 105 per well seeded, depending on cell size and growth rate) was carried out as described above.
Gel Mobility Shift Assay
Whole-cell extracts were prepared from exponentially growing cells as previously described (Mudryj et al., 1991). For gel mobility shift assays, 3 μg of protein was incubated with 32P-labeled double-stranded oligonucleotide at room temperature for 20 min in the binding buffer: 20 mM HEPES (pH 7.4), 1 mM MgCl2, 0.1 mM EDTA, 40 mM KCl, 0.5 mM dithiothreitol, 1 μg of sonicated salmon sperm DNA, 60 μg of BSA, and 1% Ficoll (Mudryj et al., 1991). DNA-protein complexes were resolved on a 4% polyacrylamide gel at 4°C. For supershift of the complexes, whole-cell extracts were preincubated with the indicated antibodies before addition of 32P-labeled oligonucleotides. The following sequences were used as the probes: CGCACGTGGG (+20 to +29; canonical E-box italicized), GCTGCGCACGTGGGAAGCCC (+16 to +35; canonical E-box italicized), GCTGCGCACCCGGGAAGCCC (+16 to +35; mutated E-box italicized), and GCGGACCCCGCCCCGTCCCG (−117 to −98; consensus Sp1 binding site italicized).
Western Blot Analysis
Forty micrograms of protein was resolved on 10% polyacrylamide gels and transferred to a nitrocellulose membrane (Hybond-ECL, Amersham Pharmacia Biotech, Inc., Piscataway, NJ) or a PVDF membrane (Immobilon P, Millipore Corp., Bedford, MA). Blocking and incubation of the membranes with primary and secondary antibodies followed the suppliers' instructions. Protein bands were detected by use of the ECL Western blotting detection system (Amersham Pharmacia Biotech, Inc.). The following antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA): c-Myc (sc-764), Mad1 (sc-222), Max (sc-197), USF1 (sc-229), and USF2 (sc-861).
RESULTS
The Sequence Downstream of the Transcription Initiation Site Is Responsible for Differential hTERT Transcription between RCC23 and RCC23+3 Cells
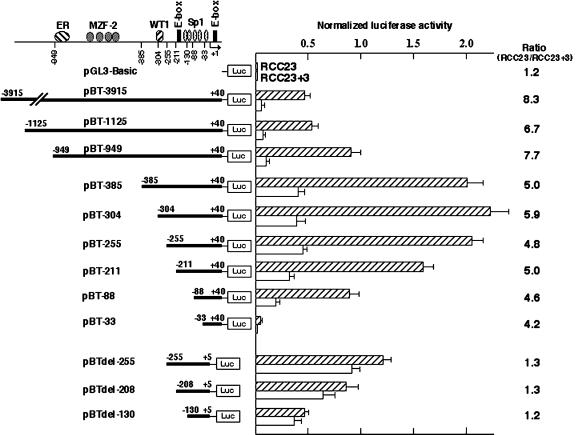
Transcriptional activity of a 3955-bp hTERT promoter fragment (−3915 to +40; construct pBT-3915) and a series of 5′-deleted fragments (from position −X to +40; constructs pBT-X's) was examined in a luciferase assay using RCC23 and RCC23+3 cells as the recipients (Figure 1). The 3955-bp fragment (pBT-3915) showed approximately eightfold higher activity in RCC23 than in RCC23+3, suggesting that the difference in hTERT mRNA expression between these two cells can be attributed largely to differential transcription from the hTERT promoter.
Figure 1.
Luciferase assay of hTERT promoter activity in RCC23 and RCC23+3. A series of hTERT promoter fragments (nucleotide positions follow Horikawa et al., 1999) were cloned upstream of the firefly luciferase reporter gene in the pGL3-Basic vector. Schematic representation of transcription factor binding sites is shown at top (ER, estrogen receptor; Misiti et al., 2000; see text for the others). The firefly luciferase activity was normalized with the Renilla reniformis luciferase activity by the cotransfected pRL-SV40. The mean and SD from at least three independent experiments are shown. For each construct, the activity in RCC23 was divided by that in RCC23+3 to determine the ratio of RCC23/RCC23+3 as an indicator of chromosome 3–mediated fold repression, shown at right.
The data from the series of 5′-deleted promoter fragments support the contributions of some known factors to hTERT transcriptional control. Specifically, the increase in the luciferase activity with the deletion of −949 to −386 (compare pBT-949 and pBT-385) is consistent with the function of MZF2 repressor and its binding sites within this region (Fujimoto et al., 2000). The marked decrease with the deletion of −211 to −34 (compare pBT-211, pBT-88, and pBT-33) can be attributed to transcriptional activation mediated by multiple Sp1 binding sites, as previously reported (Kyo et al., 2000). Interestingly, however, a significant difference between RCC23 and RCC23+3 was observed for all of the 5′-deleted promoter fragments tested, as shown by the consistently high RCC23/RCC23+3 ratio (4.2–8.3, Figure 1). These findings indicated that transcriptional regulators binding to the examined region (−3915 to −34), such as MZF2 and Sp1, control hTERT transcription in both RCC23 and RCC23+3 cells but were not critical to the differential hTERT transcription observed between the two. It is also unlikely that the upstream E-box element (−187 to −182) is responsible for the differential transcription, because the deletion containing this E-box (compare pBT-211 and pBT-88) did not abrogate the difference between RCC23 and RCC23+3.
We next tested the activity of promoter fragments with a 35-bp deletion (+6 to +40) downstream of the transcription initiation site. In all three constructs with this deletion (constructs pBTdel-255, pBTdel-208, and pBTdel-130 in Figure 1), RCC23+3 exhibited hTERT promoter activity comparable to that of RCC23, with RCC23/RCC23+3 ratios of 1.2 or 1.3, significantly lower than the ratio observed with constructs containing the 35-bp sequence. Notably, the deletion of the downstream sequence resulted in an approximately twofold increase in the transcriptional activity in RCC23+3, whereas it resulted in an ∼40% decrease in RCC23 (compare pBT-255 and pBTdel-255). These results suggest that the region downstream of the transcription initiation site contains a DNA element or elements that contribute to the differential control of hTERT transcription in RCC23 versus RCC23+3 cells.
Identification of the Downstream E-Box as a Critical DNA Element
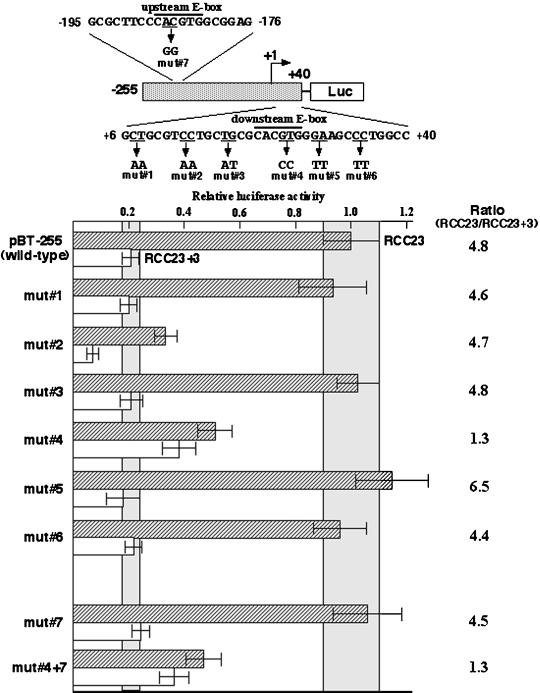
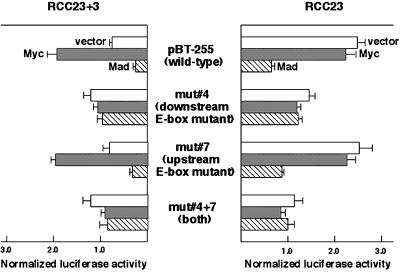
To pinpoint the critical DNA element(s), we created a series of mutations within the 35-bp downstream sequence by site-directed mutagenesis of the construct pBT-255 (mut 1–6 in Figure 2). Four of the six mutant promoter fragments (mut 1, 3, 5, and 6) showed transcriptional activities similar to that of the wild-type promoter in both RCC23 and RCC23+3 cells. In one mutant (mut 2), we observed an ∼65% decrease in the promoter activity in both RCC23 and RCC23+3, implying the presence of a novel DNA element involved in the activation of hTERT transcription; however, the difference between RCC23 and RCC23+3 was maintained in this mutant. Mutation of the downstream E-box (mut 4) resulted in a ∼50% decrease in promoter activity in RCC23 while producing a approximately twofold increase in promoter activity in RCC23+3 (RCC23/RCC23+3 ratio = 1.3), an effect similar to that observed with promoter fragments lacking the 35-bp downstream sequence. In contrast, when the upstream E-box (−187 to −182) was mutated (mut 7), no significant change in the promoter activity was observed in either RCC23 or RCC23+3, suggesting little or no contribution of the upstream E-box to the hTERT transcription in these cells. In the presence of this upstream E-box mutation, the downstream E-box mutation (mut 4 +7) again failed to show the difference between RCC23 and RCC23+3. These results identify the E-box located downstream of the transcription initiation site as a critical cis-acting DNA element in determining the differential hTERT promoter activity in our cell system and suggest that this E-box element could be involved in both activation and repression of the hTERT transcription in RCC23 and RCC23+3, respectively.
Figure 2.
Identification of DNA element responsible for the differential hTERT transcription in RCC23 and RCC23+3. Six mutations within the region downstream of the transcription initiation site (mut 1 to 6) and a mutation at the upstream E-box (mut 7) were made by site-directed mutagenesis of the construct pBT-255. The promoter activity of each fragment was measured by the luciferase assay, normalized as in Figure 1, and expressed as a value relative to the activity of the pBT-255 (wild-type) in RCC23. The mean ± SD ranges of the pBT-255 in RCC23 and RCC23+3 are highlighted for statistical comparison between this wild-type fragment and the mutant fragments. The ratio of RCC23/RCC23+3 (see Figure 1 legend) is shown on the right for each fragment.
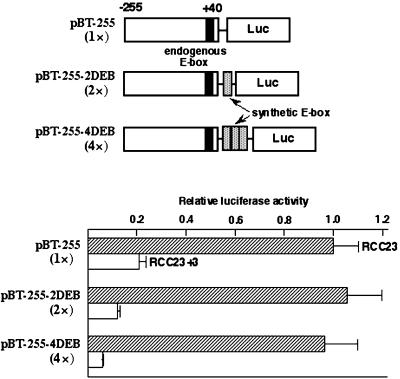
To further examine the downstream E-box–mediated regulation of the hTERT transcription, one or three copies of synthetic E-box sequences were inserted downstream of the wild-type promoter (two or four copies of downstream E-boxes in total; Figure 3). The extra copies of E-boxes did not affect the promoter activity in RCC23, implying that the E-box–mediated, activating mechanism is fully active with the single endogenous copy of E-box in this cell line. In contrast, a copy number–dependent repression of the promoter activity was observed in RCC23+3, resulting in a more obvious difference in the promoter activity between RCC23 and RCC23+3. This result does not favor the notion that an absence or inactivation of E-box–binding activator(s) is primarily responsible for the repressed hTERT transcription in RCC23+3. Instead, it supports the existence of an E-box–mediated repressive mechanism that actively functions in RCC23+3 and is defective in RCC23.
Figure 3.
Repressive effect by E-box elements in RCC23+3. One or three copies of synthetic E-box sequence were inserted downstream of the hTERT promoter in the construct pBT-255 to make the construct pBT-255–2DEB or pBT-255–4DEB, respectively (total number of downstream E-box elements is shown in parentheses). As in Figure 2, the promoter activity of each construct in RCC23 or RCC23+3 is expressed as luciferase activity relative to the pBT-255 in RCC23.
Downstream E-Box–mediated Repression Depends on the Presence of a Transferred Chromosome 3
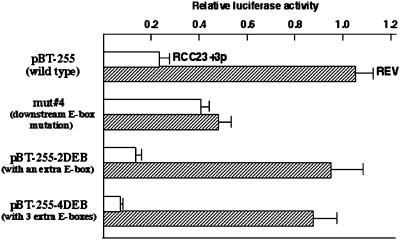
To further validate that a gene on the transferred copy of human chromosome 3 is responsible for the regulation of hTERT transcription mediated by the downstream E-box element, the activity of wild-type, E-box mutant and synthetic E-box–containing hTERT promoter fragments was examined in a second pair of RCC23-derived cells: RCC23+3p, telomerase/hTERT-negative cells with the transferred partial chromosome 3 (3pter-3q22); and REV, a telomerase/hTERT-expressing revertant clone that emerged from RCC23+3p with loss of 3p22-cen region from the transferred chromosome (Figure 4). RCC23+3p showed the same results as RCC23+3 for all the fragments examined: approximately fivefold repression compared with RCC23 in the wild-type promoter (pBT-255); approximately twofold increase with the downstream E-box mutation (mut 4); and enhancement of the repression in an E-box copy number–dependent manner (pBT-255–2DEB and pBT-255–4DEB). In contrast, the activities of these four promoter fragments in REV cells were similar to those observed in RCC23, showing ∼50% reduced activity of the E-box mutant fragment and no significant change by the addition of synthetic E-box sequences. In consequence, as observed in RCC23+3 and RCC23, the difference in hTERT promoter activity between RCC23+3p and REV was abrogated by the E-box mutation and became greater with the increased E-box copy number. These findings show that loss of the transferred chromosome 3p22-cen in the hTERT-repressed cells results in reversion to the hTERT-expressing cells, consistent with our previous mapping of the telomerase repressor gene on 3p21-p14.2 (Tanaka et al., 1998). On the basis of the reproducible results from two independent, chromosome 3–transferred clones (RCC23+3 and RCC23+3p), as well as the phenotypic reversion attributable to loss of the transferred chromosomal loci (in REV), we conclude that the downstream E-box–mediated repression of hTERT transcription depends on the function of a gene on the transferred human chromosome 3.
Figure 4.
Downstream E-box–mediated repression observed in RCC23+3p but not in a revertant clone REV. RCC23+3p and REV cells (Table 1) were used in the luciferase assay with the wild-type hTERT promoter fragment (pBT-255), the downstream E-box mutant (mut 4; see Figure 2), and the synthetic E-box–containing fragments (pBT-255–2DEB and pBT-255–4DEB; see Figure 3). The promoter activity of each construct in RCC23+3p or REV is expressed as a value relative to the activity of the pBT-255 in RCC23 (defined as 1.0 in Figures 2 and 3).
c-Myc and Mad1 Can Modulate hTERT Promoter Activity When Overexpressed but Are Not the Critical Endogenous Factors Causing Differential hTERT Transcription in RCC23 and RCC23+3
Previous work suggested that the transcription factors c-Myc and Mad1, which have an ability to bind canonical E-box elements (Sommer et al., 1998), can activate and repress hTERT promoter activity, respectively (Greenberg et al., 1999; Wu et al., 1999; Günes et al., 2000; Oh et al., 2000). The effects of these factors in RCC23 and RCC23+3 were examined by cotransfecting c-Myc and Mad1 expression plasmids with the luciferase plasmid pBT-255 or its E-box mutants. As shown in Figure 5, enforced expression of c-Myc protein enhanced the activity of the wild-type hTERT promoter in RCC23+3 but had little or no effect in RCC23. It is likely that the overexpressed c-Myc protein can abrogate the repressive mechanism functioning in RCC23+3. The inability of the overexpressed c-Myc to further enhance the promoter activity in RCC23 suggests a threshold response for the hTERT transcriptional activation. Overexpressed Mad1 protein decreased the transcriptional activity of the wild-type promoter in both RCC23 and RCC23+3 (∼70% reduction in both), consistent with its repressive effect on the hTERT transcription as suggested by others (Günes et al., 2000; Oh et al., 2000). Results from the promoter fragments mutated at either downstream or upstream E-box or at both (mut 4, 7, and 4 plus 7, respectively) showed that both activation by c-Myc expression and repression by Mad1 expression were mediated primarily by the downstream E-box element (Figure 5).
Figure 5.
Effects of c-Myc and Mad1 overexpression on hTERT promoter activity. c-Myc or Mad1 expression plasmid or vector control was cotransfected with the pBT-255 and its E-box mutants (see Figure 2). Overexpression of c-Myc or Mad1 protein was observed at similar levels in RCC23 and RCC23+3 (by Western blot analysis, not shown). Normalized luciferase activity is shown for each combination of plasmids.
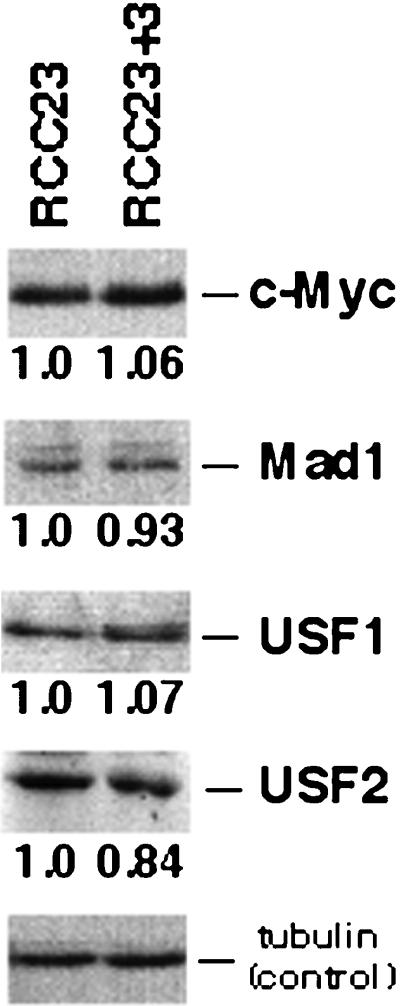
The results of overexpressing c-Myc and Mad1 proteins prompted us to examine whether endogenous c-Myc and Mad1 proteins are critical factors for the difference in hTERT transcription between RCC23 and RCC23+3 cells. Our analysis using Western blot showed that RCC23 and RCC23+3 expressed similar amounts of endogenous c-Myc and Mad1 proteins (Figure 6), consistent with the previous finding that a transferred chromosome 3 did not affect the expression levels of these proteins in 21NT breast carcinoma cells (Ducrest et al., 2001). Moreover, neither of the proteins was detected in the major E-box–binding complexes in either RCC23 or RCC23+3 under our experimental conditions (see details described below and shown in Figure 7). Thus, we have obtained no evidence that the expression level or activity of endogenous c-Myc or Mad1 is the primary determinant of the differential hTERT transcription in RCC23 and RCC23+3.
Figure 6.
Western blot analysis of E-box–binding proteins. Expression levels of representative E-box–binding proteins (c-Myc, Mad1, USF1, and USF2) and α-tubulin (a control for quantification) were measured by densitometric analysis. The value of E-box–binding proteins was normalized with that of α-tubulin. The expression level in RCC23+3 is shown relative to that in RCC23.
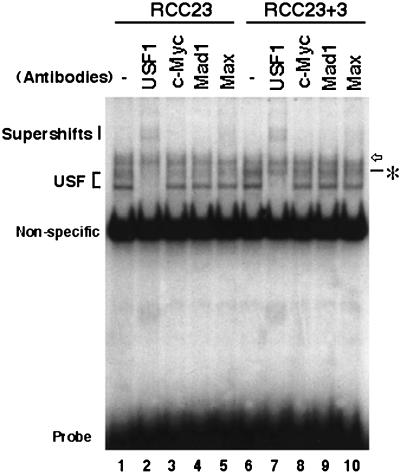
Figure 7.
Gel mobility shift assay of E-box–binding proteins in RCC23 and RCC23+3 cells. Result with use of the 10-bp probe containing a canonical E-box (+20 to +29) is shown. For lanes 2–5 and 7–10, whole-cell extracts were preincubated with the antibodies specific to the E-box–binding proteins indicated. Position of the USF complexes is shown at left. The asterisk indicates an RCC23+3-specific complex that was not supershifted or abrogated by any antibodies tested. The open arrow indicates a complex that is supershifted by the Max antibody. The supershifted bands containing the USF1 or Max antibody are indicated. These complexes were also detected by the 20-bp probe containing a canonical E-box (+16 to +35), but not by the 20-bp probe with the E-box mutated (data not shown). The strong band common to all samples was also observed with the E-box–mutated probe and unrelated sequences (e.g., the Sp1 probe; see MATERIALS AND METHODS) and represents a nonspecific binding. Bottom, free probe.
Detection of Endogenous Protein Factors That Bind the E-Box Element: USFs and a Novel RCC23+3-specific Binding Factor
To examine protein factors that bind the E-box element, a gel mobility shift assay was performed using the whole-cell extracts of RCC23 and RCC23+3. The result with the 10-bp probe containing the downstream E-box (+20 to +29) is shown in Figure 7. Antibodies to the E-box–binding proteins USF1, c-Myc, Mad1, and Max were included in the binding reactions to detect binding of these proteins. The major shifted bands were supershifted by preincubating the extracts with the USF1 antibody (lanes 2 and 7 in Figure 7). It is likely that these bands represented a USF1/USF1 homodimer and a USF1/USF2 heterodimer (Viollet et al., 1996). No significant difference was observed in the binding of USF complexes between RCC23 and RCC23+3, consistent with similar amounts of USF1 and USF2 proteins in these two cell lines as shown by Western blot analysis (Figure 6). Neither c-Myc antibody nor Mad1 antibody changed the profile of shifted bands (lanes 3, 4, 8, and 9 in Figure 7). By addition of the Max antibody, a slowly migrating, faint band was supershifted (lanes 5 and 10 in Figure 7). Thus, binding of c-Myc or Mad1 to the E-box element was not evident in either RCC23 or RCC23+3. Another E-box–binding protein, which remains to be identified, may form a complex with Max to bind the E-box element in both RCC23 and RCC23+3.
Importantly, a shifted band (marked by the asterisk in Figure 7) was observed in RCC23+3 but not in RCC23. This band was not supershifted by any of the antibodies tested and became more evident after supershift of comigrating USF complexes (compare lanes 2 and 7). This DNA-protein complex appears to be relatively unstable, because the salt concentration in the binding buffer and the electrophoresis conditions are critical to its detection. Nevertheless, the complex was observed reproducibly under our experimental conditions. The 20-bp probe containing the downstream E-box (+16 to +35), but not the 20-bp probe with the E-box mutated, detected similar profiles of binding, including the common USF complexes and the RCC23+3-specific factor (data not shown). These findings support the presence of an E-box–binding factor specific to hTERT-negative cells that plays a critical role in transcriptional control of the hTERT gene.
The Downstream E-Box Acts as a Negative Regulatory Element in Normal Human Cells but Not in Some Telomerase/hTERT-positive Cells
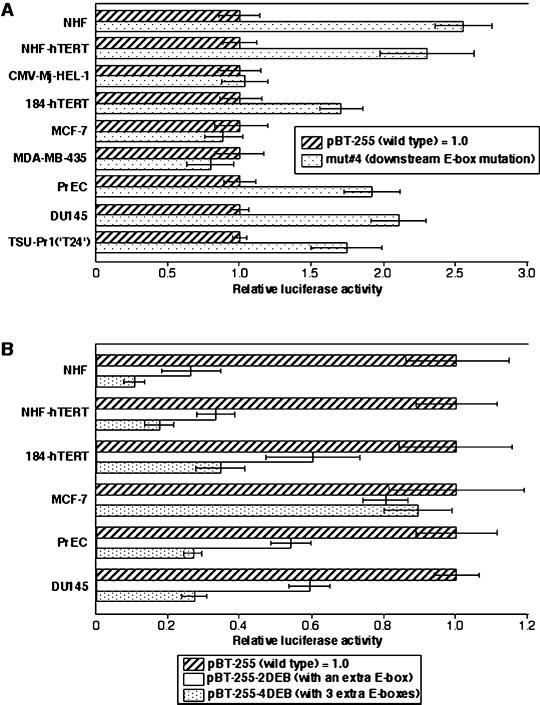
We examined whether the repressive mechanism mediated by the downstream E-box element functions in other types of normal and immortal human cells (Figure 8). In NHFs and PrECs, the mutation of the downstream E-box (mut 4) resulted in 2.5-fold and 1.9-fold increases, respectively, in hTERT promoter activity compared with the wild-type fragment (pBT-255) (Figure 8A). These data suggest that the E-box acts as a negative regulatory element in these normal human cells, as in RCC23+3. A similar increase in hTERT promoter activity with the E-box mutation was also observed in retroviral hTERT-immortalized NHFs (NHF-hTERT) and mammary epithelial cells (184-hTERT). The NHF-hTERT and 184-hTERT cells, as well as normal human cells (NHFs and PrECs), showed much lower activity (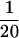 to
to 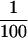 ) of the wild-type hTERT promoter than the other immortalized and cancer cell lines (see Figure 8 legend). It is therefore most likely that in these retroviral hTERT-immortalized cells, the transcription of the endogenous hTERT gene remains tightly repressed and the E-box–mediated repressive mechanism still functions. In contrast, an immortalized fibroblast cell line, CMV-Mj-HEL-1, and breast cancer cell lines MCF-7 and MDA-MB-435 showed no change or a slight decrease in the promoter activity with the E-box mutation (Figure 8A), suggesting that the E-box–mediated repressive mechanism is inactive in these immortal, endogenous telomerase/hTERT-positive cells, as in RCC23. Interestingly, however, in prostate cancer DU145 and bladder cancer TSU-Pr1(“T24”) cells, the E-box element still appeared to be able to negatively regulate hTERT transcription (Figure 8A). Thus, it is likely that the downstream E-box–mediated repressive mechanism is active in various cell types and becomes inactivated in some, but not all, cases of human cell immortalization and carcinogenesis. This notion was supported by the data obtained by use of the synthetic E-box–containing promoter fragments (Figure 8B). Normal and retroviral hTERT-immortalized cells of fibroblastic or epithelial origin (NHFs, PrECs, NHF-hTERT, and 184-hTERT) showed the enhancement of repression of the hTERT transcription in an E-box copy number–dependent manner, as observed in RCC23+3 cells. This copy number–dependent effect was not observed in MCF-7 breast cancer cells (like RCC23 and in contrast to 184-hTERT of breast epithelial origin), whereas it was evident in DU145 prostate cancer cells (like PrECs).
) of the wild-type hTERT promoter than the other immortalized and cancer cell lines (see Figure 8 legend). It is therefore most likely that in these retroviral hTERT-immortalized cells, the transcription of the endogenous hTERT gene remains tightly repressed and the E-box–mediated repressive mechanism still functions. In contrast, an immortalized fibroblast cell line, CMV-Mj-HEL-1, and breast cancer cell lines MCF-7 and MDA-MB-435 showed no change or a slight decrease in the promoter activity with the E-box mutation (Figure 8A), suggesting that the E-box–mediated repressive mechanism is inactive in these immortal, endogenous telomerase/hTERT-positive cells, as in RCC23. Interestingly, however, in prostate cancer DU145 and bladder cancer TSU-Pr1(“T24”) cells, the E-box element still appeared to be able to negatively regulate hTERT transcription (Figure 8A). Thus, it is likely that the downstream E-box–mediated repressive mechanism is active in various cell types and becomes inactivated in some, but not all, cases of human cell immortalization and carcinogenesis. This notion was supported by the data obtained by use of the synthetic E-box–containing promoter fragments (Figure 8B). Normal and retroviral hTERT-immortalized cells of fibroblastic or epithelial origin (NHFs, PrECs, NHF-hTERT, and 184-hTERT) showed the enhancement of repression of the hTERT transcription in an E-box copy number–dependent manner, as observed in RCC23+3 cells. This copy number–dependent effect was not observed in MCF-7 breast cancer cells (like RCC23 and in contrast to 184-hTERT of breast epithelial origin), whereas it was evident in DU145 prostate cancer cells (like PrECs).
Figure 8.
Examination of the downstream E-box–mediated mechanism for the hTERT repression in normal human cells, retroviral hTERT-expressing cells, and endogenous hTERT-expressing immortalized and cancer cell lines. NHF-hTERT, retroviral hTERT-expressing NHFs; CMV-Mj-HEL-1, immortalized fibroblast cell line; 184-hTERT, retroviral hTERT-expressing mammary epithelial cells 184; MCF-7, breast cancer cell line; MDA-MB-435, breast cancer cell line; DU145, prostate cancer cell line; TSU-Pr1(‘T24′), bladder cancer cell line. (A) The promoter activity of the downstream E-box–mutated fragment (mut 4; see Figure 2) was compared with that of the wild-type fragment (pBT-255), which was defined as 1.0 in each cell line. (B) The promoter activity of the synthetic E-box–containing fragments (pBT-255–2DEB and pBT-255–4DEB; see Figure 3) was compared with that of the wild-type pBT-255 [defined as 1.0 in each cell line, as in (A)]. For both A and B, note that absolute values of hTERT promoter activity in normal cells (NHFs and PrECs) and the retroviral hTERT-expressing cells (NHF-hTERT and 184-hTERT) are much lower (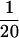 to
to 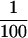 ) than in the endogenous hTERT-expressing cell lines.
) than in the endogenous hTERT-expressing cell lines.
DISCUSSION
Several known transcription factors have been reported to act as the positive or negative regulators of hTERT transcription (Poole et al., 2001). However, they may not necessarily represent physiological regulators of hTERT, because previous studies were based largely on an examination of overexpressed and/or recombinant proteins rather than endogenous proteins (Greenberg et al., 1999; Oh et al., 1999, 2000; Wu et al., 1999; Fujimoto et al., 2000; Günes et al., 2000; Kyo et al., 2000; Xu et al., 2000). In the present study, two genetically similar cell lines, with and without hTERT expression, were used in a systematic search for an endogenous factor responsible for the differential hTERT transcription. We found that the E-box element downstream of the transcription initiation site (located at +22 to +27) is responsible for the differential hTERT transcription between hTERT-positive RCC23 and hTERT-negative RCC23+3. This downstream E-box (or proximal E-box) was previously demonstrated to mediate activation and repression of the hTERT transcription by overexpressed c-Myc (Greenberg et al., 1999) and Mad1 (Günes et al., 2000), respectively. Although we also observed the downstream E-box–mediated effects of c-Myc and Mad1 overexpression (Figure 5), examination of endogenous c-Myc and Mad1 proteins by Western blot and gel mobility shift assays (Figures 6 and 7) did not show that the amount or binding activity of these endogenous proteins controlled the hTERT transcriptional level in RCC23 and RCC23+3. These results support the idea that overexpressed c-Myc is able to regulate different genes from those regulated by physiologically expressed c-Myc (Guo et al., 2000; Drissi et al., 2001), although we cannot completely rule out the possibility that a small amount of endogenous c-Myc and/or Mad1 binds to the hTERT promoter in vivo. Further analyses (e.g., chromatin immunoprecipitation) will be necessary to address this issue. The endogenous c-Myc and Mad1 proteins, in fact, play a central role in the transcriptional regulation of hTERT during cell differentiation (Nozawa et al., 2001; Xu et al., 2001). It is likely that different endogenous E-box–binding proteins regulate hTERT transcription during carcinogenic processes and cell differentiation. Mechanisms responsible for hTERT activation may also vary among cell types and individual tumors. Thus, although the Myc/Mad family did not seem critical in our cell system, our data do not necessarily exclude a role of the Myc/Mad family in hTERT activation during carcinogenic processes.
We found that the USF complex was the major protein factor binding to the E-box element within the hTERT promoter in RCC23 and RCC23+3 cells under our experimental conditions. However, no significant difference in the amount of USF binding (Figure 7) or the expression level of USF proteins (Figure 6) was observed between RCC23 and RCC23+3. Thus, the USF complex by itself does not account for the differential hTERT transcription. Whether the USF complex indeed activates hTERT transcription and whether a posttranslational modification (Cheung et al., 1999) or association with other proteins modulates the function of USF in hTERT transcription deserve further investigation.
Sp1 protein, which binds to the hTERT core promoter region, has been identified as a transcriptional activator of hTERT (Kyo et al., 2000; Poole et al., 2001). Our luciferase assay (Figure 1) suggested that the Sp1 binding sites contribute to the basal activity of the hTERT promoter in both RCC23 and RCC23+3. The gel mobility shift assay showed similar amounts of Sp1 binding in RCC23 and RCC23+3 (data not shown). It is therefore unlikely that the Sp1 binding by itself is primarily responsible for the differential promoter activity. However, a slight but reproducible difference between the two cell lines made by the fragments without a downstream E-box (i.e., pBTdel-130, pBTdel-208, and pBTdel-255 in Figure 1 and mut 4 and mut 4 plus 7 in Figure 2), which was more evident in our previous study because of the different transfection conditions (Horikawa et al., 1999; also see MATERIALS AND METHODS), may still suggest a possibility that the Sp1 binding sites and/or neighboring sequences make a minor, E-box–independent contribution to the repression by chromosome 3 transfer. It is also possible that the Sp1 and its binding sites could play a major role in hTERT activation in some immortalized and cancer cells. For example, NHFs appeared to have not only the E-box–dependent mechanism (Figure 8) but also the Sp1-mediated mechanism for tightly repressing hTERT transcription. NHFs and an hTERT-negative fibroblast cell line showed less Sp1 binding activity in the gel mobility shift assay, as well as much lower promoter activity of the fragment containing four Sp1 binding sites and no E-box (pBTdel-130) in the luciferase assay, than an hTERT-positive fibroblast cell line (Horikawa, I., unpublished data).
An important finding in this study is the evidence for the E-box–binding factor specific to the telomerase/hTERT-negative RCC23+3 cells (Figure 7). The supershift experiment suggested that this RCC23+3-specific factor was distinct from the Myc/Mad and USF families of transcription factors (Figure 7). The luciferase assay in Figure 2 showed that the downstream E-box sequence where this RCC23+3-specific factor binds can function as a negative regulatory element in RCC23+3 (because its mutation, mut 4, increased the hTERT promoter activity in RCC23+3 but not in RCC23). The negative regulatory role of this E-box element was strongly supported by the finding that additional E-box sequences repressed hTERT promoter activity in a copy number–dependent manner in RCC23+3, but not in RCC23 (Figure 3). The results from a second set of hTERT-negative and -positive cells (RCC23+3p and REV; Figure 4) were identical to those from RCC23+3 and RCC23, further validating the importance of the downstream E-box in regulation of the hTERT transcription by chromosome 3 transfer. These data lead us to the conclusion that an endogenous mechanism for the transcriptional repression of hTERT, which probably requires the binding of the RCC23+3-specific factor to the downstream E-box, actively functions in RCC23+3 and is defective in RCC23. Considering that RCC23, RCC23+3, and RCC23+3p are genetically similar except for the transferred copy of human chromosome 3 and that the reversion from RCC23+3p to REV occurred with loss of the transferred chromosome loci (Horikawa et al., 1998), the transcriptional repression mechanism can be associated with the function of a putative telomerase/hTERT repressor gene located on this chromosome. The most direct scenario is that the putative repressor gene encodes for the RCC23+3-specific E-box–binding factor detected in our gel mobility shift assay. Alternatively, a protein encoded by the putative repressor gene may either upregulate the expression of the RCC23+3-specific factor or enhance its DNA binding activity through protein-protein interaction and/or protein modification (e.g., phosphorylation). It is also possible that the USF complex participates in this repressive mechanism, because it has been suggested to act not only as a transcriptional activator but also as a repressor (Carter et al., 1997; Ghosh et al., 1997; Kiermaier et al., 1999).
The examination of a role of the downstream E-box in hTERT transcription in various types of human cells (Figure 8) suggested that the E-box–mediated repressive mechanism is also functioning in hTERT-negative normal cells and retroviral hTERT-immortalized (and most likely endogenous hTERT-repressed) cells of fibroblastic and breast and prostate epithelial origins. The breast cancer cell line MCF-7 appeared to lack the E-box–mediated repressive mechanism, as suggested by the lack of effects of either the E-box mutation or the increase in E-box copy number. Conversely, the prostate cancer cell line DU145 showed the same profile of effects of the E-box mutation and the additional E-box copies as its normal counterpart PrECs, suggesting that the hTERT activation in this cell line occurred without an inactivation of the E-box–mediated repressive mechanism. Notably, the gel mobility shift assay detected the band of interest (which was specific to RCC23+3 in Figure 7) in both MCF-7 and DU145 cell lines, as well as in normal human cells (NHFs, PrECs, and breast epithelial 184 cells) (data not shown). Taking all data together, we propose that the downstream E-box element is a target site for the negative regulatory mechanism that functions in various types of normal human cells. We hypothesize three different types of contributions of the downstream E-box element and its binding factors to hTERT transcriptional activation in human cancers: 1) an unidentified E-box–binding factor (RCC23+3-specific factor in Figure 7) may be lost or become defective in DNA binding by deletion or inactivating mutation of the repressor gene on chromosome 3 (Steenbergen et al., 1996; Tanaka et al., 1998; Cuthbert et al., 1999), thereby abrogating the E-box–mediated repressive mechanism. In this case, the E-box element may be converted to a positive regulatory element in which an activator (possibly the USF) comes to manifest its effect (for example, RCC23 renal cell carcinoma); 2) the E-box–mediated repressive mechanism may be impaired through an event other than the loss or defective DNA binding of the unidentified E-box–binding factor (for example, MCF-7 breast cancer); and 3) some mechanism independent of the downstream E-box may activate the hTERT expression even with the E-box–mediated repressive mechanism functioning (for example, DU145 prostate cancer).
It should be noted that the downstream E-box we identified as a critical element (+22 to +27), but not the upstream E-box (−187 to −182), is conserved among human, mouse, and hamster (Guo et al., 2001). It would be of interest to investigate whether the E-box–mediated mechanism also controls the transcription of mouse and hamster telomerase reverse transcriptase genes.
Our previous study by means of chromosome 3 transfer (Horikawa et al., 1998) revealed the changes in cellular phenotypes, including the endogenous hTERT expression, telomerase activity, telomere length, and cellular life span (summarized in Table 1). Our data in this study provided a molecular basis for these cellular changes. The experiments based on transient transfection of the hTERT promoter region has enabled us to identify and analyze a critical DNA element near the transcription initiation site. Although the promoter activity measured in the luciferase assay qualitatively recapitulated the expression level of endogenous hTERT mRNA, the difference between RCC23 and RCC23+3 exhibited in the luciferase assay (RCC23/RCC23+3 ratio 4.2–8.3; Figure 1) was not as obvious as the difference in expression levels of the endogenous mRNA (RCC23/RCC23+3 ratio 64 or more by Taqman assay; Table 1). This may imply that other mechanisms that are not reflected in our transient transfection-based assay also contribute to the transcriptional repression of the endogenous hTERT gene by the telomerase/hTERT repressor gene on chromosome 3. DNA sequences outside of our longest promoter fragment (−3915 to +40), e.g., a region highly conserved between human and mouse (around −5.5 kb) and minisatellite tandem repeats within the introns 2, 6, and 12 (Szutorisz et al., 2001; Leem et al., 2002), may play a supplementary role in the hTERT repression. Also possible is an involvement of DNA methylation and histone acetylation (Devereux et al., 1999; Cong and Bacchetti, 2000; Dobosy and Selker, 2001). The methylation profiles of CpG sites within the endogenous hTERT promoter region (covering approximately −500 to +100) in RCC23 and RCC23+3 were found to be the same overall, with an unmethylated CpG site of the downstream E-box in both cell lines (Devereux et al., 1999; and unpublished data). Treatment of RCC23 or RCC23+3 with the histone deacetylase inhibitor trichostatin A (300 or 500 nM for 24 h) produced no effects on the endogenous hTERT expression. Interestingly, however, the treatment with trichostatin A in combination with the DNA demethylating agent 5-aza-2′-deoxycytidine (3 μM for 96 h) resulted in a partial but significant induction of the hTERT expression in RCC23+3 cells (unpublished data). Whether a DNA methylation- and histone deacetylation–associated change in higher-order chromatin structure at the hTERT gene locus directly contributes to the hTERT repression in RCC23+3 and whether it has a functional relation to the E-box–mediated repressive mechanism remain to be examined. Our recent cloning of the whole, functional copy of the hTERT gene locus in a single bacterial artificial chromosome clone (Leem et al., 2002) should be helpful to obtain a full picture of the hTERT regulation during developmental and carcinogenic processes.
The expression profile of hTERT (i.e., expressed in most cancers and repressed in most normal somatic tissues) has given impetus to the use of the hTERT promoter as a tool for cancer-specific expression of cytotoxic genes in anticancer therapy. Indeed, the use of the hTERT promoter–driven cytotoxic gene expression system has given promising results in cell culture and animal models (Gu et al., 2000; Koga et al., 2000; Majumdar et al., 2001). A possible concern about this approach, however, is the leaky expression of cytotoxic genes in normal tissues, which may cause detrimental side effects (Dachs et al., 1997). Our results demonstrated that synthetic copies of the E-box element placed downstream of the hTERT promoter resulted in the tighter repression in the telomerase-negative RCC23+3 and normal human cells of fibroblastic and epithelial origins while maintaining the high activity in some telomerase-positive cancer cells (i.e., RCC23 and MCF-7) (Figures 3 and 8B). It is our expectation that the modified hTERT promoter (pBT-255–4DEB) should minimize the cytotoxicity in normal cells without loss of cytotoxic effect on cancer cells when it is used to drive cytotoxic genes in anticancer therapy. Further studies will be needed to prove this concept and develop a therapeutic vector and an efficient gene delivery system.
In an apparent contrast to our results, Ducrest et al. (2001) showed that the telomerase repression by human chromosome 3 in a breast cancer cell line, 21NT, was not associated with the repression of hTERT promoter activity. Our genetic complementation test by generation of somatic cell hybrids of RCC23 and 21NT cells suggests that different genes on human chromosome 3 are responsible for the telomerase repression in these two cell lines (Tanaka, H., Horikawa, I., Barrett, J.C., and Oshimura, M., manuscript in preparation). The difference in mode of effects by chromosome 3 transfer between the two cell lines (i.e., telomerase repression with or without the repression of the hTERT promoter activity) also supports the presence of two distinct telomerase repressor genes on this chromosome.
In conclusion, this study provides the first evidence for an endogenous mechanism of hTERT transcriptional repression that may be inactivated during carcinogenic processes. It also highlights the hTERT repression as a function of human tumor suppressor genes. The purification and cloning of the RCC23+3-specific E-box–binding factor will greatly facilitate understanding of telomerase regulation in normal and cancer cells and may open up a new strategy for telomerase-targeted anticancer therapy.
ACKNOWLEDGMENTS
We thank Dr. Hidetoshi Tahara for establishing NHF-hTERT and 184-hTERT cells, Dr. Theodora Devereux for CpG methylation analysis, Drs. Mitsuo Oshimura and Hiromi Tanaka for chromosomal mapping of the telomerase repressor gene, Dr. John Risinger for helpful discussion, Giannina Garcés for technical assistance, and Mary Custer for continuous encouragement.
Abbreviations used:
- hTERT
human telomerase reverse transcriptase
- USF
upstream stimulatory factors
- NHFs
normal human fibroblasts
- PrECs
prostate epithelial cells
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E01–11–0107. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E01–11–0107.
REFERENCES
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu C-P, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Cech TR. Telomerase and the maintenance of chromosome ends. Curr Opin Cell Biol. 1999;11:318–324. doi: 10.1016/S0955-0674(99)80043-X. [DOI] [PubMed] [Google Scholar]
- Carter RS, Ordentlich P, Kadesch T. Selective utilization of basic helix-loop-helix-leucine zipper proteins at the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1997;17:18–23. doi: 10.1128/mcb.17.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung E, Mayr P, Coda-Zabetta F, Woodman PG, Boam DSW. DNA-binding activity of the transcription factor upstream stimulatory factor 1 (USF-1) is regulated by cyclin-dependent phosphorylation. Biochem J. 1999;344:145–152. [PMC free article] [PubMed] [Google Scholar]
- Chiu C-P, Harley CB. Replicative senescence and cell immortality: the role of telomeres and telomerase. Proc Soc Exp Biol Med. 1997;214:99–106. doi: 10.3181/00379727-214-44075. [DOI] [PubMed] [Google Scholar]
- Cong YS, Bacchetti S. Histone deacetylation is involved in the transcriptional repression of hTERT in normal human cells. J Biol Chem. 2000;275:35665–35668. doi: 10.1074/jbc.C000637200. [DOI] [PubMed] [Google Scholar]
- Cuthbert AP, Bond J, Trott DA, Gill S, Broni J, Marriott A, Khoudoli G, Parkinson EK, Cooper CS, Newbold RF. Telomerase repressor sequences on chromosome 3 and induction of permanent growth arrest in human breast cancer cells. J Natl Cancer Inst. 1999;91:37–45. doi: 10.1093/jnci/91.1.37. [DOI] [PubMed] [Google Scholar]
- Dachs GU, Dougherty GJ, Stratford IJ, Chaplin DJ. Targeting gene therapy to cancer: a review. Oncol Res. 1997;9:313–325. [PubMed] [Google Scholar]
- Devereux TR, Horikawa I, Anna CH, Annab LA, Afshari CA, Barrett JC. DNA methylation analysis of the promoter region of the human telomerase reverse transcriptase (hTERT) gene. Cancer Res. 1999;59:6087–6090. [PubMed] [Google Scholar]
- Dobosy JR, Selker EU. Emerging connections between DNA methylation and histone acetylation. Cell Mol Life Sci. 2001;58:721–727. doi: 10.1007/PL00000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissi R, Zindy F, Roussel MF, Cleveland JL. c-Myc-mediated regulation of telomerase activity is disabled in immortalized cells. J Biol Chem. 2001;276:29994–30001. doi: 10.1074/jbc.M101899200. [DOI] [PubMed] [Google Scholar]
- Ducrest AL, Amacker M, Mathieu YD, Cuthbert AP, Trott DA, Newbold RF, Nabholz M, Lingner J. Regulation of human telomerase activity: repression by normal chromosome 3 abolishes nuclear telomerase reverse transcriptase transcripts but does not affect c-Myc activity. Cancer Res. 2001;61:7594–7602. [PubMed] [Google Scholar]
- Fujimoto K, Kyo S, Takakura M, Kanaya T, Kitagawa Y, Itoh H, Takahashi M, Inoue M. Identification and characterization of negative regulatory elements of the human telomerase catalytic subunit (hTERT) gene promoter: possible role of MZF-2 in transcriptional repression of hTERT. Nucleic Acids Res. 2000;28:2557–2562. doi: 10.1093/nar/28.13.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AK, Datta PK, Jacob ST. The dual role of helix-loop-helix-zipper protein USF in ribosomal RNA gene transcription in vivo. Oncogene. 1997;14:589–594. doi: 10.1038/sj.onc.1200866. [DOI] [PubMed] [Google Scholar]
- Greenberg RA, O'Hagan RC, Deng H, Xiao Q, Hann SR, Adams RR, Lichtsteiner S, Chin L, Morin GB, DePinho RA. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene. 1999;18:1219–1226. doi: 10.1038/sj.onc.1202669. [DOI] [PubMed] [Google Scholar]
- Gu J, Kagawa S, Takakura M, Kyo S, Inoue M, Roth JA, Fang BL. Tumor-specific transgene expression from the human telomerase reverse transcriptase promoter enables targeting of the therapeutic effects of the Bax gene to cancers. Cancer Res. 2000;60:5359–5364. [PubMed] [Google Scholar]
- Günes C, Lichtsteiner S, Vasserot AP, Englert C. Expression of the hTERT gene is regulated at the level of transcriptional initiation and repressed by Mad1. Cancer Res. 2000;60:2116–2121. [PubMed] [Google Scholar]
- Guo QM, Malek RL, Kim S, Chiao C, He M, Ruffy M, Sanka K, Lee NH, Dang CV, Liu ET. Identification of c-Myc responsive genes using rat cDNA microarray. Cancer Res. 2000;60:5922–5928. [PubMed] [Google Scholar]
- Guo W, Okamoto M, Lee YM, Baluda MA, Park NH. Enhanced activity of cloned hamster TERT gene promoter in transformed cells. Biochim Biophys Acta. 2001;1517:398–409. doi: 10.1016/s0167-4781(00)00306-7. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Stewart SA, Brooks MW, York SG, Eaton E, Kurachi A, Beijersbergen RL, Knoll JHM, Meyerson M, Weinberg RA. Inhibition of telomerase limits the growth of human cancer cells. Nat Med. 1999;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- Horikawa I, Cable PL, Afshari C, Barrett JC. Cloning and characterization of the promoter region of human telomerase reverse transcriptase gene. Cancer Res. 1999;59:826–830. [PubMed] [Google Scholar]
- Horikawa I, Oshimura M, Barrett JC. Repression of the telomerase catalytic subunit by a gene on human chromosome 3 that induces cellular senescence. Mol Carcinog. 1998;22:65–72. doi: 10.1002/(sici)1098-2744(199806)22:2<65::aid-mc1>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Horikawa I, Parker ES, Solomon GG, Barrett JC. Upregulation of the gene encoding a cytoplasmic dynein intermediate chain in senescent human cells. J Cell Biochem. 2001;82:415–421. doi: 10.1002/jcb.1169. [DOI] [PubMed] [Google Scholar]
- Kiermaier A, Gawn JM, Desbarats L, Saffrich R, Ansorge W, Farrell PJ, Eilers M, Packham G. DNA binding of USF is required for specific E-box dependent gene activation in vivo. Oncogene. 1999;18:7200–7211. doi: 10.1038/sj.onc.1203166. [DOI] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PLC, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Koga S, Hirohata S, Kondo Y, Komata T, Takakura M, Inoue M, Kyo S, Kondo S. A novel telomerase-specific gene therapy: gene transfer of caspase-8 utilizing the human telomerase catalytic subunit gene promoter. Hum Gene Ther. 2000;11:1397–1406. doi: 10.1089/10430340050057477. [DOI] [PubMed] [Google Scholar]
- König P, Rhodes D. Recognition of telomeric DNA. Trends Biochem Sci. 1997;22:43–47. doi: 10.1016/s0968-0004(97)01008-6. [DOI] [PubMed] [Google Scholar]
- Kyo S, Takakura M, Taira T, Kanaya T, Itoh H, Yutsudo M, Ariga H, Inoue M. Spl cooperates with c-Myc to activate transcription of the human telomerase reverse transcriptase gene (hTERT) Nucleic Acids Res. 2000;28:669–677. doi: 10.1093/nar/28.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem S-H, Londoño-Vallejo JA, Kim J-H, Bui H, Tubacher E, Solomon G, Park J-E, Horikawa I, Kouprina N, Barrett JC, Larionov V. The human telomerase gene: complete genomic sequence and analysis of tandem repeat polymorphisms in intronic regions. Oncogene. 2002;21:769–777. doi: 10.1038/sj.onc.1205122. [DOI] [PubMed] [Google Scholar]
- Majumdar AS, Hughes DE, Lichtsteiner SP, Wang Z, Lebkowski JS, Vasserot AP. The telomerase reverse transcriptase promoter drives efficacious tumor suicide gene therapy while preventing hepatotoxicity encountered with constitutive promoters. Gene Ther. 2001;8:568–578. doi: 10.1038/sj.gt.3301421. [DOI] [PubMed] [Google Scholar]
- Meyerson M. Role of telomerase in normal and cancer cells. J Clin Oncol. 2000;18:2626–2634. doi: 10.1200/JCO.2000.18.13.2626. [DOI] [PubMed] [Google Scholar]
- Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q, Bacchetti S, Haber DA, Weinberg RA. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- Misiti S, Nanni S, Fontemaggi G, Cong YS, Wen JP, Hirte HW, Piaggio G, Sacchi A, Pontecorvi A, Bacchetti S, Farsetti A. Induction of hT.E.R.T. expression and telomerase activity by estrogens in human ovary epithelium cells. Mol Cell Biol. 2000;20:3764–3771. doi: 10.1128/mcb.20.11.3764-3771.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudryj M, Devoto SH, Hiebert SW, Hunter T, Pines J, Nevins JR. Cell cycle regulation of the E2F transcription factor involves an interaction with cyclin A. Cell. 1991;65:1243–1253. doi: 10.1016/0092-8674(91)90019-u. [DOI] [PubMed] [Google Scholar]
- Mueller SO, Tahara H, Barrett JC, Korach KS. Immortalization of mammary cells from estrogen receptor α knock-out and wild-type mice. In Vitro Cell Dev Biol Anim. 2000;36:620–624. doi: 10.1290/1071-2690(2000)036<0620:IOMCFE>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Tahara H, Tahara E, Saito M, Ito K, Nakamura H, Nakanishi T, Tahara E, Ide T, Ishikawa F. Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nat Genet. 1998;18:65–68. doi: 10.1038/ng0198-65. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Nozawa K, Maehara K, Isobe K-i. Mechanism for the reduction of telomerase expression during muscle cell differentiation. J Biol Chem. 2001;276:22016–22023. doi: 10.1074/jbc.M011181200. [DOI] [PubMed] [Google Scholar]
- Oh S, Song Y, Yim J, Kim TK. The Wilms' tumor 1 tumor suppressor gene represses transcription of the human telomerase reverse transcriptase gene. J Biol Chem. 1999;274:37473–37478. doi: 10.1074/jbc.274.52.37473. [DOI] [PubMed] [Google Scholar]
- Oh S, Song YH, Yim J, Kim TK. Identification of Mad as a repressor of the human telomerase (hTERT) gene. Oncogene. 2000;19:1485–1490. doi: 10.1038/sj.onc.1203439. [DOI] [PubMed] [Google Scholar]
- Poole JC, Andrews LG, Tollefsbol TO. Activity, function, and gene regulation of the catalytic subunit of telomerase (hTERT) Gene. 2001;269:1–12. doi: 10.1016/s0378-1119(01)00440-1. [DOI] [PubMed] [Google Scholar]
- Sommer A, Bousset K, Kremmer E, Austen M, Luscher B. Identification and characterization of specific DNA-binding complexes containing members of the Myc/Max/Mad network of transcriptional regulators. J Biol Chem. 1998;273:6632–6642. doi: 10.1074/jbc.273.12.6632. [DOI] [PubMed] [Google Scholar]
- Stampfer MR, Garbe J, Levine G, Lichtsteiner S, Vasserot AP, Yaswen P. Expression of the telomerase catalytic subunit, hTERT, induces resistance to transforming growth factor β growth inhibition in p16INK4A(-) human mammary epithelial cells. Proc Natl Acad Sci USA. 2001;98:4498–4503. doi: 10.1073/pnas.071483998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen RDM, Walboomers JMM, Meijer CJLM, van der Raaij-Helmer EMH, Parker JN, Chow LT, Broker TR, Snijders PJF. Transition of human papillomavirus type 16 and 18 transfected human foreskin keratinocytes towards immortality: activation of telomerase and allele losses at 3p, 10p, 11q and/or 18q. Oncogene. 1996;13:1249–1257. [PubMed] [Google Scholar]
- Szutorisz H, Palmqvist R, Roos G, Stenling R, Schorderet DF, Reddell R, Lingner J, Nabholz M. Rearrangements of minisatellites in the human telomerase reverse transcriptase gene are not correlated with its expression in colon carcinomas. Oncogene. 2001;20:2600–2605. doi: 10.1038/sj.onc.1204346. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Shimizu M, Horikawa I, Kugoh H, Yokota J, Barrett JC, Oshimura M. Evidence for a putative telomerase repressor gene in the 3p14.2-p21.1 region. Genes Chromosomes Cancer. 1998;23:123–133. doi: 10.1002/(sici)1098-2264(199810)23:2<123::aid-gcc5>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- van Bokhoven A, Varella-Garcia M, Korch C, Miller GJ. TSU-Pr1 and JCA-1 cells are derivatives of T24 bladder carcinoma cells and are not of prostatic origin. Cancer Res. 2001;61:6340–6344. [PubMed] [Google Scholar]
- Viollet B, Lefrancois-Martinez AM, Henrion A, Kahn A, Raymondjean M, Martinez A. Immunochemical characterization and transacting properties of upstream stimulatory factor isoforms. J Biol Chem. 1996;271:1405–1415. doi: 10.1074/jbc.271.3.1405. [DOI] [PubMed] [Google Scholar]
- Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- Xu D, Popov N, Hou M, Wang Q, Bjorkholm M, Gruber A, Menkel AR, Henriksson M. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc Natl Acad Sci USA. 2001;98:3826–3831. doi: 10.1073/pnas.071043198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu DW, Wang Q, Gruber A, Bjorkholm M, Chen ZG, Zaid A, Selivanova G, Peterson C, Wiman KG, Pisa P. Downregulation of telomerase reverse transcriptase mRNA expression by wild type p53 in human tumor cells. Oncogene. 2000;19:5123–5133. doi: 10.1038/sj.onc.1203890. [DOI] [PubMed] [Google Scholar]