Abstract
Evidence is presented that mitochondria are implicated in the previously described programmed cell death (PCD) process induced by acetic acid in Saccharomyces cerevisiae. In yeast cells undergoing a PCD process induced by acetic acid, translocation of cytochrome c (CytC) to the cytosol and reactive oxygen species production, two events known to be proapoptotic in mammals, were observed. Associated with these events, reduction in oxygen consumption and in mitochondrial membrane potential was found. Enzymatic assays showed that the activity of complex bc1 was normal, whereas that of cytochrome c oxidase (COX) was strongly decreased. This decrease is in accordance with the observed reduction in the amounts of COX II subunit and of cytochromes a+a3. The acetic acid-induced PCD process was found to be independent of oxidative phosphorylation because it was not inhibited by oligomycin treatment. The inability of S. cerevisiae mutant strains (lacking mitochondrial DNA, heme lyase, or ATPase) to undergo acetic acid-induced PCD and in the ATPase mutant (knockout in ATP10) the absence of CytC release provides further evidence that the process is mediated by a mitochondria-dependent apoptotic pathway. The understanding of the involvement of a mitochondria-dependent apoptotic pathway in S. cerevisiae PCD process will be most useful in the further elucidation of an ancestral pathway common to PCD in metazoans.
INTRODUCTION
Programmed cell death (PCD), of which apoptosis is the most common morphological expression, is described as an orchestrated collapse of the cell. This process plays an important role in the normal development and homeostasis mechanisms of multicellular organisms. At least two major apoptotic pathways have been described in mammalian cells. One requiring the participation of mitochondria, called “intrinsic pathway,” and another one in which mitochondria are bypassed and caspases are activated directly, called “extrinsic pathway” (Hengartner, 2000; Matsuyama et al., 2000). Regarding the mitochondrial pathway, two main events have been proposed as integral control elements in the cell's decision to dye, namely, the release of apoptogenic factors such as cytochrome c (CytC) and the production of reactive oxygen species (ROS) (Liu et al., 1996; Kluck et al., 1997; Pham et al., 2000). Release of CytC to the cytosol drives the assembly of a high-molecular-weight complex, the mitochondrial apoptosome that activates caspases (Adrian and Martin, 2001). Translocation of CytC to the cytosol is, therefore, a pivotal event in apoptosis. CytC is a soluble protein loosely bound to the outer face of the inner mitochondrial membrane, and its release is associated with an interruption of the normal electron flow at the complex III site of the respiratory chain, that could divert electron transfer to the generation of superoxide (Cai and Jones, 1998). The mechanism by which CytC is released from mitochondria during apoptosis remains unknown. However, two competing models have been proposed, a volume-dependent mechanism, involving mitochondrial swelling and the rupture of the outer membrane, and a volume-independent mechanism, in which the permeability of the outer membrane is selectively altered (Matsuyama et al., 2000). Pavlov et al. (2001) reported a new high conductance channel named mitochondrial apoptosis-induced channel linked to apoptosis in mammalian cells and Bax expression in yeast, which is in agreement with the latter model. The authors propose this channel as a candidate for the outer mitochondrial membrane pore through which CytC and possible other factors exit mitochondria during apoptosis.
Recently, we have shown that acetic acid (20–80 mM) induces death in exponential cells of Saccharomyces cerevisiae which displays the most common PCD hallmarks such as chromatin condensation along the nuclear envelope, exposure of phosphatidylserine on the outer surface of the cytoplasmic membrane, and occurrence of DNA fragmentation (Ludovico et al., 2001a). It was also shown that similar to hydrogen peroxide (Madeo et al., 1999), acetic acid at high doses (>120 mM) induces cell morphological changes typical of necrosis without apoptotic markers such as terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL)-positive phenotype. Acetic acid is a normal end product of fermentation carried out by S. cerevisiae and may be produced by contaminating acetic acid bacteria being therefore quite familiar to the yeast environment. In this context the occurrence of acetic acid-induced PCD points to a physiological role for such a cell death in yeast.
In the past few years the occurrence of a PCD process independent of Bax-expression and with an apoptotic phenotype in S. cerevisiae has been reported (Madeo et al., 1997, 1999; Laun et al., 2001; Ludovico et al., 2001a; Yamaki et al., 2001; Carratore et al., 2002). However, only Yamaki et al. (2001) studied the involvement of mitochondria in such a cell death pathway. The authors observed that cell death mediated by deletion of histone chaperone ASF1/CIA1 is associated with a decrease in mitochondrial membrane potential, dysfunction of mitochondrial ATPase, and release of CytC to the cytoplasm, a phenotype that largely resembles mammalian apoptosis. Although controversial, there is an interpretation that mitochondria seem to be involved in Bax-induced cell death in yeast. Matsuyama et al. (1998) reported that ATP4, which is a nuclear gene encoding subunit 4 of the yeast mitochondrial ATPase complex, is required for yeast death induced by Bax-expression. Additionally, as in mammalian cells, Bax-expression leads to CytC release from yeast mitochondria (Manon et al., 1997; Kluck et al., 2000). Manon et al. (1997) described that Bax-expression in yeast cells stimulates the release of CytC to the cytosol and causes a concomitant decrease in the amount of COX complex.
In the present article a large body of evidence supporting the involvement of mitochondria in the S. cerevisiae PCD process triggered by acetic acid is presented, indicating that, like in mammalian cells, the PCD in yeast can be mediated by a mitochondria-dependent apoptotic pathway.
MATERIALS AND METHODS
Microorganisms and Growth Conditions
The yeast S. cerevisiae W303-1A (MAT a ade2 leu2 his3 trp1 ura3), the respective knockout in ATP10 and CYC3 genes and the isogenic derivative ρ0 strain (lacking mitochondrial DNA) were used. These organisms were maintained on YEPD agar slants containing glucose (2%, wt/vol), yeast extract (1%, wt/vol), peptone (2%, wt/vol), and agar (2%, wt/vol). In experiments, the yeast cells were subcultured in liquid YEPD medium. The growth experiments were performed in 250-ml flasks containing a ratio 2:1 of air-to-liquid phase, and incubated on a mechanical shaker (150 rpm) at 26°C.
Treatments with Acetic Acid and Inhibition of Protein Synthesis or Oxidative Phosphorylation
Stationary phase cells were harvested and suspended (107 cells/ml) in treatment medium (YEPD, pH 3.0, set with HCl) containing 0, 120, 140, 180, 200, and 240 mM acetic acid. The treatments were carried out for 200 min at 26°C with magnetic stirring (150 rpm). Inhibition of protein synthesis or oxidative phophorylation was performed by adding 50 μg/ml cycloheximide (Merck, Whitehouse Station, NJ) or 10 μM oligomycin (Matsuyama et al., 1998), respectively, at the same time as the different acetic acid concentrations tested. At these concentrations cycloheximide and oligomycin were not cytotoxic after 200-min incubation, as assessed by counting colony-forming units (cfu). Viability was determined by cfu counts after 2 d of incubation at 26°C on YEPD agar plates. No further colonies appeared after that incubation period. In all the above-mentioned experiments, the extracellular pH did not change during the incubations.
TUNEL and Propidium Iodide Staining
DNA strand breaks were demonstrated by TUNEL with the In Situ Cell Death Detection Kit, Fluorescein (Roche Applied Science, Indianapolis, IN) as described previously (Ludovico et al., 2001a).
Isolation of Yeast Mitochondria and Mitochondrial Respiratory Chain Activity Measurements
Mitochondria were isolated from S. cerevisiae (W303-1A and ATP10 mutant) cells grown to stationary phase, harvested, and resuspended in YEPD, pH 3.0, in the absence or presence of 140 mM acetic acid for 200 min. Mitochondria were prepared essentially as described by Faye et al. (1974), except that Zymolyase 20,000 instead of Glusulase was used to digest the cell wall. The postmitochondrial supernatant (supernatant after the first 12,000-rpm centrifugation) was kept for the detection of CytC. Mitochondria were washed twice with 0.5 M sorbitol at 12,000 rpm for 15 min and suspended in 0.5 M sorbitol. Protein concentration was ascertained by the method published by Lowry et al. (1951).
Oxygen utilization in isolated mitochondria was measured polarographically in 1 ml of standard medium (0.5 M sorbitol, 20 mM K2PO4 pH 7.4, and 1 mM EDTA) with a Clark oxygen electrode (model 5300; Yellow Springs Instruments, Yellow Springs, OH), in a water-jacketed cell, magnetically stirred at room temperature. Twenty micrograms of mitochondrial protein was used to assay for NADH oxidase activity. Oxygen consumption was monitored using NADH (0.8 mM) as a substrate. In a set of reactions, the effect of exogenous CytC on the oxidative rate was checked out by adding 10 μM horse CytC (Sigma-Aldrich, St. Louis, MO) to the chamber. Respiration was inhibited by addition of 700 μM KCN.
Measurement of respiratory chain enzymatic activities in isolated mitochondria was performed essentially as described by Tzagoloff et al. (1975). Cytochrome c oxidase (COX) activity was measured at room temperature in 20 mM K2HPO4, pH 7.5, containing 65 μM reduced CytC. COX activity was assayed in mitochondria (10 μg of protein) permeabilized with potassium deoxycholate to maximize the access of the substrate to the enzyme, by measuring oxidation of ferrocytochrome c at 550 nm. NADH-cytochrome c reductase activity was measured in potassium deoxycholate permeabilized mitochondria at room temperature in 10 mM K2HPO4, pH 7.5, containing 100 μM KCN by following the reduction of CytC (65 μM) at 550 nm in the presence of 1 M NADH.
Cytochromes Spectra
Optical absorption spectra of mitochondrial cytochromes were obtained on an Aminco DW-2A dual wavelength scanning spectrophotometer. The spectrophotometer was operated in the split beam mode with 1-nm band pass. Mitochondria were extracted at a protein concentration of 5 mg/ml with potassium deoxycholate under conditions that quantitatively solubilize all the cytochromes (Tzagoloff et al., 1975). Difference spectra of the reduced (sodium dithionite) vs. oxidized (potassium ferricyanide) extracts were recorded at room temperature. The α absorption bands corresponding to cytochromes a and a3 have maxima at 603 nm. The maxima for cytochrome b and for cytochrome c and c1 are 560 and 550 nm, respectively.
Cytochrome c Distribution
The release of CytC from mitochondria to cytoplasm when S. cerevisiae (W303-1A and ATP10 mutant) cells were grown in the presence or absence of acetic acid was checked out by Western blot detection of CytC in the subcellular compartments. Total mitochondrial proteins (5, 10, or 20 μg) and the postmitochondrial supernatant (PMS) were electrophoretically separated on 12.0% SDS-polyacrylamide gels (Laemmli, 1970) and transferred to 0.2-μm nitrocellulose membranes. The membranes were blocked with 5% (wt/vol) milk powder for at least 1 h followed by the incubation for another hour with anti-yeast CytC polyclonal antibody (kind gift from Dr. R. Lill). The blots were then incubated in goat anti-rabbit IgG conjugated to horseradish peroxidase (Sigma-Aldrich). The SuperSignal chemiluminescent substrate kit (Pierce Chemical, Rockford, IL) was used for the final detection.
Steady-State Levels of bc1, Complex COX, and ATPase Subunits
The detection of cytochrome b, COX subunit II and V, and ATPase subunit 6 were performed by Western blotting as described above for CytC detection, and by using primary polyclonal antibodies against the yeast forms of the mentioned subunits. All the antibodies used were a kind gift of Dr. Alexander Tzagoloff (Columbia University, New York, NY).
Assessment of Mitochondrial Membrane Potential (ΔΨm) and of ROS Production
Assessment of ΔΨm in whole cells was performed by flow cytometry and epifluorescence microscopy as described by Ludovico et al. (2001b). The control suspensions of killed cells used in the assessment of ΔΨm and ROS production were prepared by boiling the cell suspensions for 10 min as described by Ludovico et al. (2001b).
ROS production by mitochondria was monitored by using the MitoTracker Red CM-H2XRos staining (Molecular Probes, Eugene, OR). The reduced version of MitoTracker Red CMXRos does not fluoresce until entering an actively respiring cell, where it is oxidized by ROS to a red fluorescent compound, which is sequestered in the mitochondria. Cells were harvested and then suspended in the treatment medium (107 cells/ml). Before the addition of acetic acid, cells were preloaded with 50 μg/ml dye for 20 min at 37°C. After the preloading, the different acetic acid concentrations were added and the fluorescence was detected by flow cytometry and epifluorescence microscopy.
Flow cytometric analysis was performed in a basic FACSCalibur (BD Biosciences, Franklin Lakes, NJ) flow cytometer equipped with an argon-ion laser emitting a 488-nm beam at 15 mW. Red fluorescence was collected through a 560-nm short-pass dichroic, a 640-nm long-pass, and another 670-nm long-pass. Twenty thousand cells per sample were analyzed at low flow rate. An acquisition protocol was defined to measure forward scatter (FS Log), side scatter (SS Log), and red fluorescence (FL3 Log) on a four-decade logarithmic scale. Data were acquired and analyzed with CELLQuest PRO 3.3 (BD Biosciences).
RESULTS
Wild-Type S. cerevisiae Stationary Cells Undergo a PCD Process Induced by Acetic Acid
The study was focused on the role of mitochondria in the previously reported acetic acid-induced PCD process in S. cerevisiae W303-1A (Ludovico et al., 2001a). Stationary cells rather than exponential cells were used for that purpose. Cells from stationary growth phase are more advantageous because they possess fully active mitochondria and display a higher mitochondrial mass. When exposed to different acetic acid concentrations at pH 3.0, and in the presence of glucose, S. cerevisiae stationary cells also committed to a PCD process. This conclusion is based on the detection of cell death accompanied by DNA strand breaks evaluated by the TUNEL assay (our unpublished data), an evident apoptotic marker, and by the fact that cycloheximide inhibited cell death (Figure 1). Consistent with the recognized higher resistance of stationary cells to different stress agents, the effect was only observed for higher acetic acid concentrations. Actually, although acetic acid concentrations >120 mM are necrotic for exponential cells, they induce a PCD process in stationary cells. In fact, 140 mM acetic acid concentration induces, after 200 min, ∼50% of loss of cell viability evaluated by cfu and ∼30% of TUNEL-positive cells. This concentration (140 mM) was selected to perform mitochondrial function analysis. Incubation with acetic acid concentrations >200 mM resulted in no detectable TUNEL staining (our unpublished data).
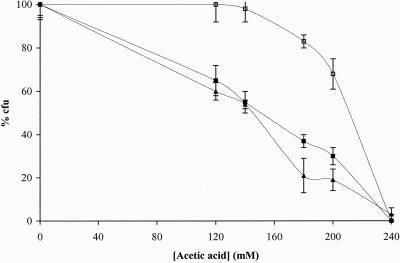
Figure 1.
Programmed cell death of S. cerevisiae W303-1A stationary cells induced by acetic acid is partially inhibited by cycloheximide and is independent of oxidative phosphorylation. Relative survival (percentage of cfu on YEPD agar plates; 100% corresponds to the number of cfu at time 0) of cells incubated for 200 min with 140 mM acetic acid in the absence (▪) or presence of cycloheximide (□) or oligomycin (▴).
To evaluate whether the acetic acid-induced PCD in S. cerevisiae is affected by the inhibition of oxidative phosphorylation, the treatment with acetic acid was carried out in the presence of oligomycin. Figure 1 shows that cell death was not affected by the drug.
Cytochrome c Is Translocated from Mitochondria to Cytosol during Acetic Acid-induced PCD Process
To check whether acetic acid-induced PCD process was accompanied by a release of CytC from mitochondria to cytosol, the levels of CytC in mitochondria and in the PMS (containing soluble cytosolic proteins) from S. cerevisiae W303-1A cells undergoing a acid-induced PCD were detected by Western blot analysis. The amount of CytC present in mitochondria of cells treated with 140 mM acetic acid was decreased by two- to threefold compared with the mitochondria from untreated cells (Figure 2). This portion of “lost” CytC in mitochondria from acetic acid treated cells was detected in its PMS, whereas no CytC was detected in the PMS of untreated control cells (Figure 2). Levels of other mitochondrial proteins, such as COX subunits II and V were not detected in PMS (our unpublished data), although COX II was found to be reduced in mitochondria (described below). Decrease of the CytC amount in mitochondria was confirmed by cytochrome spectra analysis of mitochondria from treated and untreated cells. As shown in Figure 3A, there was some decrease in the amount of cytochromes c+c1 extracted from the mitochondrial membranes.
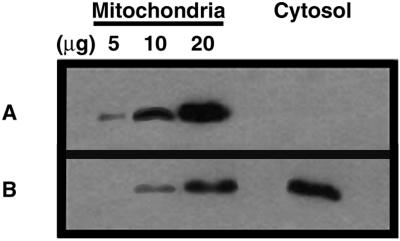
Figure 2.
In S. cerevisiae W303-1A cytochrome c is released from mitochondria to the cytosol during PCD induced by acetic acid. CytC was detected by immunodetection in mitochondria (5, 10, and 20 μg of protein) and postmitochondrial supernatants obtained from S. cerevisiae stationary untreated cells (A) or cells treated with 140 mM acetic acid (B).
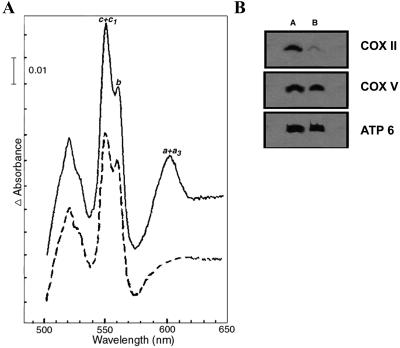
Figure 3.
Release of CytC parallels a reduction of cytochrome c oxidase in acid-treated cells. (A) Cytochrome spectra. Cytochrome spectra of S. cerevisiae mitochondria isolated from untreated cells (full line) or from cells treated with 140 mM acetic acid (dashed line). The α absorption bands corresponding to cytochromes a+a3 have maxima at 603 nm. The corresponding maximum for cytochrome b is 560 nm and for cytochrome c+c1, 550 nm. (B) Steady-state levels of COX II subunit are decreased in mitochondria from acid-treated cells. Immunodetection of COX II and V and ATPase 6 subunits in mitochondrial membranes of S. cerevisiae untreated cells (A) or cells treated with 140 mM acetic acid (B).
The levels of CytC were also evaluated in mitochondria and in the PMS from S. cerevisiae ATP10 mutant cells, which do not undergo a PCD process induced by acetic acid as shown hereafter. CytC was not detected in PMS and it was found to be at a normal level in mitochondria, compared with untreated cells (our unpublished data). These results indicate that CytC is specifically translocated from mitochondria to the cytosol during the acid-induced PCD.
ROS Are Produced in Mitochondria during Acetic Acid-induced PCD Process
S. cerevisiae W303-1A stationary cells stained with MitoTracker Red CM-H2Xros and treated with 140 mM acetic acid were analyzed by flow cytometry. An increase in the red fluorescence indicative of cells with an increased mitochondrial ROS production could only be detected after 100 min of treatment (our unpublished data). The results obtained after 200 min of treatment showed a heterogeneous population and a second subpopulation (∼45%) with a higher mean fluorescence intensity (Figure 4). The epifluorescence analysis showed that this subpopulation display a bright red fluorescence localized in mitochondria (our unpublished data). Moreover, killed cells displayed a highest red fluorescence corresponding to an unspecific cell staining (our unpublished data).
Figure 4.
ROS are produced in mitochondria of S. cerevisiae W303-1A cells treated with acetic acid. Overlay of red fluorescence histograms obtained for cells stained with MitoTracker Red CM-H2XRos. Untreated cells (thin gray line), or treated cells with 140 mM acetic acid (thick black line).
Acetic Acid-induced PCD Is Accompanied by Mitochondrial Alterations
The respiratory capacity of mitochondria isolated from control and acetic acid-treated cells was assayed polarographically by measuring oxygen uptake with NADH as substrate. The effect on respiration of exogenously added CytC was evaluated in a set of experiments. Results presented in Table 1 show that mitochondria isolated from cells treated with 140 mM acetic acid have a dramatically reduced oxygen consumption, with a decrease of nearly 75% in NADH oxidase activity. Although an increase of ∼2.8-fold of the oxygen consumption was observed after addition of CytC, the inability to fully restore the respiration rate with this addition suggested that an enzymatic portion of the respiratory chain could be intrinsically affected.
Table 1.
Respiratory activities of mitochondria from control and acetic acid treated cells Mitochondria were assayed polarographically for NADH oxidase. The specific activities reported (expressed as nanomoles of O2 per minute per milligram of protein) were corrected for KCN-insensitive respiration. Cytochrome oxidase activity (expressed as micromoles of cytochrome c oxidized per minute per milligram of mitochondrial protein) was assayed spectrophotometrically in mitochondria permeabilized with potassium deoxycholate by measuring oxidation of ferrocytochrome c at 550 nm. NADH cytochrome c reductase (expressed as micromoles of cytochrome c reduced per minute per milligram of mitochondrial protein) was also measured spectrophotometrically as described under MATERIALS AND METHODS. The rates measured in at least two independent assays did not differ by >10%. The values reported are the average of the two assays.
| Acetic acid treatment (mM) | NADH oxidase
|
NADH CytC reductase | Cytochrome oxidase | |
|---|---|---|---|---|
| − CytC | +CytC | |||
| 0 | 367 | 550 | 0.602 | 2.925 |
| 140 | 68 | 192 | 0.645 | 1.552 |
To establish the biochemical basis for the decrease in the respiratory activity of mitochondria, the NADH-CytC reductase and COX activities of isolated mitochondria from untreated and acid-treated cells, were measured. As shown in Table 1, the treatment with 140 mM acetic acid resulted in a decrease of 50% of COX activity in mitochondrial membranes, whereas NADH-CytC reductase activity was essentially identical to that obtained with mitochondria from untreated cells. These results confirm that COX complex was affected, whereas complex bc1 was unaffected by the treatment. Cytochromes spectra were recorded in isolated mitochondria (Figure 3A) to clarify the observed respiratory chain alterations (Table 1). In agreement with the observed lower COX activity, a decrease in the amount of cytochromes a+a3 in mitochondrial membranes of cells treated with acetic acid was observed, whereas the levels of cytochrome b were not affected (Figure 3A).
When the COX subunits II and V were analyzed by immunodetection of mitochondrial proteins obtained from cells treated with 140 mM acetic acid, the amount of COX II protein but not COX V was found to be lower than in the control (Figure 3B). Additionally, the amount of cytochrome b from bc1 complex and of subunit 6 of ATPase remained identical to control (Figure 3B).
Some studies on mammalian apoptosis reported an increase in ΔΨm after a lethal stimulus, with ΔΨm decreasing later in the death process (Vander Heiden et al., 1997, 1999). Consistently with such observations, in our study, acetic acid treatment induced a transient slight hyperpolarization followed by a depolarization (our unpublished data). However, although with a lower ΔΨm, cells maintained the specific mitochondria staining indicating that mitochondria membrane integrity is still preserved (our unpublished data).
Mitochondrial Respiration Is Essential for S. cerevisiae to Undergo a PCD Process Induced by Acetic Acid
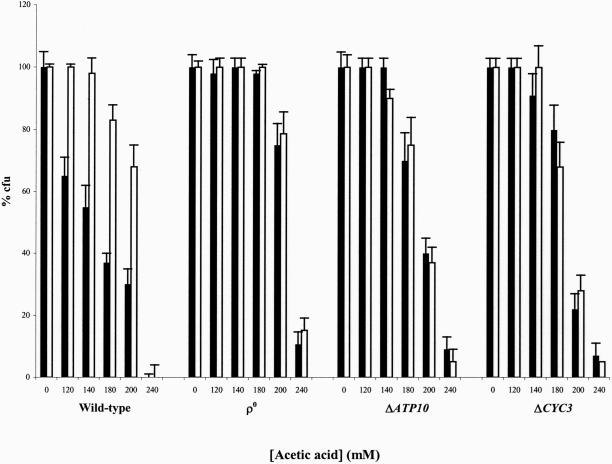
The requirement of mitochondria function in the PCD process induced by acetic acid was analyzed by the study of three S. cerevisiae W303-1A mutant strains, namely, the ρ0, lacking mitochondrial DNA; the null ATP10 mutant, deleted in an assembly factor of mitochondrial ATPase; and the null CYC3 mutant, deleted in the gene encoding a heme lyase, essential for the covalent binding of the heme group to isoform 1 and 2 of apocytochrome c (Pearce and Sherman, 1995). It was observed that these mutant strains were more resistant to death induced by acetic acid, comparatively to the wild-type strain. In addition, cycloheximide had no effect on survival (Figure 5) and no TUNEL-positive cells were found, for any of the acetic acid concentrations tested, even for concentrations inducing a percentage of dead cells identical to that obtained for the wild type (our unpublished data).
Figure 5.
Respiratory-deficient mutant cells of S. cerevisiae W303-1A are more resistant to acetic acid and acid-induced death is not inhibited by cycloheximide. Relative survival (percentage of cfu on YEPD agar plates; 100% corresponds to the number of cfu at time 0) of wild-type, ρ0 (lacking mitochondrial DNA), ΔATP10 (depleted of ATPase), and ΔCYC3 (depleted of CytC) cells of S. cerevisiae incubated for 200 min with acetic acid in the absence (black bars) or presence (white bars) of cycloheximide.
DISCUSSION
Mitochondria have been implicated in mammalian PCD processes (Skulachev, 1996, 1998, 2000, 2001; Kluck et al., 1997; Yang et al., 1997; Green and Reed, 1998) being called the “cell death organelle.” Data reported in this article allowed us to conclude that mitochondria are, as well, implicated in the active cell death of S. cerevisiae W303-1A induced by acetic acid. The strategy followed to analyze the possible role of mitochondria in that PCD process involved the use of stationary cells of S. cerevisiae, which have a high mass of fully active mitochondria, and of selected S. cerevisiae strains with mutations affecting mitochondria. Regarding the first strategy, we found that S. cerevisiae W303-1A stationary cells, like exponential cells (Ludovico et al., 2001a), can organize its own death in response to acetic acid. Using those cells, the possibility was addressed that CytC could be in this unicellular eukaryont, as in mammalian cells, a direct or indirect apoptogenic factor. The analysis of CytC localization by immunodetection revealed that acetic acid triggers CytC release from mitochondria to the cytosol in S. cerevisiae W303-1A under PCD. CytC extrusion from mitochondria was also concluded from the results of cytochromes spectra analysis. Recently, Yamaki et al. (2001) observed a release of CytC to the cytoplasm associated to a phenotype that largely resembles mammalian apoptosis in S. cerevisiae cells lacking the histone chaperone ASF1/CIA1. The human homolog of this chaperone seems to be involved in the regulation of apoptosis. Manon et al. (1997), using heterologous expression of the proapoptotic protein Bax, had previously described CytC release in S. cerevisiae. In this context, because CytC release was found in the apoptosis-like cell death induced by Bax-expression, deletion of the histone chaperone ASF1/CIA1, and acetic acid treatment (Ligr et al., 1998; Ludovico et al., 2001a; Yamaki et al., 2001), we have an explanation why the apoptotic phenotype is identical in those three situations. It is conceivable that when in the cytosol after its translocation from mitochondria, yeast CytC could work as an apoptogenic factor, as reported for mammalian cells. In these cells the proapoptotic activity of cytoplasmic CytC is due to the activation of a caspase cascade. Madeo et al. (2002) reported a caspase-related protease that regulates yeast apoptosis, named yeast caspase-1 (YCA-1). These authors showed that the disruption of YCA-1 gene resulted in a higher survival to acetic acid treatment compared with the wild type, whereas its overexpression led to the enhancement of cell death. In this context, when in cytosol, CytC may also work as an apoptogenic factor in S. cerevisiae through the activation of YCA-1, but further research is needed to get a complete frame of the interaction between these two proteins.
On the other hand, the release of CytC from mitochondria to the cytosol, being accompanied by the loss of the molecule at the mitochondria, leads to the occurrence of another proapoptotic event, namely, the production of ROS. Indeed, the observed increase in ROS production in S. cerevisiae W303-1A cells exposed to acetic acid may well be the consequence of the reduction in COX activity due to the loss of CytC from mitochondria. Whether loss of COX activity is a consequence of CytC release or a primary effect of acetic acid on the assembly of the enzyme remains to be studied, although we favor the first possibility. The observed deficiency in COX activity could be responsible for an inhibition of the electron transfer with the accumulation of reducing equivalents in the middle portion of the electron transfer chain, and thus directing one-electron transfer to O2, resulting in the production of superoxide (Cai and Jones, 1998). ROS production seems to be an event common to all S. cerevisiae apoptotic phenotypes reported so far (Madeo et al., 1997, 1999; Fröhlich and Madeo 2000; Laun et al., 2001; Levine et al., 2001; Narasimhan et al., 2001; this study).
In mitochondria, CytC is an essential component of the respiratory chain acting as an electron carrier between complex bc1 and COX complex. Therefore, the decrease in the amount of CytC in mitochondria of S. cerevisiae exposed to acetic acid must have other functional implications. The biochemical basis for the observed decrease in oxygen consumption and in ΔΨm is the important reduction in COX activity. Such a reduction could be due to an inefficient COX assembly in the absence of CytC. The decrease of steady-state levels of COX II protein observed in mitochondria from acetic acid-treated cells that have lost CytC and the impossibility to detect it in PMS indicate that, probably a degradation of the subunit occurred, as has been reported in mutants lacking CytC (Pearce and Sherman, 1995). In fact, COX II participates in CytC binding (Bisson et al., 1977) and the absence of CytC seems to destabilize COX II, making it susceptible to degradation (Pearce and Sherman, 1995). Manon et al. (2001) reported very recently that Bax-induced CytC release is directly involved in the decrease of COX activity by activating the mitochondrial AAA-type protease Yme1p, which leads to COX II degradation.
A pattern of mitochondrial dysfunction identical to that discussed above was observed in S. cerevisiae cells expressing Bax (Manon et al., 1997). With this new insight, a link between major alterations in the respiratory chain, namely, decrease in the amount of CytC and reduction of the COX activity, and the PCD process in yeast, can be envisaged, being the first time that the apoptotic mitochondrial pathway was found to be activated in yeast wild-type cells independent of the heterologous expression of Bax.
The comparison between main structural and functional cellular changes, including mitochocondrial changes, associated to S. cerevisiae programmed cell death induced by Bax-expression, by deletion of histone chaperone ASF1/CIA1, or by acetic acid treatment is outlined in Table 2.
Table 2.
Involvement of mitochondria in yeast programmed cell death Differences between S. cerevisiae programmed cell death induced by Bax-expression, mediated by deletion of histone chaperone ASF1/CIA1 (Yamaki et al., 2001), and induced by acetic acid treatment (Ludovico et al., 2001a; this study): main structural and functional cellular changes associated.
| Cellular changes | Bax-expression | asf1/cia1 | Acetic acid |
|---|---|---|---|
| Exposition of phosphatidylserine on the outer | 213 | nd | 213 |
| surface of the plasma membrane | Ligr et al., 1998 | ||
| Chromatin condensation | 213 | 213 | 213 |
| Ligr et al., 1998 | |||
| DNA fragmentation | 213 | nd | 213 |
| Ligr et al., 1998 | |||
| Fragmentation of nuclei | nd | 213 | − |
| Fragmentation of chromatin | nd | 213 | 213 |
| Mitochondrial membrane potential | Hyperpolarization | Depolarization | Hyperpolarization followed |
| Gross et al., 2000 | by depolarization | ||
| Dysfunction of mitochondrial proton pump | nd | 213 | nd |
| Release of cytochrome c | 213 | 213 | 213 |
| Manon et al., 1997 | |||
| ROS production | 213 | nd | 213 |
| Gross et al., 2000 | |||
| Decrease of cytochrome c oxidase | 213 | nd | 213 |
| Manon et al., 1997 | |||
| Mitochondrial apoptosis-induced channel activity | 213 | nd | nd |
| Pavlov et al., 2001 |
, present; −, absent; nd, not determined.
The above-discussed results, obtained with experiments using stationary cells of S. cerevisiae W303-1A, indicated that mitochondria would be involved in the PCD process induced by acetic acid through the release of CytC and ROS production. The use of three S. cerevisiae strains with mutations affecting mitochondrial respiratory chain function led to results that further indicate that mitochondria are required for that process. Although the literature concerning the mitochondria involvement in Bax-induced cell death is controversial (Greenhalf et al., 1996; Kissova et al., 2000), our results clearly demonstrate that a yeast cell depleted of mitochondrial DNA does not undergo a PCD process in response to acetic acid.
Supporting the interpretation that CytC exhibits apoptogenic properties in S. cerevisiae, the mutant in CYC3 gene, which encodes a heme lyase essential for the heme binding to apocytochrome c (isoform 1 and 2) (Pearce and Sherman, 1995), did not display PCD when exposed to acetic acid. It is interesting to notice that in ρo strains, the CYC1 gene (coding for iso-1-CytC) is four- to sixfold repressed, probably due to the lack of mitochondrial DNA and of mitochondrial respiratory activity (Epstein et al., 2001), perhaps explaining why those strains do not undergo a PCD induced by acetic acid.
Matsuyama et al. (1998) related cell death induced by Bax-expression with the mitochondrial ATPase complex, namely, with ATP subunit 4. Our results regarding the inability of ATP10 mutant (ATP10 codes a mitochondrial protein encoded by nuclear DNA that acts as an ATPase assembly factor; Paul et al., 2000) to undergo PCD by exposure to acetic acid indicate that a fully assembled F0F1-ATPase is required for the PCD process. Moreover, and because it has been proposed that ATPase complex could be involved in the mechanism of CytC release in apoptotic mammalian cells (Matsuyama et al., 2000), our results with ATP10 null mutant showing that treatment with 140 mM acetic acid does not result in PCD nor CytC release suggest that the ATPase complex would also be involved in the mechanism of CytC release from mitochondria during PCD induced by acetic acid in S. cerevisiae. Because of the absence of oxidative phosphorylation in the ρ0 and ATP10 mutant strains, it could be argued that the inability of those strains to develop the acid-induced PCD process could be due to the failure in mitochondrial ATP generation. Nevertheless, this cannot be the explanation because cells of wild-type strain are able to induce PCD even when mitochondrial ATP synthesis is inhibited by oligomycin.
In summary, the experimental evidence herein strongly supports that the PCD process induced by acetic acid in S. cerevisiae is mediated by a mitochondria-dependent apoptotic pathway. Therefore, in yeast the pivotal role of mitochondria is related not only to the exogeneous cell death induced by Bax-expression but also, and more relevantly, to the endogenous PCD induced with acetic acid. These advances surpass the contemporary knowledge on PCD in unicellular eukaryotes and raise new questions regarding PCD in yeast, such as 1) is CytC an activating factor of YCA-1; and 2) is CytC release an event common to all endogenous PCD processes? The answers to these questions will be most useful in the further elucidation of ancestral pathway(s) common to PCD in metazoans and will validate the yeast cell as the tool of choice in apoptosis research.
ACKNOWLEDGMENTS
The stay of P.L. in Department of Biology at Columbia University was supported in part by “Fundação Luso-Americana para o Desenvolvimento.” P.L. has a fellowship from PRAXIS XXI (Fundação da Ciência e Tecnologia, Portugal). A.B. is a recipient of a grant MDACU01991001 from the Muscular Dystrophy Association. We thank Prof. Vladimir Skulachev and Dr. Frank Madeo for critical reading of the manuscript and all the helpful comments and suggestions.
Abbreviations used:
- COX
cytochrome c oxidase
- CytC
cytochrome c
- ΔΨm
mitochondrial membrane potential
- PCD
programmed cell death
- PMS
postmitochondrial supernatant
- ROS
reactive oxygen species
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick-end labeling
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E01–12–0161. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E01–12–0161.
REFERENCES
- Adrian C, Martin SJ. The mitochondrial apoptosome: a killer unleashed by the cytochrome seas. Trends Biochem Sci. 2001;26:390–397. doi: 10.1016/s0968-0004(01)01844-8. [DOI] [PubMed] [Google Scholar]
- Bisson R, Gutweniger H, Montecucco C, Colonna R, Zanotti A, Azzi A. Covalent binding of arylazido derivatives of cytochrome c to cytochrome oxidase. FEBS Lett. 1977;81:147–150. doi: 10.1016/0014-5793(77)80948-4. [DOI] [PubMed] [Google Scholar]
- Cai J, Jones DP. Superoxide in apoptosis. Mitochondrial generation triggered by cytochrome c loss. J Biol Chem. 1998;273:11401–11404. doi: 10.1074/jbc.273.19.11401. [DOI] [PubMed] [Google Scholar]
- Carratore MR, Croce C, Simili M, Taccini E, Scavuzzo M, Sbrana S. Cell cycle and morphological alterations as indicative of apoptosis promoted by UV irradiation in S. cerevisiae. Mutat Res. 2002;513:183–191. doi: 10.1016/s1383-5718(01)00310-2. [DOI] [PubMed] [Google Scholar]
- Epstein C, Waddle JA, Hale IV W, Davé V, Thornton J, Macatee TL, Garner HR, Butow R. Genome-wide responses to mitochondrial dysfunction. Mol Biol Cell. 2001;12:297–308. doi: 10.1091/mbc.12.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye G, Kujawa C, Fukuhara H. Physical and genetic organization of petite and grande yeast mitochondrial DNA. IV. In vivo transcription products of mitochondrial DNA and localization of 23 S ribosomal RNA in petite mutants of Saccharomyces cerevisiae. J Mol Biol. 1974;88:185–203. doi: 10.1016/0022-2836(74)90304-0. [DOI] [PubMed] [Google Scholar]
- Fröhlich KU, Madeo F. Apoptosis in yeast – a monocellular organism exhibits altruistic behavior. FEBS Lett. 2000;473:6–9. doi: 10.1016/s0014-5793(00)01474-5. [DOI] [PubMed] [Google Scholar]
- Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- Greenhalf W, Stephan C, Chaudhuri B. Role of mitochondria and C-terminal membrane anchor of Bcl-2 in Bax induced growth arrest and mortality in Saccharomyces cerevisiae. FEBS Lett. 1996;380:169–175. doi: 10.1016/0014-5793(96)00044-0. [DOI] [PubMed] [Google Scholar]
- Gross A, Pilcher K, Blachly-Dyson E, Basso E, Jockel J, Bassik M, Korsmeyer S, Forte M. Biochemical and genetic analysis of the mitochondrial response of yeast to BAX and BCL-XL. Mol Cell Biol. 2000;20:3125–3136. doi: 10.1128/mcb.20.9.3125-3136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- Kissova I, Polcic P, Kempna P, Zeman I, Sabova L, Kolarov J. The cytotoxic action of Bax on yeast cells does not require mitochondrial ADP/ATP carrier but may be related to its import to the mitochondria. FEBS Lett. 2000;471:113–118. doi: 10.1016/s0014-5793(00)01379-x. [DOI] [PubMed] [Google Scholar]
- Kluck R, Bossy-Wetzel E, Green DR, Newmeyer DD. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- Kluck R, Ellerby LM, Ellerby HM, Naiem S, Yaffe M, Margoliash E, Bredesen D, Mauk AG, Sherman F, Newmeyer D. Determinants of cytochrome c pro-apoptotic activity –The role of lysine 72 trimethylation. J Biol Chem. 2000;275:16127–16133. doi: 10.1074/jbc.275.21.16127. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laun P, Pichova A, Madeo F, Fuchs J, Ellinger A, Kohlwein S, Dawes I, Frohlich KU, Breitenbach M. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol Microbiol. 2001;39:1166–1173. [PubMed] [Google Scholar]
- Levine A, Belenghi B, Damari-Weisler H, Granot D. Vesicle-associated membrane protein of Arabidopsis Suppresses Bax-induced Apoptosis in Yeast downstream of oxidative burst. J Biol Chem. 2001;276:46284–46289. doi: 10.1074/jbc.M107375200. [DOI] [PubMed] [Google Scholar]
- Ligr M, Madeo F, Frohlich E, Hilt W, Frohlich KU, Wolf DH. Mammalian Bax triggers apoptotic changes in yeast. FEBS Lett. 1998;438:61–65. doi: 10.1016/s0014-5793(98)01227-7. [DOI] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Ludovico P, Sansonetty F, Côrte-Real M. Assessment of mitochondrial membrane potential in yeast cell populations by flow cytometry. Microbiology. 2001b;147:3335–3343. doi: 10.1099/00221287-147-12-3335. [DOI] [PubMed] [Google Scholar]
- Ludovico P, Sousa MJ, Silva MT, Leão C, Côrte-Real M. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology. 2001a;147:2409–2415. doi: 10.1099/00221287-147-9-2409. [DOI] [PubMed] [Google Scholar]
- Madeo F, Fröhlich E, Fröhlich KU. A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol. 1997;139:729–734. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Fröhlich E, Ligr M, Gray M, Sigrist SJ, Wolf DH, Fröhlich KU. Oxygen stress: a regulator of apoptosis in yeast. J Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Herker E, Maldener C, Wissing S, Lächelt S, Herlan M, Fehr M, Lauber K, Sigrist S, Wesselborg S, Fröhlich K-U. A caspase-related protease regulates apoptosis in yeast. Mol Cell. 2002;9:1–20. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- Manon S, Chaudhuri B, Guérin M. Release of cytochrome c and decrease of cytochrome c oxidase in Bax-expressing cells, and prevention of these effects by coexpression of Bcl-xL. FEBS Lett. 1997;415:29–32. doi: 10.1016/s0014-5793(97)01087-9. [DOI] [PubMed] [Google Scholar]
- Manon S, Priault M, Camougrand N. Mitochondrial AAA-Type protease Yme1p is involved in Bax effects on cytochrome c oxidase. Biochem Biophys Res Commun. 2001;289:1314–1319. doi: 10.1006/bbrc.2001.6120. [DOI] [PubMed] [Google Scholar]
- Matsuyama S, Llopis J, Deveraux QL, Tsien R, Reed JC. Changes in mitochondrial and cytosolic pH: early events that modulate caspase activation during apoptosis. Nat Cell Biol. 2000;2:318–325. doi: 10.1038/35014006. [DOI] [PubMed] [Google Scholar]
- Matsuyama S, Xu Q, Velours J, Reed JC. The mitochondrial F0F1-ATPase proton pump is required for function of the proapoptotic protein Bax in yeast and mammalian cells. Mol Cell. 1998;1:327–336. doi: 10.1016/s1097-2765(00)80033-7. [DOI] [PubMed] [Google Scholar]
- Narasimhan ML, Damsz B, Coca MA, Ibeas JI, Yun DJ, Pardo JM, Hasegawa PM, Bressan RA. A plant defense response effector induces microbial apoptosis. Mol Cell. 2001;8:921–30. doi: 10.1016/s1097-2765(01)00365-3. [DOI] [PubMed] [Google Scholar]
- Paul MF, Barrientos A, Tzagoloff A. A single amino acid change in subunit 6 of the yeast mitochondrial ATPase suppresses a null mutation in ATP10. J Biol Chem. 2000;275:29238–29243. doi: 10.1074/jbc.M004546200. [DOI] [PubMed] [Google Scholar]
- Pavlov E, Priault M, Pietkiewicz D, Cheng E, Antonsson B, Manon S, Korsmeyer S, Mannella C, Kinnally K. A novel, high conductance channel of mitochondria linked to apoptosis in mammalian cells and Bax expression in yeast. J Cell Biol. 2001;155:725–731. doi: 10.1083/jcb.200107057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce D, Sherman F. Degradation of cytochrome oxidase subunits in mutants of yeast lacking cytochrome c and suppression of the degradation by mutation of yme1. J Biol Chem. 1995;270:20879–20882. doi: 10.1074/jbc.270.36.20879. [DOI] [PubMed] [Google Scholar]
- Pham N, Robison B, Hedley D. Simultaneous detection of mitochondrial respiration chain activity and reactive oxygen in digitonin-permeabilized cells using flow cytometry. Cytometry. 2000;41:245–251. doi: 10.1002/1097-0320(20001201)41:4<245::aid-cyto2>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Skulachev VP. Why are mitochondria involved in apoptosis? Permeability transition pores and apoptosis as selective mechanisms to eliminate superoxide-producing mitochondria and cell. FEBS Lett. 1996;397:710–717. doi: 10.1016/0014-5793(96)00989-1. [DOI] [PubMed] [Google Scholar]
- Skulachev VP. Cytochrome c in the apoptotic and antioxidant cascades. FEBS Lett. 1998;423:275–280. doi: 10.1016/s0014-5793(98)00061-1. [DOI] [PubMed] [Google Scholar]
- Skulachev VP. Mitochondria in the programmed death phenomena; a principle of biology: “It is better to die than to be wrong.”. IUBMB Life. 2000;49:365–373. doi: 10.1080/152165400410209. [DOI] [PubMed] [Google Scholar]
- Skulachev VP. The programmed death phenomena, aging, and the Samurai law of biology. Exp Gerontol. 2001;36:995–1024. doi: 10.1016/s0531-5565(01)00109-7. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A, Akai A, Needleman RB. Assembly of the mitochondrial membrane system. Characterization of nuclear mutants of Saccharomyces cerevisiae with defects in mitochondrial ATPase and respiratory enzymes. J Biol Chem. 1975;250:8228–8235. [PubMed] [Google Scholar]
- Vander Heiden MG, Chandel NS, Williamson EK, Schumacker PT, Thompson CB. Bcl-xL regulates the membrane potential and volume homeostasis of mitochondria. Cell. 1997;91:627–637. doi: 10.1016/s0092-8674(00)80450-x. [DOI] [PubMed] [Google Scholar]
- Yamaki M, Umehara T, Chimura T, Horikoshi M. Cell death with predominant apoptotic features in Saccharomyces cerevisiae mediated by deletion of the histone chaperone ASF1/CIA1. Genes Cells. 2001;6:1043–1054. doi: 10.1046/j.1365-2443.2001.00487.x. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Chandel NS, Schumacker PT, Thompson CB. Bcl-xL prevents cell death following growth factor withdrawal by facilitating mitochondrial ATP/ADP exchange. Mol Cell. 1999;3:159–167. doi: 10.1016/s1097-2765(00)80307-x. [DOI] [PubMed] [Google Scholar]
- Yang J, Liu X, Bhalla K, Kim N, Ibrado AM, Cai J, Peng T, Jones D, Wang X. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]