Abstract
The Saccharomyces cerevisiae heat-shock protein (Hsp)40, Ydj1p, is involved in a variety of cellular activities that control polypeptide fate, such as folding and translocation across intracellular membranes. To elucidate the mechanism of Ydj1p action, and to identify functional partners, we screened for multicopy suppressors of the temperature-sensitive ydj1-151 mutant and identified a yeast Hsp110, SSE1. Overexpression of Sse1p also suppressed the folding defect of v-Src kinase in the ydj1-151 mutant and partially reversed the α-factor translocation defect. SSE1-dependent suppression of ydj1-151 thermosensitivity required the wild-type ATP-binding domain of Sse1p. However, the Sse1p mutants maintained heat-denatured firefly luciferase in a folding-competent state in vitro and restored human androgen receptor folding in sse1 mutant cells. Because the folding of both v-Src kinase and human androgen receptor in yeast requires the Hsp90 complex, these data suggest that Ydj1p and Sse1p are interacting cochaperones in the Hsp90 complex and facilitate Hsp90-dependent activity.
INTRODUCTION
Molecular chaperones are required to catalyze a number of cellular activities, including protein folding, transport, and degradation. These activities can be highly interrelated. For example, chaperones that fail to refold denatured proteins may redirect the polypeptide to proteolytic machines in the cell (reviewed in Wickner et al., 1999). In addition, chaperones can prevent premature folding or aggregation to deliver nascent polypeptides to translocation complexes localized at the endoplasmic reticulum (ER) or mitochondrial membranes (reviewed in Rassow and Pfanner, 1996; Rapoport et al., 1999). Finally, specific molecular chaperones may deliver protein substrates to other chaperones. Notably, chaperone “bucket-brigades” were observed when the protein-folding activities of the DnaK-DnaJ-GrpE and GroEL/GroES complexes were first uncovered, and during the translocation and subsequent folding of proteins in the mitochondria (Manning-Krieg et al., 1991; Langer et al., 1992; reviewed in Frydman, 2001). In another example, the mammalian heat-shock protein (Hsp)110 molecular chaperone acts as a “holdase,” binding to unfolded proteins and preventing their aggregation until ATP-dependent refolding is catalyzed by the Hsp70 and Hsp40 chaperones (Oh et al., 1997). Not surprisingly, the expression of mammalian Hsp110 is induced by heat, and overexpression of Hsp110 confers thermotolerance to Rat-1 and HeLa cells. Subsequent studies determined that the holdase activity of mammalian Hsp110 requires its putative peptide-binding and C-terminal domains, but not the ATP-binding domain (Oh et al., 1999).
The cytoplasmic Ydj1 protein in the yeast Saccharomyces cerevisiae is a member of the Hsp40 family of molecular chaperones (Caplan and Douglas, 1991; Atencio and Yaffe, 1992) that activate the ATPase activities of cognate Hsp70 chaperones through the J-domain, an ∼70-amino acid motif that interacts directly with Hsp70 (reviewed in Cheetham and Caplan, 1998; Kelley, 1998). Regulation of Hsp70 ATPase activity in turn modulates the affinity and on/off rates of Hsp70 polypeptide clients. Some Hsp40s, like Ydj1p, can also bind directly to polypeptides (Cyr, 1995). Because of their multiple partners and biochemical activities, it is anticipated that a specific DnaJ homolog plays many critical roles in the cell. Consistent with this notion, yeast containing the ydj1-151 temperature-sensitive mutation accumulate untranslocated preproteins in the cytoplasm at the nonpermissive temperature (Caplan et al., 1992) and are defective for the translation of heterologous proteins (Brodsky et al., 1998) and cyclin-dependent phosphorylation (Yaglom et al., 1996). Ydj1 also appears to be important for Hsp90 function. For example, mutations in YDJ1 compromise v-Src folding and the activity of steroid hormone receptors, both of which also require Hsp90 activity (reviewed in Caplan, 1999). Furthermore, mutation of both YDJ1- and Hsp90-encoding genes in the same cell results in a synthetic lethal defect (Kimura et al., 1995), which suggests a functional interaction between the chaperones. Because Ydj1p and Hsp90 do not appear to form a stable bimolecular complex (Kimura et al., 1995; Chang et al., 1997), Hsp70 and other chaperones may “bridge” Hsp90 and Ydj1p. Consistent with this view, Hsp90 is recruited to some polypeptides via Hop, which binds to the ADP-bound form of Hsp70, and to which Ydj1p is also bound (Kosano et al., 1998). Additionally, in recent work it has been shown that entry of the avian progesterone receptor into the Hsp90 chaperoning pathway is initiated by binding of the Hsp40 homologs Ydj1p or Hdj2p (Hernández et al., 2002).
Whereas much information on the mechanism of Ydj1p action has been obtained from these genetic and biochemical studies, the Hsp110 family of molecular chaperones in yeast has been relatively ill-defined. Deletion of one of the genes encoding an Hsp110 chaperone in S. cerevisiae, known as SSE1, results in poor viability at every temperature examined (Mukai et al., 1993; Shirayama et al., 1993). Although it is not clear why Sse1p supports optimal cell growth, purified yeast Sse1p, like mammalian Hsp110, exhibits holdase activity (Brodsky et al., 1999). In addition, Sse1p interacts biochemically and genetically with the yeast Hsp90 complex (Liu et al., 1999), and yeast lacking the Hsp90 homologs Hsc82 (constitutively expressed at high levels and moderately stress inducible) and Hsp82 (constitutively expressed at low levels but heat inducible) are inviable (Borkovich et al., 1989; Nathan and Lindquist, 1995). However, it is unknown whether holdase activity and/or the action of Sse1p on Hsp90-dependent functions are essential for cell growth.
To begin to address this question and to better characterize the yeast homolog of the Hsp110 family (reviewed in Easton et al., 2000), we used both genetic and biochemical methods. First, we found that overexpression of Sse1p rescues the thermosensitivity of yeast containing the ydj1-151 mutation. Purification and functional analyses of wild-type and mutant forms of Sse1p indicate that the presumptive ATP-binding domain is dispensable for the in vitro holdase activity of the chaperone. However, yeast overexpressing holdase-proficient Sse1p mutants cannot rescue the ydj1-151 growth defect. These results indicate that the contribution of Sse1p to cell growth arises at least in part from facilitating Hsp90-dependent functions, and more generally that demonstrations of chaperone holdase activity might not correspond to an essential in vivo activity.
MATERIALS AND METHODS
Yeast Strains and Culture Conditions
Yeast strains used in this study were as follows: RSY607, MATα ura3-52 leu2-3,112 PEP4::URA3; W303, MATα ade2-1 his3-11 leu2-3,112 ura3-1 trp1-1 can1-100; ACY17b, MATα ade2-1 his3-11 leu2-3,112 ura3-1 trp1-1 can 1-100 ydj1-2::HIS3 LEU2::ydj1-151 (Caplan et al., 1992); E0020, MATα his3 leu2 ura3 trp1 sse1::HIS3; E0030, MATa/MATα his3/his3 leu2/leu2 ura3/ura3 trp1/trp1 sse1::HIS3/sse1::URA3 (Shirayama et al., 1993); and JGY014a, a haploid spore of E0030, MATa his3 leu2 ura3 trp1 sse1::URA3 (this study). Standard media and methods for the growth, manipulation, and transformation of yeast were used (Kaiser et al., 1994). For serial dilution assays, saturated liquid cultures grown at 26°C in synthetic complete medium lacking uracil (SC-ura) were adjusted to an optical density measured at 600 nm (OD600) of 3.2, serially diluted 10-fold in sterile water, plated on SC-ura plates containing either galactose or glucose as the carbon source, and incubated at the indicated temperatures for 4 d. Yeast lysates for immunoblot analysis were prepared as described previously (Brodsky et al., 1998).
Genetic and Molecular Methods
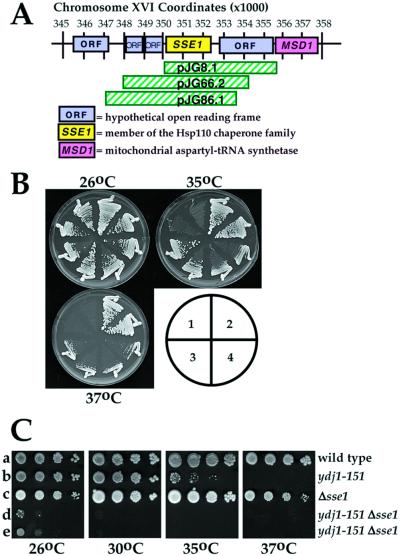
To screen for multicopy suppressors of the ydj1-151 temperature-sensitive phenotype, strain ACY17b was transformed by the lithium-acetate method with a URA3-marked yeast genomic library in plasmid YEp24 (Carlson and Botstein, 1982), obtained from Dr. John Woolford (Carnegie Mellon University, Pittsburgh, PA). Transformants were selected for growth on SC-ura at 26°C and replicates were incubated at either 26 or 37°C for 2 d. Approximately 22,000 transformants were screened, and plasmids from 60 strains in which suppression of the temperature sensitivity of ydj1-151 was apparent were reisolated and retransformed into ACY17b to verify suppression. Of these, 13 plasmids were obtained that suppressed the ydj1-151 temperature sensitivity upon retransformation and that fell into three distinct groups as assayed by restriction digest. DNA sequence analysis of a representative insert from each group was performed using the Sequenase kit (Amersham Biosciences, Piscataway, NJ) and revealed that the common open reading frame (ORF) in each clone was SSE1 (Figure 1A). Suppression of ydj1-151 temperature sensitivity by SSE1 was verified by subcloning a BamHI-ClaI fragment containing SSE1 from one of the original, isolated clones (pJG8.1) into the high copy vector pRS426 (Sikorski and Hieter, 1989) to create pJG816. Retransformation of pJG816 into yeast strain ACY17b resulted in growth at permissive and semipermissive temperatures (Figure 1B). SSE1 on a single copy CEN vector was created by introducing the ∼2.1-kb BamHI-SalI fragment from pJG816 into pRS314 (Sikorski and Hieter, 1989).
Figure 1.
SSE1 is a multicopy suppressor of ydj1-151 temperature sensitivity and interacts with YDJ1. (A) Map of chromosome XVI in S. cerevisiae depicting the approximate positions of the DNA inserts that rescued ydj1-151 thermosensitivity, all of which included SSE1. (B) Ydj1-151 cells were transformed with a multicopy YEp24 yeast vector either 1) lacking or 2) containing YDJ1, 3) pJG8.1 (original complementing clone), or 4) pJG816 (SSE1) were struck onto SC-ura medium and were incubated at the indicated temperatures for 4 d. The growth of two transformants from each strain is shown. (C) Serial dilutions of W303 wild-type parental yeast (a), ACY17b ydj1-151 cells (b), JGY014a sse1Δ cells (c), or yeast deleted for SSE1 and containing the ydj1-151 allele (d and e) were spotted onto complete medium and incubated at the indicated temperatures for 4 d.
To construct plasmids for the expression and purification of wild-type and mutant hexahistidine-tagged forms of Sse1p, an SSE1-containing DNA fragment flanked by BamHI and XbaI sites was generated by polymerase chain reaction (PCR) amplification from pJG816 by using primers 5′ CATTTGGATCCAAGATGAGTACTCCATTTGGTTTAG 3′ and 5′ GCTGCCTGCAGATTAGTCCATGTCAAC 3′, the PCR product was treated with BamHI and XbaI, gel-purified using the QiaQuick kit (QIAGEN, Valencia, CA), and the fragment was cloned into pTrcHisA (Invitrogen, Carlsbad, CA), resulting in addition of a hexhistidine (His6) tag to the N-terminus of SSE1. The tagged SSE1 was then excised from pTrcHisA by digestion with NcoI, the overhangs were filled in with Klenow (Roche Applied Science, Indianapolis, IN), and the DNA was digested with KpnI and gel purified as described above. The resulting DNA fragment was ligated into the high copy, galactose-inducible expression vector pYES2 (Invitrogen) that had been digested with HindIII, filled in with Klenow, and similarly digested with KpnI. This resulted in the construction of pJG010 (PGAL-His6SSE1).
Plasmids containing site-directed mutations in SSE1 were PCR amplified from pJG010 by using the QuickChange kit (Stratagene, La Jolla, CA). The primers used to construct pJG015 (PGAL-His6SSE1–1) were 5′ CCAAACCAATAATTCTTTGCAAGTTGGCGACAGTG 3′ and 5′ CACTGTCGCCAACTTGCAAAGAATTATTGGTTTGG 3′, resulting in amino acid substitution K69Q. To construct pJG026 (PGAL-His6SSE1-2) primers 5′CGAAGTCCCTACCATCAAAATGCTTGTCGCAGGC 3′ and 5′GCCTGCGACAAGCATTTTGATGGTAGGGACTTCCG 3′ were used, resulting in amino acid substitution G233D. Primers 5′ CGCCTCTAGATTAGTCCATGTCAACATCACC 3′ and 5′ GCGGATCCATGCATCATCATCATCATCATGGGTGCCGCCTTTATTTGCGCCATTCACTCTCC 3′ were used to construct the His6-tagged truncation mutant (Sse375-694p) lacking the ATP binding domain (pSse CTD). The resulting mutations were confirmed by DNA sequence analysis.
Diploid yeast containing one copy of the ydj1-151 allele and deleted for one copy of SSE1 were obtained from a cross between ACY17b and JGY014a. Sporulation was induced and haploid progeny were obtained and analyzed genetically using established methods (Kaiser et al., 1994).
Antibodies
An anti-Sse1p antiserum was generated by Cocalico Biologicals (Reamstown, PA) against a peptide corresponding to amino acids 663–684 in Sse1p and that included a cysteine added at the C terminus (AAMAEKLAAQRKAEAEKKEEKKC; synthesized by the University of Pittsburgh Peptide Synthesis Facility, Pittsburgh, PA) and to which bovine serum albumin was conjugated using 4succinimidyloxycarbonyl-methyl-α[2-pyridyldithio]toluene (Pierce Chemical, Rockford, IL). Antibodies against prepro-α factor (ppαF) were obtained from Dr. Randy Schekman (University of California, Berkeley, Berkeley, CA), and antibodies against yeast BiP (Brodsky and Schekman, 1993), Ydj1p (Caplan and Douglas, 1991), and Sec61p (Stirling et al., 1992) were described previously. Polyclonal antibody against Hsp90 was a kind gift from Dr. Susan Lindquist (Massachusetts Institute of Technology, Cambridge, MA); anti-penta-histidine antibody was from QIAGEN, and anti-phosphotyrosine and anti-Src antibodies were obtained from Upstate Biotechnology (Lake Placid, NY).
Pulse-Chase and Immunoprecipitation Assays
The accumulation of the precursor form of the yeast mating prepheromone (ppαF) was used to assay for a posttranslational translocation defect (Deshaies and Schekman, 1987; Caplan et al., 1992). In brief, ydj1-151 mutant yeast were transformed with either vector (pRS426) or a high copy plasmid containing SSE1 (pJG816), grown at 26°C, and then radiolabeled with 0.1 mCi/ml of “EXPRE35S35S” 35S-Methionine and Cysteine Labeling Mix (PerkinElmer Life Sciences, Boston, MA) at either 26 or 37°C for 10 min. Labeling was stopped by the addition of cycloheximide to a final concentration of 0.2 mg/ml, aliquots were removed at the indicated time points and extracts were prepared as described previously (Morrow and Brodsky, 2001). Anti-ppαF or anti-Kar2p (BiP) antibody was added to extracts containing ∼5 × 106 cpm of 35S, and the mixture was incubated overnight at 4°C before immune complexes were precipitated for 2 h at room temperature with protein A-Sepharose CL-4B (Amersham Biosciences) in IP wash buffer (50 mM Tris-Cl pH 7.4, 0.2% SDS, 150 mM NaCl, 1% Triton X-100, and 5 mM EDTA). The resulting immunoprecipitates were washed (Rothblatt et al., 1989), the proteins were resolved by SDS-PAGE, and results were obtained by PhosphorImager analysis (Fuji Medical Systems, Stamford, CT).
Analysis of v-Src Activity in Yeast
v-Src activity was assayed essentially as described (Dey et al., 1996). Yeast cultures (100 ml) were grown at 30°C in minimal medium containing 2% raffinose (wt/vol) to OD600 = 0.2, and v-Src expression was induced by the addition of 2% galactose (wt/vol). After a 6-h induction period, cells were harvested and resuspended in protein extraction buffer (20 mM HEPES pH 7.5, 100 mM KCl, 0.1 mM EDTA pH 7.5, 5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 2 μg/ml protease inhibitor cocktail). The resuspended cells were broken in the presence of 1.5 g of 0.4-mm-diameter glass beads by two 60-s bursts in a mini-BeadBeater (Biospec Products, Bartlesville, OK) at 4°C, with cooling on ice for 60 s between bursts. The crude lysate was cleared at 14,000 × g for 10 min at 4°C. Protein concentrations were measured using the Bio-Rad Protein Assay reagent (Bio-Rad, Hercules, CA).
The levels of tyrosine phosphorylation and v-Src protein were assayed using anti-phosphotyrosine and anti-Src monoclonal antibodies. Lysates (20 μg of total protein) were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membrane. Filters were washed briefly with Tween 20/Tris-buffered saline (TTBS) (20 mM Tri-HCl pH 7.5, 0.5 M NaCl, and 0.05% Tween 20) and blocked overnight with TTBS containing 5% nonfat dry milk. Filters were incubated with the respective primary antibodies diluted 1:1000 in antibody dilution buffer (1× phosphate-buffered saline, 3% bovine serum albumin, 0.05% Tween 20, and 0.1% thimerosal) for 2 h. Filters were washed three times for 10 min in TTBS. Filters were then treated with secondary antibody (horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G [for anti-phosphotyrosine and anti-Src] or goat anti-rabbit [for anti-Ydj1]) for 1 h, and washed and treated with the Super Signal West Pico chemiluminescence reagent (Pierce Chemical) and exposed to x-ray film.
Viability of the strains expressing galactose-regulated v-Src was analyzed by inoculating a sample of log phase cultures onto medium containing either glucose or galactose and incubating the plates at 30°C for 3 d.
Androgen Receptor-Hormone Binding Assay
Yeast cells were grown in selective medium containing 2% galactose (wt/vol) to an OD600 = 0.2 at 30°C, and 1-ml aliquots were used for each assay. The binding was performed essentially as described (Caplan et al., 1995). In brief, [3H]R1881 (DuPont, Wilmington, DE) diluted 1:5 with unlabeled R1881 to a final concentration of 100 nM plus or minus 20 μM of unlabeled R1881 (200-fold excess) was added to the cells, which were incubated for 90 min at 25°C with shaking, washed three times with ice-cold water, and the amount of associated radioactivity was assessed in 10 ml of scintillation fluid. Nonspecific counts were calculated from the samples containing 200-fold excess of unlabeled R1881 and subtracted from the radioactivity for samples incubated in the absence of excess hormone.
Purification of Sse1p
Sse1p and the mutant and truncated versions of this protein were purified from yeast by using the galactose-inducible expression vectors described above. Yeast strain W303 was transformed with the appropriate plasmid, cells were grown overnight at 26°C in 1 liter of SC-ura containing 2% raffinose as the carbon source, washed, and then transferred to 4 liters of SC-ura containing 2% galactose. Cells were harvested in mid/late-log phase (OD600 of ∼2.5) and were converted to spheroplasts (Brodsky and Schekman, 1993). Spheroplasts were resuspended in 25 ml of ice-cold lysis buffer (50 mM HEPES pH 7.4, 300 mM NaCl, 10 mM imidazole, 5 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml each pepstatin A and leupeptin), an equal volume of glass beads was added, and the cells were agitated with four 1-min pulses on a Vortex mixer at the highest setting with cooling of the sample on ice between each pulse. The lysate was cleared by a 5-min centrifugation at 5,000 × g, followed by a 10-min centrifugation at 11,000 × g, and the final supernatant was applied to a 5-ml Ni2+-nitrilotriacetic acid-agarose column (QIAGEN). The column was washed with 5 volumes of lysis buffer and then sequentially with 5 volumes of lysis buffer containing each of the following: 1) 1% Triton, 2) 5 mM ATP, and 3) 1 M NaCl. Bound proteins were eluted with lysis buffer containing 500 mM imidazole. Peak fractions containing His6-tagged protein as assessed by SDS-PAGE and staining with Coomassie brilliant blue, and by immunoblot analysis using anti-Sse1p antibody were diluted 1:3 with buffer 88 (20 mM HEPES pH 6.8, 150 mM KOAc, 5 mM MgOAc, and 250 mM sorbitol) and applied to a 5-ml Q-Sepharose column (Amersham Biosciences). The column was washed with buffer 88 containing 0.4 M KOAc and eluted with a 12- × 12-ml gradient of buffer 88 containing 0.4 M KOAc to buffer 88 containing 2 M KOAc. Fractions containing the purified proteins as determined by SDS-PAGE and staining with Coomassie brilliant blue were dialyzed against 1000 volumes of dialysis buffer (50 mM Tris-Cl pH 7.4, 50 mM NaCl, 0.8 mM dithiothreitol, 2 mM MgCl2, and 5% vol/vol glycerol) overnight at 4°C, and the protein in the dialysate was concentrated in Centricon 30 microconcentrators (Millipore, Bedford, MA) following the manufacturer's instructions. Protein concentration was determined using the Bio-Rad Protein Assay reagent, with bovine serum albumin as the standard. The purity of each protein was estimated to be >95% and aliquots of the purified proteins were snap-frozen in liquid nitrogen and stored at −80°C.
In Vitro Luciferase Refolding and Aggregation Assays
The holdase activity of Sse1p was assessed both by following its ability to retain thermally denatured firefly luciferase in a folding competent conformation, which upon addition of cytosol and ATP resulted in active luciferase (Oh et al., 1997; Brodsky et al., 1999), and by its ability to temper the aggregation of thermally inactivated luciferase as assessed by spectroscopy (Oh et al., 1997). In brief, the folding assay was assembled by denaturing 150 nM of firefly luciferase (Sigma-Aldrich, St. Louis, MO) in luciferase assay buffer (25 mM HEPES pH 7.8, 5 mM MgOAc, 50 mM KCl, 5 mM β-mercaptoethanol, and 1 mM ATP) by heating to 42°C in the absence or presence of the indicated molar ratio of purified wild-type or mutant Sse1p for 30 min. Refolding was then assayed in an Analytical Luminescence Laboratory luminometer (BD PharMingen, San Diego, CA) with luciferin (BD PharMingen) as the substrate upon the addition of an ATP-regenerating system and cytosol at 26°C as published (Brodsky et al., 1999). Triplicate aliquots were removed at the indicated time points, and a parallel reaction was “mock denatured” at 26°C and the level of luciferase activity in this control reaction was set to 100%. To assay luciferase aggregation, a final concentration of 1.9 μM luciferase was preincubated in luciferase assay buffer or at a 1:5 molar ratio of Sse1p in buffer at room temperature for 15 min before it was diluted 12.5-fold into prewarmed (42°C) luciferase assay buffer and stirred. The absorbance was measured at 320 nm on a 14DS UV-VIS-IR spectrophotometer (AVIV, Lakewood, NJ) with a water-jacketed cuvette holder equilibrated to 42°C.
RESULTS
SSE1 Overexpression Suppresses the ydj1-151 Temperature-sensitive Growth Defect
To better understand how Ydj1p exerts its pleiotropic effects in the cell, we initiated a genetic screen to isolate multicopy suppressors of the ydj1-151 temperature-sensitive phenotype. As described in MATERIALS AND METHODS, three classes of complementing clones were isolated, each containing the SSE1 ORF; retransformation of only the SSE1 ORF on a multicopy plasmid also rescued ydj1-151 thermosensitivity (Figure 1B), as did the introduction of SSE1 on a CEN single copy vector (our unpublished data). In either case, the expression of Sse1p was approximately three- to fourfold greater than endogenous levels as determined using anti-Sse1p antiserum (our unpublished data). SSE1 encodes one of the two yeast Hsp110s, but only Sse1p is required for optimal cell growth at all temperatures (Mukai et al., 1993; Shirayama et al., 1993). Pertinent to this study, Sse1p has been found associated with the yeast Hsp90 complex (Caplan, 1999; Liu et al., 1999), and strains deleted for SSE1 are hypersensitive to Hsp90 inhibitors and defective for glucocorticoid receptor function (Liu et al., 1999).
To examine whether SSE1 and YDJ1 interact, we mated a strain lacking the Sse1p-encoding gene (“sse1Δ”) with ydj1-151 cells, selected for diploids, and then dissected haploid progeny after nitrogen starvation. We then compared the growth characteristics of an isogenic wild-type strain, sse1Δ and ydj1-151 cells, and two progeny containing both mutations (Figure 1C). At every temperature examined, a strong synthetic effect on the growth of sse1Δ ydj1-151 cells compared with the single mutant and wild-type strains was observed. Such synthetic interactions suggest that the YDJ1 and SSE1 gene products interact and/or that they function in similar pathways.
Effect of Sse1p Overexpression on Prepro-α Factor Translocation and v-Src Folding in ydj1-151 Yeast
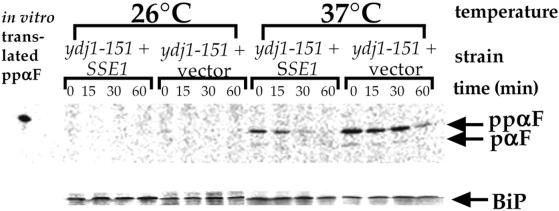
Previous studies showed that Ydj1p is important for the posttranslational translocation of the yeast mating prepheromone ppαF into the ER (Caplan et al., 1992). Thus, we analyzed whether overexpression of Sse1p in ydj1-151 yeast suppressed the ppαF translocation defect by using a pulse-chase and immunoprecipitation approach (see MATERIALS AND METHODS). A pulse-chase analysis was conducted in ydj1-151 cells containing either the Sse1p overexpression vector or a vector lacking insert at permissive (26°C) and nonpermissive (37°C) temperatures, and the fate of ppαF was followed. As shown in Figure 2, ppαF accumulated in the ydj1-151 strain shifted to 37°C regardless of whether or not Sse1p was overexpressed. However, the level of ppαF that accumulated was lowered two- to eightfold when the SSE1 encoding vector was present, suggesting a limited rescue of this phenotype. The defect in ppαF translocation in yeast containing a temperature-sensitive mutation in Ssa1p, ssa1-45 (Becker et al., 1996), was not rescued by overexpression of Sse1p (our unpublished data).
Figure 2.
SSE1 partially suppresses the ppαF translocation defect in ydj1-151 yeast. Ydj1-151 yeast containing either pJG816 or a vector lacking insert were labeled with [35S]methionine and chased for the indicated times at either 26 or 37°C, and ppαF and BiP were immunoprecipitated from cell extracts as described in MATERIALS AND METHODS. The positions of ppαF that migrates similarly to in vitro translated ppαF in the first lane and pαF are indicated. BiP serves as a control for the efficiencies of the radiolabeling and immunoprecipitation in this experiment. The decrease in ppαF intensity over time arises from translocation and intracellular degradation of the accumulated material.
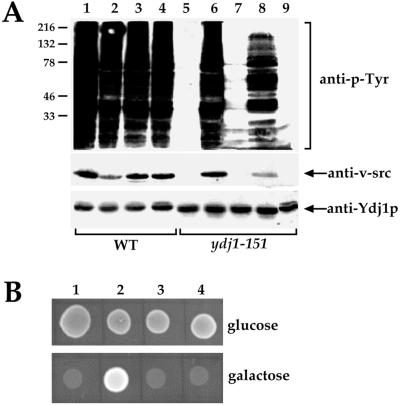
We also tested whether Sse1p overexpression rescued the v-Src folding defect in ydj1-151 yeast. Expression of v-Src protein leads to tyrosine phosphorylation of many yeast proteins, but this is completely abolished in the ydj1-151 and several other ydj1 mutants (Kimura et al., 1995; Dey et al., 1996). Because the endogenous levels of phosphotyrosine are low in yeast, v-Src activity can be measured by immunoblot analysis by using an anti-phosphotyrosine monoclonal antibody. In the absence of v-Src expression, this antibody appears unreactive when incubated with a whole cell lysate (Figure 3A, lane 9). In contrast, upon v-Src expression in wild-type cells, many yeast proteins become phosphorylated on tyrosine, resulting in the presence of multiple, phosphorylated substrates (Figure 3A, lanes 1–4). In the ydj1-151 strain, there is no visible phosphotyrosine activity and v-Src is undetectable (Figure 3A, lane 5). However, the introduction of SSE1-encoding plasmids completely restored both v-Src levels and the phosphotyrosine activity (Figure 3A, e.g. compare lanes 5 and 6). We conclude that Sse1p overexpression rescues the v-Src folding defect in the ydj1-151 mutant.
Figure 3.
SSE1 rescues v-Src activity in the ydj1-151 mutant. (A) Wild-type and ydj1-151 cells were transformed with a single copy vector (pRS314) either harboring (lanes 2 and 6) or lacking SSE1 (lanes 1 and 5), or a multicopy vector (pRS424) either harboring (lanes 4 and 8) or lacking SSE1 (lanes 3 and 7). All cells also contained a galactose-inducible v-Src expression vector, except those in lane 9, which contained only pRS424-SSE1. Cells were grown at 30°C, and extracts were prepared and analyzed for phosphotyrosine (anti-p-Tyr) and v-Src (anti-v-src) levels as described in MATERIALS AND METHODS. Ydj1p serves as a loading control. (B) ydj1-151 (lanes 1 and 2) or wild-type (lanes 3 and 4) yeast transformed with the pRS314-SSE1 expression vector (lanes 1 and 3) or the vector lacking the SSE1 insert (lanes 2 and 4) and containing the galactose-regulated v-Src expression vector were inoculated onto glucose- or galactose-containing medium and incubated at 30°C for 3 d.
We corroborated these data by examining the effect of v-Src expression on the viability of the ydj1-151 strain (Dey et al., 1996). Expression of v-Src in wild-type cells is lethal regardless of whether Sse1p is overexpressed (Figure 3B, lanes 3 and 4). In contrast, growth is apparent in ydj1-151 yeast expressing v-Src when the SSE1-encoding plasmid is lacking (Figure 3B, lane 2), but expression of v-Src is lethal when the SSE1-encoding plasmid is present (Figure 3B, lane 1), indicating further that overexpression of Sse1p rescues a ydj1-151 physiological defect.
Rescue of ydj1-151 Temperature Sensitivity Requires the ATP-binding Domain of Sse1p
The data presented above show that SSE1 rescues the Ydj1 requirement for v-Src activity but is less able to suppress a posttranslational translocation defect. We suggest that Sse1p substitutes for Ydj1p in a restricted manner but does not globally bypass Ydj1p function. Consistent with this view is the finding that SSE1 overexpression cannot suppress the temperature-sensitive lethal phenotype of a ydj1Δ mutant (Fewell and Brodsky, unpublished data). Because Sse1 is an Hsp70-like chaperone that interacts with Hsp90, the suppression of the ydj1-151 mutant defects may be restricted to Hsp90-dependent function (Figure 3). In one scenario, Ydj1-151p might become stabilized or functional in the Hsp90 complex only when multiple copies of Sse1p are present. Furthermore, by virtue of its homology to the Ssa1p hsp70, which interacts with Ydj1p in an ATP-dependent manner (Caplan et al., 1992; Cyr and Douglas, 1994; Becker et al., 1996; Lu and Cyr, 1998; McClellan and Brodsky, 2000), the ATP-binding domain of Sse1p may also be essential for ydj1-151 growth at elevated temperatures.
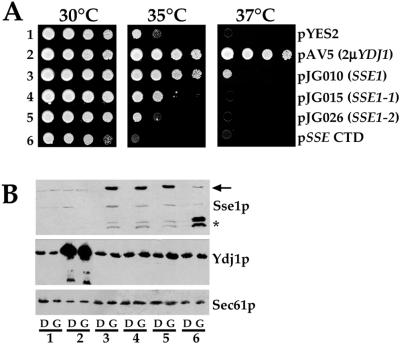
We have been unable to show that Sse1p exhibits ATPase activity (our unpublished data), although Sse1p and mammalian Hsp110 are predicted to possess the structural elements necessary for ATP binding. However, these motifs are poorly conserved with respect to Hsp70 and Grp170, and it remains unclear whether the ATP-binding domain is required for all Sse1p/Hsp110 activities (Chen et al., 1996; Oh et al., 1999; Easton et al., 2000). Thus, to examine whether the nucleotide-binding domain of Sse1p is important for the suppression of ydj1-151 thermosensitivity, we site-directed amino acid substitutions at conserved residues found in Sse1p (K69Q and G223D) and the yeast cytoplasmic and luminal Hsp70 molecular chaperones Ssa1p and BiP, respectively (Figure 4; Craven et al., 1997; Easton et al., 2000); strains expressing Ssa1p and BiP containing these mutations are defective for ATP hydrolysis and accumulate untranslocated ppαF, and expression of the BiP mutants is dominantly lethal (McClellan et al., 1998; McClellan and Brodsky, 2000). We also deleted the ATP-binding domain (residues 1–374) in Sse1p and introduced a new translation start site to express the C-terminal domain (CTD) of Sse1p. Galactose-inducible expression constructs either lacking or containing wild type or the Sse1p mutants were transformed into the ydj1-151 mutant, transformants were restruck, and overnight cultures were serially diluted onto selective medium containing galactose (Figure 5A). As anticipated, ydj1-151 yeast grew poorly at 37°C (Figure 5A, row 1) unless they contained a multicopy vector that constitutively expressed wild-type Ydj1p (Figure 5A, row 2). We found that cells containing the wild-type SSE1 construct, but not the sse1 mutant constructs, grew significantly better on galactose-containing medium at 35 and 37°C (Figure 5A, compare row 3 with rows 4–6). To verify that the wild-type and mutant proteins were expressed, total protein extracts were prepared from each strain and the levels of Sse1p, Ydj1p, and the ER membrane protein Sec61p (as a loading control) were determined by immunoblot analysis by using anti-Sse1p, anti-Ydj1p, and anti-Sec61p antisera, respectively. As displayed in Figure 5B, endogenous Sse1p was barely visible unless cells contained wild-type (lane 3) or the Sse1p mutants (lanes 4–6) on the galactose-inducible expression vector and were grown on galactose (G). In this experiment, the steady-state levels of Ydj1-151p in the cells appeared constant, regardless of whether wild-type or mutant Sse1p was expressed. In other experiments, expression of the Sse1p mutants reduced the steady-state levels of Ydj1-151p approximately twofold (our unpublished data; see DISCUSSION); as anticipated, significantly greater amounts of Ydj1p were present when a constitutive 2-μ (YEp24-based) Ydj1p-expression vector was present (Figure 5B, lane 2).
Figure 4.
Amino acid sequence alignment of the Kar2p, Ssa1p, and Sse1p molecular chaperones. Protein coding sequences were aligned using CLUSTALW (Thompson et al., 1994), and blackened and shaded residues correspond to positions containing at least two identities or two similarities, respectively. Dashes indicate gaps in the sequence alignment. The sse1-1 mutation is K69Q in the SSE1 numbering system, the sse1-2 mutation is G233D, and the Sse1p CTD starts at position 375.
Figure 5.
sse1 mutants are unable to rescue ydj1-151 thermosensitivity. (A) Cultures of ydj1-151 yeast transformed with a galactose-inducible expression vector lacking an insert (1), or containing the gene encoding wild-type Sse1p (3), Sse1-1p (4), Sse1-2p (5), or the Sse1p-CTD (6) were spotted onto SC-ura medium supplemented with galactose and incubated for 4 d at the indicated temperatures. Lane 2 contains ydj1-151 yeast with a multicopy, constitutive YDJ1 expression vector (pAV5). (B) Overnight cultures of each strain were grown in SC-ura supplemented with either glucose (D) or galactose (G) at 26°C, cell extracts were prepared, and immunoblots were analyzed using anti-Sse1p, anti-Ydj1p, or anti-Sec61p antiserum. The arrow indicates the migration of full-length Sse1p, and the asterisk denotes the positions of Sse1p CTD (upper band) and a degradation product (lower band). The numbers correspond to those in A.
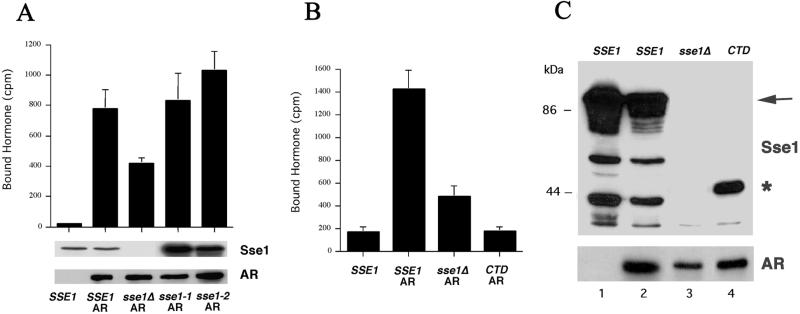
Although the Sse1-1 (K69Q), Sse1-2 (G233D), and CTD Sse1p mutant proteins were expressed in vivo, it was possible that they were inactive. To examine whether this was the case, we asked whether expression of the mutants in an sse1Δ background rescued the binding of a synthetic hormone to the androgen receptor heterologously expressed in yeast, because androgen receptor folding in yeast requires Hsp90 complex activity (Fang et al., 1996). First, we showed that wild-type cells lacking androgen receptor exhibited background levels of hormone binding (Figure 6, lane 1), whereas sse1Δ yeast expressing wild-type SSE1 bound significant amounts of hormone when the receptor was present (lane 2). In contrast, sse1Δ mutant cells containing a vector lacking the insert showed an approximate twofold decrease in hormone binding, but maximal binding was restored by the expression of the Sse1-1p and Sse1-2p mutants. We note that the steady-state levels of the Sse1p mutants in this experiment were higher than wild type; thus, it is possible that restoration of maximal hormone binding required greater amounts of the mutant than wild-type protein. Nevertheless, because the Sse1-1 and Sse1-2 mutant proteins were unable to rescue ydj1-151 thermosensitivity when the same expression system was used (Figure 5A), we conclude that the mutant proteins are at least partially active in vivo, but that this activity is insufficient to rescue ydj1-151 defects. In contrast, expression of the Sse1p CTD failed to restore hormone binding in sse1Δ cells (Figure 6B), and ydj1-151 yeast expressing the CTD fragment grew less well than ydj1-151 yeast containing an empty vector (Figure 5A), suggesting that this mutant may be partially dominant in these backgrounds. Overall, these data suggest that a wild-type ATP-binding domain of Sse1p is important for restoration of growth of ydj1-151 yeast at the nonpermissive temperature, but that the ATP-binding domain may not be required for Hsp90 complex function.
Figure 6.
Sse1p mutants restore hormone binding to sse1Δ cells expressing androgen receptor. (A) Wild-type (SSE1) or sse1Δ mutant cells either lacking or containing the androgen receptor expression vector (AR) and/or galactose-inducible expression vectors for wild-type Sse1p (SSE1), or the Sse1-1 (sse1-1), Sse1-2 (sse1-2), or Sse1 CTD (CTD) proteins, or the vector lacking an insert (sse1Δ) were assayed for binding to synthetic hormone (R1881) as described in MATERIALS AND METHODS. Top, data represent the means of three independent determinations, ± SD. Bottom, immunoblot analyses using anti-Sse1p and anti-androgen receptor antisera. (B) In a separate experiment, the binding of hormone to sse1Δ cells expressing wild-type SSE1 or the Sse1p CTD was examined. Here, the CTD exerted a negative effect on hormone binding (binding was lower than in yeast lacking Sse1p). (C) An immunoblot of extracts from the strains used in B probed with anti-Sse1p (top) and anti-AR (bottom) antisera. The arrow indicates the migration of full-length Sse1p, and the asterisk denotes the position of Sse1p CTD.
Sse1 Mutant Proteins Retain a Thermally Denatured Polypeptide in a Folding-competent Conformation
We previously demonstrated that purified, wild-type Sse1p “holds” thermally denatured firefly luciferase in a conformation that facilitates cytosol-dependent refolding (Brodsky et al., 1999). Others have found that both wild-type and the C-terminal halves of the mammalian Hsp110, which lacks the ATP-binding domain, exhibit holdase activity (Oh et al., 1997, 1999), suggesting that the ATP-binding domain is dispensable. To determine whether our Sse1 mutant proteins were similarly holdase proficient, galactose-inducible, hexahistidine-tagged forms of wild-type and the mutant Sse1 proteins were expressed in yeast and purified as described in the MATERIALS AND METHODS.
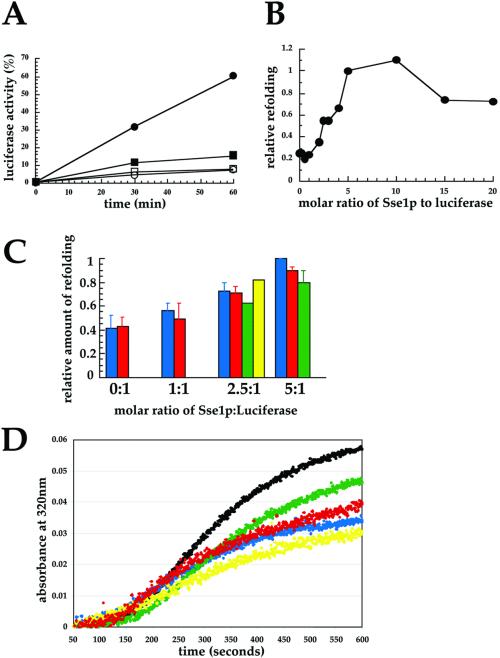
Although we had determined that Sse1p holds thermally denatured firefly luciferase in a folding-competent conformation (Brodsky et al., 1999), the ATP dependence of refolding and at what molar ratio maximal holdase activity was evident were not explored. As shown in Figure 7A, we found that addition of yeast cytosol and an ATP-regenerating system permitted ∼60% of the luciferase to refold that had first been denatured at 42°C in the presence of a 10-fold molar excess of Sse1p. In contrast, minimal refolding was apparent either if Sse1p or ATP was absent from the thermal denaturation or during the refolding reaction, respectively. The requirement for ATP is consistent with our previous data indicating that ssa1 mutant cytosol is defective for luciferase refolding (Brodsky et al., 1999) and that mammalian Hsc70 (an Ssa1p homolog) and Hdj-1 (a DnaJ homolog) in the presence of ATP are sufficient to refold Hsp110-held luciferase (Oh et al., 1997).
Figure 7.
Sse1p ATP-binding domain mutants are proficient for in vitro holdase activity. (A) In vitro Sse1p holdase activity was assayed as described in MATERIALS AND METHODS. ●, Reactions containing Sse1p at a 10:1 molar ratio of chaperone to luciferase during luciferase denaturation and chased with cytosol and ATP; ▪, reactions lacking Sse1p but chased with cytosol and ATP; ○, reactions containing Sse1p and chased with cytosol but no ATP; □, reactions lacking Sse1p and chased with cytosol but no ATP. Refolding activity (100%) represents the amount of luminescence when samples containing luciferase were mock denatured and then incubated with cytosol and ATP. (B) Reactions containing the indicated molar ratio of Sse1p to luciferase during thermal denaturation were chased with cytosol and ATP and luciferase refolding was assayed. The amount of refolding at a 5:1 ratio of Sse1p to luciferase was set to 1 in each experiment. Data represent the means of at least three independent experiments. (C) Relative amounts of refolding at the indicated molar ratios of wild-type Sse1p (blue), Sse1-1p (red), Sse1-2p (yellow), and the Sse1p CTD (green) to luciferase were assayed after the addition of cytosol and ATP. When error bars are present, the data represent the means of at least three independent experiments, ± SD. The amount of luciferase refolding in reactions containing a 5:1 M ratio of wild-type Sse1p to luciferase was always set to 1. (D) Luciferase aggregation over time was assayed upon thermal denaturation in the presence of a 5:1 M ratio of wild-type or the mutant Sse1p derivatives as described in MATERIALS AND METHODS. The black line indicates luciferase denatured in the absence of Sse1p, and the colored lines correspond to the protein preparations described in C.
Next, we titrated the molar ratio of Sse1p to luciferase during the 42°C denaturation before cytosol and ATP were added and found that maximal refolding occurred at an Sse1p:luciferase ratio of 5–10:1 (Figure 7B). We then compared the activities of the Sse1-1p (K69Q), Sse1-2p (G233D), CTD Sse1p mutant, to wild-type Sse1p in this assay. As shown in Figure 7C, we found that the activities of the mutant proteins were indistinguishable from wild-type Sse1p. Finally, we assayed whether wild-type Sse1p, Sse1-1p, Sse1-2p, and the CTD of Sse1p reduced firefly luciferase aggregation upon thermal denaturation. Addition of Sse1p to the cuvette at a 5:1 molar ratio to luciferase resulted in approximately a 41% reduction relative to a reaction lacking Sse1p, and the Sse1-1 and Sse1-2 mutant proteins yielded values of 35 and 48%, respectively; surprisingly, the Sse1p CTD was less efficient at preventing luciferase aggregation than the full-length proteins (17%; Figure 7D), even though it acted as an efficient holdase (Figure 7C). An explanation for this phenomenon is provided in DISCUSSION. As a negative control, we found that bovine serum albumin was without effect in this assay (our unpublished data). These combined data indicate that the ATP-binding domain of the yeast Hsp110 homolog is dispensable for in vitro holdase activity (Figure 7), although this domain is required to suppress ydj1-151 thermosensitivity (Figure 5). These data suggest that the rescue of Hsp90-dependent activity in the ydj1-151 mutant is more elaborate than simply increasing the cell's capacity to prevent protein aggregation.
DISCUSSION
We report herein that overexpression of Sse1p, the more abundant of two yeast Hsp110s, rescues the temperature-sensitive growth defect of the ydj1-151 mutant. One established biochemical function of the Hsp110s is that they exhibit holdase activity (Oh et al., 1997, 1999; Brodsky et al., 1999), i.e., they can retain a thermally or chemically denatured polypeptide in a refolding-competent conformation. Our in vitro and in vivo analyses of wild-type and mutant derivatives of yeast Sse1p suggest that holdase activity may be insufficient to restore growth of the ydj1-151 strain at elevated temperatures. This result was initially somewhat surprising as overexpression of Hsp110 in mammalian cells confers thermotolerance (Oh et al., 1997). However, we also show that overexpression of Sse1p rescues an Hsp90-mediated defect in the ydj1-151 strain (Figure 3). Because Hsp90 itself, and other components of the Hsp90 complex, exhibit holdase activity (Bose et al., 1996; Freeman and Morimoto, 1996; Freeman et al., 1996) and are required for the maturation and folding of many cellular substrates (reviewed in Caplan, 1999), our data suggest instead that Sse1p repairs a ydj1-151–induced defect in the Hsp90 complex. Consistent with this hypothesis, Sse1p associates with the Hsp90 complex, and sse1Δ cells exhibit defects ascribed to Hsp90-dependent phenomena (Liu et al., 1999; Figure 6). Furthermore, when we precipitated overexpressed wild-type or mutant Sse1p from transformed ydj1-151 cells, we found that the amount of coprecipitated Ydj1-151p was lower when the Sse1p mutant proteins were overexpressed (our unpublished data). Clearly, a more detailed mapping and examination of the physical interactions and communications between these and other chaperones associated with the Hsp90 complex is warranted.
In contrast to the pronounced rescue of v-Src activity upon Sse1p overexpression in the ydj1-151 mutant (Figure 3), the ability of SSE1 to rescue the ydj1-151 translocation defect was subtle (Figure 2). We suggest that this modest level of rescue is independent of Hsp90 action for the following reasons. First, Hsp90 mutant cells do not exhibit a defect in ppαF translocation (our unpublished data), and second, based on models for Hsp70/Ssa1p-promotion of posttranslational translocation (reviewed in Fewell et al., 2001), increasing the concentration and thus the extent of Sse1p-mediated holdase activity in the cell might help retain ppαF in a translocation-competent conformation. Because an Ssa1p-Ydj1p interaction is required for translocation (Caplan et al., 1992; Becker et al., 1996; McClellan and Brodsky, 2000), and because overexpression of Sse1p fails to rescue the ppαF translocation defect in an ssa1 mutant, it will be interesting in the future to explore whether Sse1p affects the interaction of Ssa1p and/or Ydj1p with posttranslationally translocated precursor proteins (Chirico, 1992).
Sse1p is one of two hsp110s in yeast, and it is curious that we only uncovered SSE1 as a multicopy suppressor of ydj1-151. One trivial explanation for this result is that the library lacked SSE2, which may result from the fact that SSE2 mRNAs are rare, although the transcription of SSE2 is induced upon heat shock (Mukai et al., 1993; Shirayama et al., 1993). However, it is important to note that cells deleted for SSE2 grow as well as wild-type yeast, even at high temperature, but that the sse1Δ strain grows poorly. This observation may arise from the fact that Sse1p, but not Sse2p, interacts with the Hsp90 complex (Liu et al., 1999), and is consistent with the isolation of only SSE1 from the screen described in this study.
When examining the ability of the Sse1p mutants to exhibit in vitro holdase activity and the suppression of luciferase aggregation (Figure 7) we found that the CTD of Sse1p, although being almost as active as wild-type Sse1p for its ability to retain luciferase in a folding-competent conformation, exhibited a reduced ability to prevent luciferase aggregation during thermal denaturation. One explanation for these data is that not all luciferase “aggregates” that form during thermal denaturation will be unable to fold upon the subsequent addition of cytosol and ATP, and indeed, we observe some luciferase refolding in reactions lacking Sse1p. Another possibility is that the aggregates observed in the spectrophotometer represent both luciferase and the Sse1p-CTD, which may be aggregation prone, but the luciferase “held” by the CTD can still refold when cytosol and ATP are present; this scenario is also consistent with our observations that the Sse1p CTD acts as a dominant negative mutant for growth and for restoration of hormone binding (see RESULTS). More generally, the data presented in this report caution that the use of in vitro assays comparing wild-type and mutant chaperones might neglect more subtle, genetic consequences of the mutants in the crowded cellular milieu. This issue is particularly important since most chaperones exist in multimeric protein complexes, such as the Hsp90 complex.
ACKNOWLEDGMENTS
We thank Dr. S. Fewell for comments on the manuscript and for the construction of the CEN-SSE1 plasmid; Drs. S. Lindquist, J. Woolford, J. Subjeck, D. Easton, H. Oh, K. Morano, and D. Thiele for generously supplying reagents and advice; and Dr. A. Toh-e for yeast strains. We are also grateful to Dr. R. Duda for help with the AVIV spectrophotometer and to David Hornack for excellent technical assistance. This work was supported by grant MCB-9904575 from the National Science Foundation (to J.L.B.) and the Irma T. Hirschl Career Science Award (to A.J.C.).
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.02–04–0051. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.02–04–0051.
REFERENCES
- Atencio DP, Yaffe MP. MAS5, a yeast homolog of DnaJ involved in mitochondrial protein import. Mol Cell Biol. 1992;12:283–291. doi: 10.1128/mcb.12.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Walter W, Yan W, Craig EA. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S, Weikl T, Bugl H, Buchner J. Chaperone function of Hsp90-associated proteins. Science. 1996;274:1715–1717. doi: 10.1126/science.274.5293.1715. [DOI] [PubMed] [Google Scholar]
- Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Lawrence JG, Caplan AJ. Mutations in the cytosolic DnaJ homologue, YDJ1, delay and compromise the efficient translation of heterologous proteins in yeast. Biochemistry. 1998;37:18045–18055. doi: 10.1021/bi980900g. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Schekman R. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Werner ED, Dubas ME, Goeckeler JL, Kruse KB, McCracken AA. The requirement for molecular chaperones during endoplasmic reticulum- associated protein degradation demonstrates that protein export and import are mechanistically distinct. J Biol Chem. 1999;274:3453–3460. doi: 10.1074/jbc.274.6.3453. [DOI] [PubMed] [Google Scholar]
- Caplan AJ. Hsp90's secrets unfold: new insights from structural and functional studies. Trends Cell Biol. 1999;9:262–268. doi: 10.1016/s0962-8924(99)01580-9. [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Cyr DM, Douglas MG. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992;71:1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Douglas MG. Characterization of YDJ1: a yeast homologue of the bacterial dnaJ protein. J Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan AJ, Langley E, Wilson EM, Vidal J. Hormone-dependent transactivation by the human androgen receptor is regulated by a DnaJ protein. J Biol Chem. 1995;270:5251–5257. doi: 10.1074/jbc.270.10.5251. [DOI] [PubMed] [Google Scholar]
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Chang HC, Nathan DF, Lindquist S. In vivo analysis of the Hsp90 cochaperone Sti1 (p60) Mol Cell Biol. 1997;17:318–325. doi: 10.1128/mcb.17.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Easton D, Oh H, Lee-Yoon D, Liu X, Subjeck J. The 170 kDA glucose regulated stress protein is a large HSP70-, HSP110-like protein of the endoplasmic reticulum. FEBS Lett. 1996;380:68–72. doi: 10.1016/0014-5793(96)00011-7. [DOI] [PubMed] [Google Scholar]
- Chirico WJ. Dissociation of complexes between 70 kDa stress proteins and presecretory proteins is facilitated by a cytosolic factor. Biochem Biophys Res Commun. 1992;189:1150–1156. doi: 10.1016/0006-291x(92)92324-q. [DOI] [PubMed] [Google Scholar]
- Craven RA, Tyson JR, Stirling CJ. A novel subfamily of Hsp70 in the endoplasmic reticulum. Trends Cell Biol. 1997;7:277–282. doi: 10.1016/S0962-8924(97)01079-9. [DOI] [PubMed] [Google Scholar]
- Cyr DM. Cooperation of the molecular chaperone Ydj1 with specific Hsp70 homologs to suppress protein aggregation. FEBS Lett. 1995;359:129–132. doi: 10.1016/0014-5793(95)00024-4. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Douglas MG. Differential regulation of Hsp70 subfamilies by the eukaryotic DnaJ homologue YDJ1. J Biol Chem. 1994;269:9798–9804. [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R. A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J Cell Biol. 1987;105:633–645. doi: 10.1083/jcb.105.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B, Caplan AJ, Boschelli F. The Ydj1 molecular chaperone facilitates formation of active p60v-src in yeast. Mol Biol Cell. 1996;7:91–100. doi: 10.1091/mbc.7.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DP, Kaneko Y, Subjeck JR. The hsp110 and Grp1 70 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 2000;5:276–290. doi: 10.1379/1466-1268(2000)005<0276:thagsp>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Fliss AE, Robins DM, Caplan AJ. Hsp90 regulates androgen receptor hormone binding affinity in vivo. J Biol Chem. 1996;271:28697–28702. doi: 10.1074/jbc.271.45.28697. [DOI] [PubMed] [Google Scholar]
- Fewell SW, Travers KJ, Weissman JS, Brodsky JL. The action of molecular chaperones in the early secretory pathway. Annu Rev Genet. 2001;35:149–191. doi: 10.1146/annurev.genet.35.102401.090313. [DOI] [PubMed] [Google Scholar]
- Freeman BC, Morimoto RI. The human cytosolic molecular chaperones hsp90, hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 1996;15:2969–2979. [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Toft DO, Morimoto RI. Molecular chaperone machines: chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor-associated protein p23. Science. 1996;274:1718–1720. doi: 10.1126/science.274.5293.1718. [DOI] [PubMed] [Google Scholar]
- Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- Hernández MP, Chaldi A, Toft DO. hsp40 binding is the first step in the hsp90 chaperoning pathway for the progesterone receptor. J Biol Chem. 2002;277:11873–11881. doi: 10.1074/jbc.M111445200. [DOI] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kelley WL. The J-domain family and the recruitment of chaperone power. Trends Biochem Sci. 1998;23:222–227. doi: 10.1016/s0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Yahara I, Lindquist S. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- Kosano H, Stensgard B, Charlesworth MC, McMahon N, Toft D. The assembly of progesterone receptor-hsp90 complexes using purified proteins. J Biol Chem. 1998;273:32973–32979. doi: 10.1074/jbc.273.49.32973. [DOI] [PubMed] [Google Scholar]
- Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- Liu XD, Morano KA, Thiele DJ. The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J Biol Chem. 1999;274:26654–26660. doi: 10.1074/jbc.274.38.26654. [DOI] [PubMed] [Google Scholar]
- Lu Z, Cyr DM. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- Manning-Krieg UC, Scherer PE, Schatz G. Sequential action of mitochondrial chaperones in protein import into the matrix. EMBO J. 1991;10:3273–3280. doi: 10.1002/j.1460-2075.1991.tb04891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan AJ, Brodsky JL. Mutation of the ATP-binding pocket of SSA1 indicates that a functional interaction between Ssa1p and Ydj1p is required for post-translational translocation into the yeast endoplasmic reticulum. Genetics. 2000;156:501–512. doi: 10.1093/genetics/156.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan AJ, Endres JB, Vogel JP, Palazzi D, Rose MD, Brodsky JL. Specific molecular chaperone interactions and an ATP-dependent conformational change are required during posttranslational protein translocation into the yeast ER. Mol Biol Cell. 1998;9:3533–3545. doi: 10.1091/mbc.9.12.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow MW, Brodsky JL. Yeast ribosomes bind to highly purified reconstituted Sec61p complex and to mammalian p180. Traffic. 2001;2:705–716. doi: 10.1034/j.1600-0854.2001.21005.x. [DOI] [PubMed] [Google Scholar]
- Mukai H, Kuno T, Tanaka H, Hirata D, Miyakawa T, Tanaka C. Isolation and characterization of SSE1 and SSE2, new members of the yeast HSP70 multigene family. Gene. 1993;132:57–66. doi: 10.1016/0378-1119(93)90514-4. [DOI] [PubMed] [Google Scholar]
- Nathan DF, Lindquist S. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh HJ, Chen X, Subjeck JR. Hsp110 protects heat-denatured proteins and confers cellular thermoresistance. J Biol Chem. 1997;272:31636–31640. doi: 10.1074/jbc.272.50.31636. [DOI] [PubMed] [Google Scholar]
- Oh HJ, Easton D, Murawski M, Kaneko Y, Subjeck JR. The chaperoning activity of hsp110. Identification of functional domains by use of targeted deletions. J Biol Chem. 1999;274:15712–15718. doi: 10.1074/jbc.274.22.15712. [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Matlack KE, Plath K, Misselwitz B, Staeck O. Posttranslational protein translocation across the membrane of the endoplasmic reticulum. Biol Chem. 1999;380:1143–1150. doi: 10.1515/BC.1999.145. [DOI] [PubMed] [Google Scholar]
- Rassow J, Pfanner N. Protein biogenesis: chaperones for nascent polypeptides. Curr Biol. 1996;6:115–118. doi: 10.1016/s0960-9822(02)00437-2. [DOI] [PubMed] [Google Scholar]
- Rothblatt JA, Deshaies RJ, Sanders SL, Daum G, Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J Cell Biol. 1989;109:2641–2652. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Kawakami K, Matsui Y, Tanaka K, Toh-e A. MSI3, a multicopy suppressor of mutants hyperactivated in the RAS-cAMP pathway, encodes a novel HSP70 protein of Saccharomyces cerevisiae. Mol Gen Genet. 1993;240:323–332. doi: 10.1007/BF00280382. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling CJ, Rothblatt J, Hosobuchi M, Deshaies R, Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol Biol Cell. 1992;3:129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- Yaglom JA, Goldberg AL, Finley D, Sherman MY. The molecular chaperone Ydj1 is required for the p34CDC28-dependent phosphorylation of the cyclin Cln3 that signals its degradation. Mol Cell Biol. 1996;16:3679–3684. doi: 10.1128/mcb.16.7.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]