Abstract
Nuclear bodies represent a heterogeneous class of nuclear structures. Herein, we describe that a subset of nuclear bodies is highly enriched in components of the ubiquitin–proteasome pathway of proteolysis. We coined the term clastosome (from the Greek klastos, broken and soma, body) to refer to this type of nuclear body. Clastosomes contain a high concentration of 1) ubiquitin conjugates, 2) the proteolytically active 20S core and the 19S regulatory complexes of the 26S proteasome, and 3) protein substrates of the proteasome. Although detected in a variety of cell types, clastosomes are scarce under normal conditions; however, they become more abundant when proteasomal activity is stimulated. In contrast, clastosomes disappear when cells are treated with proteasome inhibitors. Protein substrates of the proteasome that are found concentrated in clastosomes include the short-lived transcription factors c-Fos and c-Jun, adenovirus E1A proteins, and the PML protein. We propose that clastosomes are sites where proteolysis of a variety of protein substrates is taking place.
INTRODUCTION
The proteasome is a large proteolytic complex involved in regulated degradation of intracellular proteins (Coux et al., 1996; Baumeister et al., 1998; Voges et al., 1999). Short-lived proteins such as cell cycle regulators and transcription factors are specifically targeted for proteasome degradation by the ubiquitin-conjugation pathway. Other substrates for this pathway include abnormal soluble proteins from the nucleus and the cytosol, and proteins of the endoplasmic reticulum that have been retro-translocated to the cytosol (Bonifacino and Weissman, 1998). Most protein substrates of the proteasome are marked by covalent ligation to ubiquitin, which then acts as a signal to target the modified substrate for proteolytic degradation.
The ubiquitin signal is generated by a series of dedicated enzymes that recognize the target proteins (Hershko and Ciechanover, 1998). Conjugation of ubiquitin involves the formation of an isopeptide bond between the C-terminal glycine residue of ubiquitin and the ε-amino group of a lysine residue of the target protein. Ubiquitination begins with an activating enzyme, E1, that hydrolyses ATP and forms a high-energy thioester between a cysteine of its active site and the C terminus of ubiquitin. The ligation of ubiquitin to the substrate is then carried out by a complex composed of a ubiquitin-protein ligase (E3) and a ubiquitin-conjugating enzyme (E2), with E3 being the major determinant of substrate specificity. Conjugated ubiquitin can be a substrate for further ubiquitination reactions and, indeed, most substrates are modified by multiubiquitin chains in which single ubiquitin molecules are linked via isopeptide bonds (Thrower et al., 2000).
The proteasome is a 26S multimeric enzyme composed of two subcomplexes, the 20S proteasome and the 19S regulator (Bochtler et al., 1999; Voges et al., 1999; Zwickl et al., 2000). The 20S proteasome forms the proteolytic core, whereas the 19S regulator (or PA700) confers ATP dependency and ubiquitinated substrate specificity on the enzyme. Current models propose a multistep mechanism for protein degradation by the proteasome. The substrate is initially tethered to the proteasome by specific recognition of its ubiquitin tag; this is followed by interaction of the substrate with the regulatory particle, which exhibits chaperone-like activity and promotes substrate unfolding (Braun et al., 1999); the unfolded substrate is then translocated through a narrow channel to gain access to the proteolytic active sites of the enzyme located within the lumen of the core particle.
Although it has been traditionally thought that multiubiquitin chains are virtually essential to target protein substrates for degradation by the eukaryotic 26S proteasome, recent work unraveled a class of proteins that can be degraded by the proteasome in vivo independently of ubiquitin (Verma and Deshaies, 2000). These include ornithine decarboxylase and the cyclin-dependent kinase inhibitor p21Cip1. More recently, the PML protein was shown to be degraded by the proteasome (Zhu et al., 2001), and whether this process depends on ubiquitination remains an open question.
Immunolocalization and biochemical fractionation studies showed that proteasomes are localized both in the nucleus and in the cytoplasm of eukaryotic cells (Brooks et al., 2000, and references therein). In living yeast cells, 26S proteasomes accumulate predominantly in the nuclear envelope-endoplasmic reticulum network (Enenkel et al., 1998; Wilkinson et al., 1998), and recent data suggest that proteasome localization at these sites is important for proper function (Tatebe and Yanagida, 2000). In contrast, green fluorescent protein-tagged proteasomes appear uniformly distributed throughout the cytoplasm and the nucleoplasm in live human cells (Reits et al., 1997). Within these two compartments proteasomes diffuse rapidly, suggesting that recognition and degradation of substrates result from random collisions between the proteasome and intracellular proteins (Reits et al., 1997). Contrasting with this view, dividing cells show a preferential concentration of proteasomes around spindle microtubules (Amsterdam et al., 1993). This pattern is reminiscent of the behavior of cyclin B, which is degraded by the ubiquitin–proteasome pathway and disappears after a wave that starts at the spindle poles and spreads to the spindle equator (Huang and Raff, 1999). This immediately raises the question of whether association of proteasomes with the spindle contributes to the spatially regulated disappearance of cyclin B at the end of mitosis.
During interphase, proteasomes are responsible for the selective degradation of a number of short-lived nuclear transcription factors whose activity must be tightly regulated, such as nuclear factor-κB, STAT1, and Fos/Jun (Ciechanover et al., 1991; Palombella et al., 1994; Kim and Maniatis, 1996). In face of the increasing evidence showing that compartmentalization of regulatory factors by sequestration plays a key role in nuclear function, we decided to investigate the subnuclear localization of the 26S proteasome. Our results show that although proteasomes are most often diffusely distributed throughout the nucleoplasm, they occasionally concentrate in discrete structures. Electron microscopic analysis revealed that these structures correspond to complex doughnut-shaped nuclear bodies. In addition to 19S and 20S proteasomes, the bodies contain ubiquitin, and a number of proteasome substrates, including the adenoviral protein E1A proteins, c-Fos and c-Jun, and PML. The results further show that proteasome-containing nuclear bodies are dynamic structures, which assemble transiently under conditions of high proteolysis in the nucleus. We propose the name clastosome (from the Greek klastos, broken and soma, body) to refer to this nuclear domain.
MATERIALS AND METHODS
Cell Culture
HeLa cells (human epitheloid carcinoma, cervix), and WI-38 cells (human female lung diploid fibroblasts) were grown as monolayers in minimum essential medium supplemented with 2 mM l-glutamine, 50 IU/ml penicillin, 50 mg/ml streptomycin, and 10% fetal calf serum (Invitrogen, Carlsbad, CA). Primary cultures of Schwann cells were prepared as described by Brockes et al. (1979). Briefly, sciatic nerves were dissected from 3-d-old Sprague-Dawley rats, and mixed cultures of Schwann cells and endoneurial fibroblasts were maintained in minimum essential medium supplemented with 10% fetal bovine serum for 1 d. To reduce fibroblast contamination, the culture was then incubated for 2 d in the presence of 10−5 M cytosine arabinose. On the 4th d the cells were transferred to polylysine-coated coverslips and maintained for further 3 d in the presence of 10% fetal bovine serum, 2 μM forskolin, and 20 μg/ml bovine pituitary extract.
Animals
Male, 3-mo-old rats of the Sprague-Dawley strain were kept on a 12-h day, 12-h night lighting regime with lights out at 8:00 AM. All animals were sacrificed by overdose of pentobarbital. To induce an osmotic stress, animals were given a single intraperitoneal injection of 1.5 M NaCl (18 ml/kg), as described previously (Lafarga et al., 1998).
Immunofluorescence
For indirect immunofluorescence, cultured cells were grown on 10- × 10-mm glass coverslips. The cells were washed twice in phosphate-buffered saline (PBS), fixed with 3.7% formaldehyde (freshly prepared from paraformaldehyde) in PBS for 10 min at room temperature, and subsequently permeabilized with 0.5% Triton X-100 in PBS for 20 min at room temperature. Rats were perfused through the ascending aorta with 3.7% formaldehyde in PBS, pH 7.4, for 15 min at room temperature. Supraoptic nuclei were dissected out of 500-μm-thick hypothalamic slices. These tissue fragments were washed in PBS for 1 h and individually transferred to a drop of PBS on a siliconized slide. Then, a coverslip was applied on top of the slide and the tissue was squashed by percussion with a histological needle. The preparation was then frozen in dry ice, and the coverslip was removed using a razor blade. The slides with adhered neurons were sequentially dehydrated in 96 and 70% ethanol at 4°C for 10 min and rinsed in PBS. Before immunostaining, the samples were sequentially incubated with 0.5% Triton X-100 in PBS for 10 min, 0.1 M glycine in PBS for 30 min, and 0.01% Tween 20 in PBS for 5 min. For immunolabeling, cells were rinsed in PBS containing 0.05% Tween 20, incubated for 1 h with primary antibodies diluted in PBS, washed in PBS containing 0.05% Tween 20, and incubated for 45 min with the appropriate secondary antibodies conjugated to fluorescein (fluorescein isothiocyanate), or Texas Red (Jackson Immunoresearch Laboratories, West Grove, PA). Finally, the coverslips were mounted in VectaShield (Vector Laboratories, Peterborough, United Kingdom) and sealed with nail polish. Confocal microscopy was performed with either a laser scanning microscope (LSM 510; Carl Zeiss, Göttingen, Germany) or an MRC-1024 (Bio-Rad, Hercules, CA) by using excitation wavelengths of 488 nm (for fluorescein isothiocyanate) and 543 nm (for Texas Red). Each channel was recorded independently, and pseudocolor images were generated and superimposed.
Immunoelectron Microscopy
Immunoelectron microscopy was performed on cultured Schwann cells or tissue fragments from rat sciatic nerve. Samples were fixed with 4% formaldehyde (freshly prepared from paraformaldehyde) in 0.12 M phosphate buffer, pH 7.4, for 20 min at room temperature, rinsed in phosphate buffer, dehydrated in increasing concentrations of methanol, and embedded in Lowicryl K4 M at −20°C. Ultrathin sections on gold grids were sequentially incubated with 0.1 M glycine in PBS (15 min), 3% bovine serum albumin (BSA) in PBS (15 min), and finally the primary antibody diluted in 1% BSA, 0.1 M glycin in PBS (1 h at room temperature). After washing, the sections were incubated with appropriate secondary antibody conjugated to either 5- or 10-nm gold particles (BioCell, Cardiff, United Kingdom) diluted 1:25 in 0.1% BSA in PBS (45 min at room temperature). Finally, the sections were washed and stained with 10% aqueous uranyl acetate. As controls, sections were treated as described but omitting the primary antibody. Ultrathin sections were examined with an EM208 electron microscope (Philips, Eindhoven, the Netherlands) operated at 60 kV.
Antibodies
Proteasomes were detected using polyclonal antibodies directed against the 20S catalytic core (Arribas et al., 1994; Menqual et al., 1996), and the 19S regulator ATPase subunit 6b (Tbp7; Affiniti Research Products, Exeter, United Kingdom). Ubiquitin-conjugates were labeled with antibody Z 0458 from DAKO (Glostrup, Denmark), and PML was detected with monoclonal antibody (mAb) PG-M3 from Santa Cruz Biotechology (Santa Cruz, CA). Additionally, the following antibodies were used: mAb 3A3 directed against Hsp70/Hsc70 (MA3-006; Affinity Bioreagents, Golden, CO); mAb M73 raised against E1A proteins (Harlow et al., 1985); mAb 2G9C3 directed against c-Fos (Oncogene Science, Cambridge, MA); mAb anti-c-Jun (J31920; Transduction Laboratories, Lexington, KY); polyclonal antibodies anti-c-Jun (PC06; Oncogene Science); mAb 4G3 (Euro Diagnostica, The Netherlands) directed against the U2B" snRNP protein (Habets et al., 1989); rabbit polyclonal serum 204.3 anti-coilin (Bohmann et al., 1995); mAb 2B1 anti-SMN (Liu and Dreyfuss, 1996); and rabbit polyclonal antibody C-20 (sc-333; Santa Cruz Biotechnology) directed against Sam68 (Chen et al., 1999). Specificity of antibodies used was confirmed by Western blotting.
Proteolysis Inhibitors
HeLa cells were exposed for 3 h to the proteasomal inhibitors MG132 (50 μM; Calbiochem, San Diego, CA), lactacystin (10 μM; Calbiochem), or the lysosomal protease inhibitor E64 (10 μM; Sigma-Aldrich, St. Louis, MO). Each of these compounds was dissolved in dimethyl sulfoxide (DMSO), and the final concentration of DMSO in the culture medium was 0.25%.
Plasmids and Transfections
Plasmid pE1A contains nucleotides 1–1773 of genomic Ad2 sequences (pML 005 in Bondesson et al., 1994). Plasmids encoding human c-Jun (pMT35), human c-Jun with a deletion of the δ domain (pMT140), and chicken v-Jun (pMT59) were described previously (Treier et al., 1994). HeLa cells were transiently transfected using FuGene 6 (Roche Applied Science, Indianapolis, IN), according to the manufacturer's instructions.
RESULTS
Mammalian Cell Nuclei Contain Domains Enriched in Proteasomes, Ubiquitin, and Short-lived Transcription Factors
To study the subcellular distribution of proteasomes in mammalian cells, immunofluorescence was performed using polyclonal antibodies specific for the 20S core catalytic component of the 26S proteasome. Characterization of these antibodies was described previously (Arribas et al., 1994; Mengual et al., 1996).
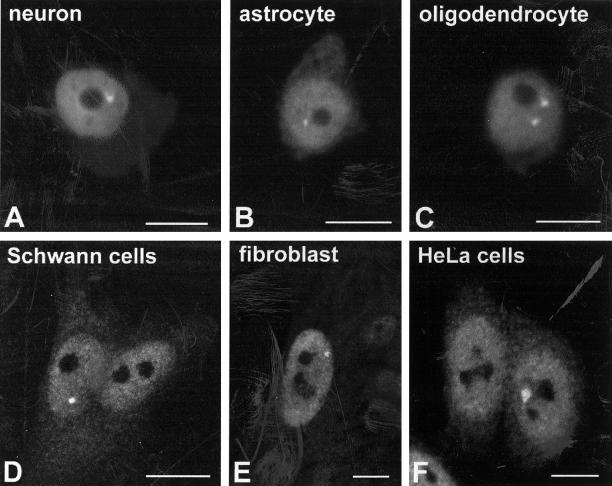
In agreement with other studies, anti-20S antibodies label both the nucleus and the cytoplasm with a diffuse pattern. Similar results were observed in a variety of cell types, including neurons, astrocytes, and oligodendrocytes from rat brain, primary cultured rat Schwann cells, primary human fibroblasts, and HeLa cells. Surprisingly, a fraction of all cellular populations analyzed contained in addition at least one brightly labeled discrete structure in the nucleoplasm (Figure 1). These structures are heterogeneous in size and shape, and although they are occasionally observed at the periphery of the nucleolus, they appear randomly located in the interior of the nucleus.
Figure 1.
Mammalian cell nuclei contain domains highly enriched in proteasomes. Antibodies directed against the 20S catalytic subunit of the proteasome were used to perform immunofluorescence on a variety of mammalian cell types. These included neurosecretory neurons of the supraoptic nucleus (A), astrocytes (B), and oligodendrocytes (C) obtained from squash preparations of rat hypothalamus, and primary cultures of Schwann cells prepared from rat sciatic nerve (D), human fibroblasts WI-38 (E), and HeLa cells (F). Although anti-20S proteasome antibodies label both nucleus and cytoplasm, staining of the nucleoplasm is more intense. Note the presence of brightly labeled intranuclear structures, of heterogeneous size and shape. Bar, 10 μm.
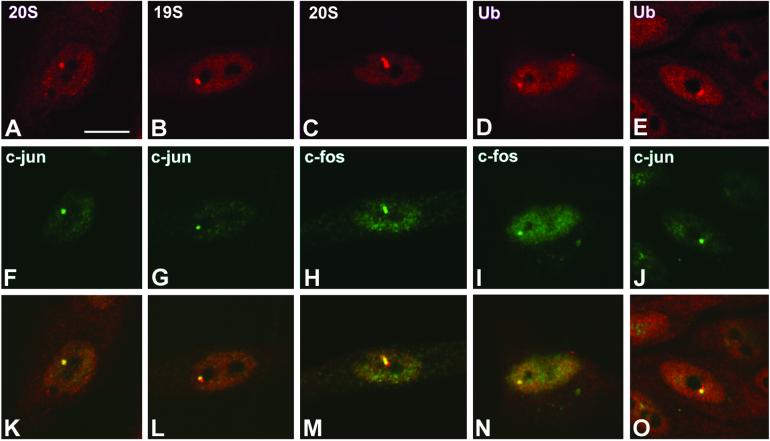
Double-labeling experiments revealed that 20S, 19S proteasomes and ubiquitin-conjugates colocalize in the brightly labeled nuclear structures (Figure 2). In addition, both c-Jun and c-Fos (two short-lived transcription factors degraded by the ubiquitin–proteasome pathway) accumulate in the proteasome-enriched structures. Proteins that concentrate in discrete nuclear domains but do not colocalize with the proteasome-enriched structures include coilin (Gall, 2000), splicing factors (Misteli, 2000), SMN (Liu and Dreyfuss, 1996), and Sam68 (Chen et al., 1999) (our unpublished data).
Figure 2.
Ubiquitin and proteins degraded by the proteasome colocalize in proteasome-domains. Primary Schwann cells were double-labeled with antibodies directed against 20S proteasome and c-Jun (A and F), 19S proteasome and c-Jun (B and G), 20S proteasome and c-Fos (C and H), ubiquitin and c-fos (D and I), and ubiquitin and c-Jun (E and J). (K–O) Overlay of red and green images. Overlapping of red and green staining produces a yellowish color. Bar, 10 μm.
Proteasome-enriched Nuclear Domains Are Dynamic Structures
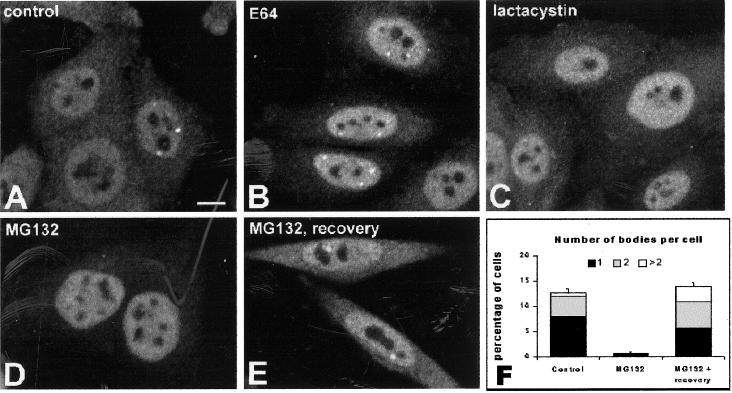
Proteasomal function can be inhibited pharmacologically by covalent or noncovalent modification of its β-subunits. Commonly used proteasome inhibitors include peptide aldehydes and lactacystin. Peptide aldehydes act by forming a reversible hemiacetal adduct with Thr1 on catalytically active β-subunits (Rock et al., 1994; Löwe et al., 1995; Seemüller et al., 1995), whereas the natural product lactacystin irreversibly alkylates Thr1 (Omura et al., 1991; Fenteany et al., 1995; Dick et al., 1996; Groll et al., 1997). To investigate how proteasome inhibitors affect proteasome-enriched nuclear domains, HeLa cells were treated for 12 h with either the peptide aldehyde MG132; lactacystin; a protease inhibitor specific for lysosomal cysteine proteases (E64); or the solvent in which the inhibitors were dissolved (DMSO). Treated cells were fixed and immunolabeled with anti-20S proteasome polyclonal antibodies (Figure 3, A–E). The proportion of cells containing proteasome-enriched domains (or bodies), and the number of bodies present per nucleus was then estimated (Figure 3F).
Figure 3.
Proteasome-enriched domains disappear in the presence of proteasome inhibitors. HeLa cells were either mock treated (A) or incubated for 3 h in the presence of E64 (B), lactacystin (C), and MG132 (D). The cells depicted in E were allowed to recover for 12 h after treatment with MG132. All cells were immunolabeled with antibodies directed against the 20S catalytic core of the proteasome. Bar, 10 μm. (F) Proportion of cells containing proteasome-enriched domains and the number of bodies per cell were estimated. After treatment with E64, the proportion of cells that contain bodies does not significantly differ from the control population. The graph depicts means ± SEs (three separate experiments were performed and a total of 300 cells analyzed for each drug treatment).
Incubation with MG132 caused a dramatic disappearance of proteasome-enriched bodies, compared with control cells treated with either solvent only or E64 (a protease inhibitor that does not interfere with proteasome activity). Because MG132 causes a reversible inhibition of the proteasome, cells treated with this peptide for 12 h were washed and allowed to recover for 12 h. After this recovery period, the proportion of cells containing at least one body was similar to that observed in control cells, but the number of bodies per nucleus was higher. Thus, inhibition of proteasomal function induces the disassembly of proteasome-containing nuclear domains, whereas withdrawal of the inhibition leads to reassembly of these structures. The higher number of bodies observed after inhibitor withdrawal may be the result of increased proteasomal activity on proteins targeted for degradation that accumulated during the period of treatment with the inhibitor.
If the assembly of proteasome-enriched bodies depends on proteasomal activity, it is expected that increasing the cellular levels of proteasome substrates should stimulate the formation of these structures. We therefore thought to estimate the number of bodies in cells under conditions that are known to trigger proteasome-dependent protein degradation.
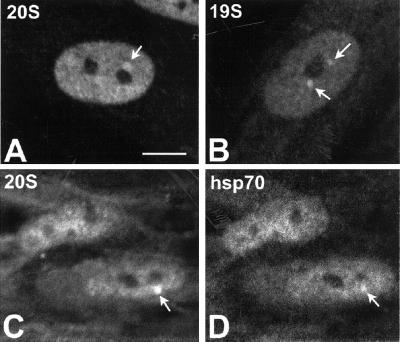
Transient expression of immediate-early genes such as c-fos is highly inducible as a result of a specific response to growth factors or serum (Bravo et al., 1986). This prompted us to investigate the distribution of proteasomes in response to serum stimulation of cultured cells. Primary human fibroblasts (WI38) were cultured in the presence of 0.1% fetal calf serum for 4 d, transferred to fresh medium containing 20% serum, and analyzed 3 h later. In clear contrast with unstimulated cells, which were largely devoid of proteasome-containing bodies (our unpublished data), cells incubated with serum for 3 h contained one or more bodies (Figure 4, A–D). As depicted in Figure 4, C and D, these nuclear bodies are also labeled by an mAb (3A3) directed against Hsp70 and Hsc70. Several lines of evidence indicate that the molecular chaperones Hsp70 and Hsc70 interact with the proteasome and are required for the degradation of some proteins (Bercovich et al., 1997; Fischer et al., 1997; Luders et al., 2000; Verma et al., 2000). In fact, substrate unfolding is a kinetically dominant step in proteolysis, and at least certain substrates require extraproteasomal chaperones for efficient degradation (Thrower et al., 2000). Thus, the finding that chaperones Hsp70/Hsc70 localize in proteasome-enriched bodies supports the view that these structures contain functional proteasomes.
Figure 4.
Serum stimulation of fibroblasts induces the appearance of proteasome-enriched domains. Primary human fibroblasts (WI-38) grown in the presence of 0.1% fetal calf serum (FCS) for 4 d were stimulated for 75 min with 20% FCS. Immunofluorescence was performed using antibodies directed against 20S proteasome (A) or 19S proteasome (B). Cells were additionally double-labeled with anti-20S proteasome and anti-Hsp70/Hsc70 antibodies (C and D). Bar, 10 μm.
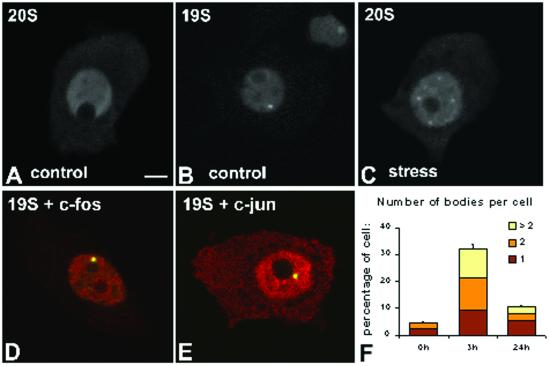
It is well established that administration of hypertonic NaCl solutions by intraperitoneal injection in rats causes both osmotic and stressful effects (Ceccateli et al., 1989; Sharp et al., 1991; Xiong and Hatton, 1996). This type of stimulus evokes a transient expression of immediate-early genes such as c-fos and c-jun in hypothalamic magnocellular neurosecretory neurons (Wang et al., 1997, and references therein). This is followed by an up-regulated synthesis of antidiuretic hormone, a major regulator of body fluid balance (Sherman et al., 1986; Herman et al., 1991; Ding et al., 1994). We have previously shown that in unstimulated animals, the neurosecretory neurons in hypothalamic supraoptic nucleus (SON) are devoid of Fos immunoreactivity. At 30 min after hypertonic saline injection, c-Fos is detected in most neuronal nuclei. Expression of Fos is maximal at 2 h after injection, decreases drastically at 12 h and is no longer detected at 24 h (Lafarga et al., 1998). Consistent with these results, administration of hypertonic saline was shown to induce a transient synthesis of c-fos mRNA in SON neurons, which occurs within 5 min and peaks at 30–60 min, whereas expression of Fos protein is maximal at 2 h and decreases thereafter (Sharp et al., 1991; Xiong and Hatton, 1996). In addition to Fos and Jun, a number of other stress-induced proteins are likely to be degraded by the ubiquitin–proteasome pathway in these cells. We therefore counted the number of proteasome-containing bodies in SON neurons from control (unstimulated) animals, and animals sacrificed 3 h after hypertonic saline injection (i.e., when proteolytic destruction of stress-induced proteins is high). The results show that at 3 h after injection there is a drastic increase in the proportion of neurons containing at least one body, as well as a significant increase in the number of bodies per nucleus (Figure 5, A–F). Both Fos and Jun proteins colocalize with 19S and 20S proteasomes in the nuclear body (Figure 5, D and E). At 24 h after injection the number of bodies decreased significantly, although they were still more abundant than in control cells (Figure 5F).
Figure 5.
Stress induces the formation of proteasome-enriched domains in rat brain neurons. Hypotalamic neurosecretory neurons were obtained from control animals (A and B) and animals sacrificed 3 h after intraperitoneal injection of an hypertonic saline solution to induce osmotic stress (C–E). Cells were immunolabeled with antibodies directed against 20S proteasome (A and C) or 19S proteasome (B). Cells were additionally double-labeled with either anti-19S proteasome (D, red staining) and anti-c-Fos antibodies (D, green staining), or anti-19S proteasome (E, red staining) and anti-c-Jun antibodies (E, green staining). (D and E) Overlay of red and green images; colocalization produces a yellowish color. Bar, 10 μm. (F) Proportion of neurons containing proteasome-enriched domains and the number of bodies per cell were estimated at 0, 3, and 24 h after saline injection. The graph depicts means ± SEs (for each time point four animals were sacrificed, and from each animal ∼150 neurons were analyzed).
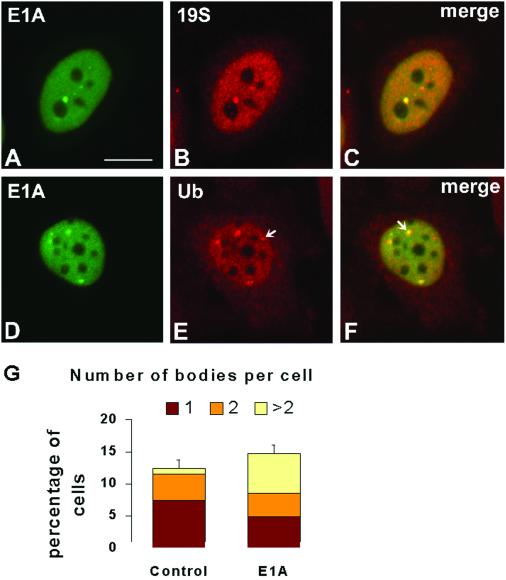
Additional regulators of proteasomal activity are the adenovirus early region 1 transforming proteins. Adenovirus E1A interacts with regulatory components of the 19S proteasome and inhibits the degradation of p53 and E2F transcription factors (Hateboer et al., 1996; Nakajima et al., 1998; Grand et al., 1999; Turnell et al., 2000). Furthermore, E1A is itself a substrate for proteasomal-mediated degradation (Turnell et al., 2000). After transient expression of E1A in HeLa cells, the protein appears exclusively localized in the nucleoplasm with higher concentration in structures of irregular shape and size (Figure 6, A and D). Double-labeling experiments indicate that E1A, proteasomes, and ubiquitin colocalize in the same structures (Figure 6, A–F). Although the proportion of cells containing labeled nuclear bodies is not significantly affected by E1A expression, cells transfected with E1A have more bodies per nucleus than cells that were either nontransfected (Figure 6G) or transfected with control plasmids (our unpublished data). Noteworthy, colocalization of E1A with proteasome and ubiquitin is not complete. There are nuclear bodies labeled by proteasome or ubiquitin antibodies that do not contain E1A (Figure 6E, arrow), and E1A proteins do not always occupy the entire proteasome/ubiquitin-enriched structure (Figure 6F, arrow).
Figure 6.
Adenovirus E1A oncoproteins localize in proteasome-enriched domains and stimulate their assembly. HeLa cells transiently transfected with plasmid pE1A, which expresses adenovirus E1A proteins, were double-labeled with anti-E1A antibody M73 (green staining) and either anti-19S proteasome (B and C, red staining) or anti-ubiquitin antibodies (E and F, red staining). (C and F) Overlay of red and green images; colocalization produces a yellowish color. Note that in some cases the E1A proteins occupy only a fraction of the ubiquitin-labeled structures (F, arrow). In E, the arrow indicates a domain enriched in ubiquitin-conjugates but devoid of E1A. Bar, 10 μm. (G) Proportion of transfected cells containing proteasome-enriched domains and the number of bodies per cell were estimated in a total of 300 randomly selected cells. The graph depicts means ± SEs.
In summary, our data show that the proteasome-enriched nuclear domains contain proteins that are either proteasome substrates or interact with proteasome components. These nuclear domains or bodies disappear when cells are treated with proteasome inhibitors and they form transiently in response to an up-regulation of proteasome activity in the nucleus. Taken together, the data suggest that these structures represent sites where proteasome-dependent proteolysis is taking place. We will refer to them as clastosomes.
Viewed with the Electron Microscope, Clastosomes Correspond to Complex Nuclear Bodies
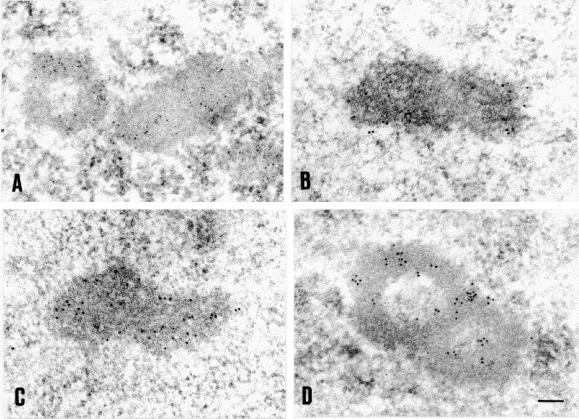
Anti-20S proteasome and anti-c-Jun antibodies were used to perform immunoelectron microscopy on ultrathin sections of rat brain neurons. As depicted in Figure 7, immunogold particles specifically decorate defined subnuclear structures. These are elongated structures with 0.2–0.5 μm in diameter and 0.5–0.9 μm in length. In cross sections these structures present a characteristic ring-like or doughnut-shaped profile, and occasionally they appear in pairs (Figure 7, A and D). Although no obvious links to nucleoli, chromatin, or any other subnuclear compartment were detected, the proteasome-containing structures are often found in proximity to a nucleolus. On the basis of these morphological features, the proteasome-containing structures correspond to the so-called complex nuclear bodies characterized by the presence of a ring-like “capsule” surrounding a central core with heterogeneous content (Bouteille et al., 1974).
Figure 7.
Electron microscopy reveals that proteasome-domains correspond to complex nuclear bodies. Ultrathin sections from primary Schwann cells were immunogold-labeled with antibodies directed against 20S proteasomes (A, 5-nm gold particles), 19S proteasomes (B, 10-nm gold particles), ubiquitin (C, 10-nm gold particles), and c-Jun (D, 10-nm gold particles). Bar, 0.1 μm.
Clastosomes Contain PML
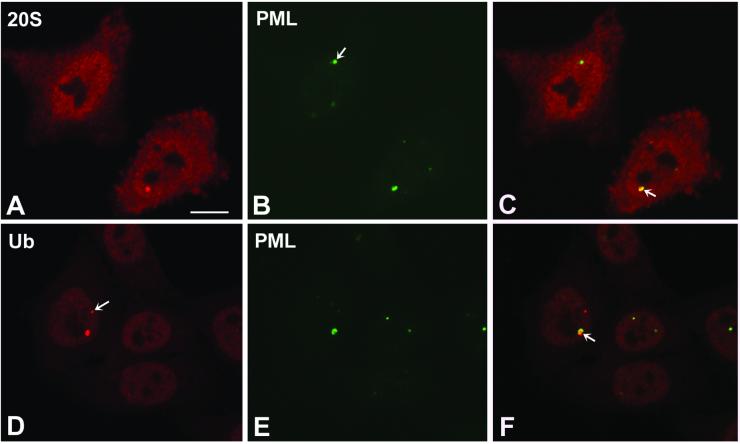
Immunoelectron microscopic analysis with anti-PML antibodies has revealed that nuclear bodies associated with this protein are shaped like a doughnut (Zhong et al., 2000). Given this morphological similarity with clastosomes, we performed double-labeling experiments with anti-PML and either anti-proteasome or anti-ubiquitin antibodies. As illustrated in Figure 8, PML is detected in clastosomes. As observed with E1A proteins (Figure 6), this colocalization is not always perfect. PML tends to occupy only part of a clastosome, indicating that other components are present in the structure. Furthermore, several PML bodies do not stain with proteasome or ubiquitin antibodies. In good agreement with these data, it was reported that at the electron microscopic level PML bodies devoid of proteasomes appear as spheroid aggregates, whereas PML bodies containing proteasomes have the characteristic shell-like or doughnut-shaped structure (Lallemand-Breitenbach et al., 2001).
Figure 8.
PML protein is present in clastosomes. HeLa cells were double-labeled with anti-PML antibody (green staining) and either anti-20S proteasome (A and C, red staining) or anti-ubiquitin antibodies (D and F, red staining). (C and F) Overlay of red and green images; colocalization produces a yellowish color. Note that PML occupies only a fraction of large clastosomes (C and F, arrow). The arrow in B points to a PML body that does not concentrate proteasomes. In D, the arrow points to a clastosome that contains little, if any, PML. Bar, 10 μm.
Clastosomes Are Distinct from Proteasome-containing Nuclear Inclusions
In addition to clastosomes, some cells contain nuclear inclusions intensely labeled by anti-proteasome and anti-ubiquitin antibodies. Nuclear inclusions are very diverse in appearance from nuclear bodies and may consist of amorphous, filamentous, or membranous material (Moneron and Bernhard, 1969). In particular, degenerative diseases are often associated with the presence of intranuclear inclusions formed by deposits of aggregated protein in affected neurons and muscle (Galvão et al., 2001; Orr, 2001). Although these deposits are highly enriched in ubiquitylated proteins and components of the proteasome, they are structurally distinct from nuclear bodies and hence they should not be confused with clastosomes. Recent evidence suggests that proteasomes are recruited to these abnormal protein aggregates but because the substrate proteins are tangled into a clump they cannot be efficiently degraded. As a result, the proteasome possibly remains hold onto its prey and consequently fails to degrade the normal protein targets that are constantly being produced in the cell (Bence et al., 2001).
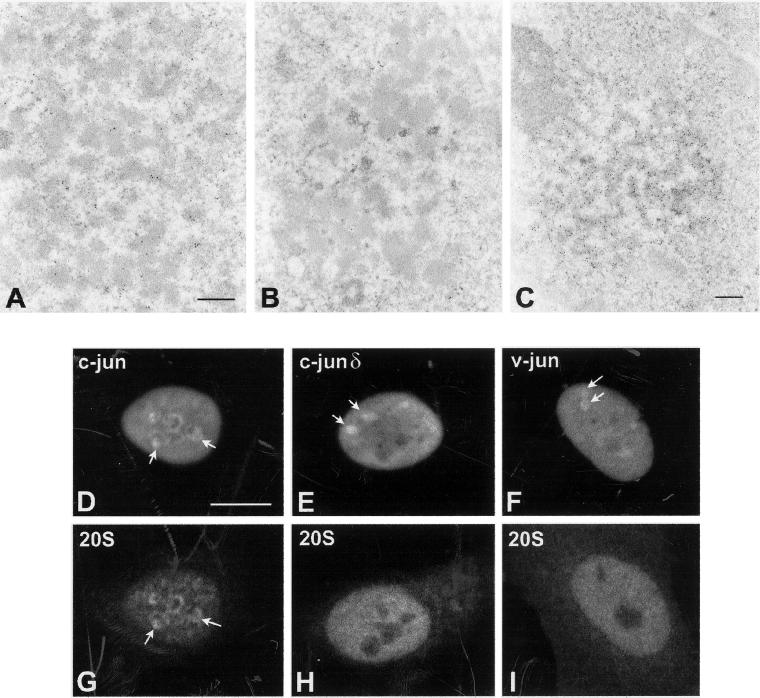
To experimentally address whether nonphysiological deposits of aggregated and ubiquitylated proteins can recruit proteasomes onto intranuclear inclusions, we analyzed the distribution of proteasomes in HeLa cells overexpressing the transcription factor c-Jun. c-Jun is a short-lived protein degraded in the nucleus by the ubiquitin–proteasome pathway (Treier et al., 1994). The protein contains a 27-amino acid segment, the δ domain, which is required for efficient ubiquitination. When the δ domain is deleted, c-Jun is neither ubiquitinated nor degraded (Treier et al., 1994). The oncogenic retroviral counterpart of c-Jun, v-Jun, lacks the δ domain and is not ubiquitinated in vivo. Consequently, v-Jun escapes the mechanism of down-regulation by degradation and the resulting increased stability of the protein most likely contributes to its oncogenicity (Treier et al., 1994).
The overexpressed c-Jun protein forms deposits at the periphery of nucleoli, which are labeled by anti-proteasome antibodies. Viewed with the electron microscope, c-Jun and components of the proteasome are detected in amorphous aggregates that form tangles around nucleoli (Figure 9, A–C). Discrete spheroidal or doughnut-shaped structures containing the exogenously expressed protein are not observed, indicating that overexpressed c-Jun does not assemble into clastosomes. After treatment with proteasome inhibitors, c-Jun is still detected over the tangles of amorphous aggregates (Figure 9C), but labeling of these structures by anti-proteasome antibodies is greatly reduced (Figure 9B). This suggests that in the presence of inhibitors the proteasome is no longer recruited to the c-Jun protein aggregates. We next examined the distribution of proteasomes in HeLa cells overexpressing a deletion mutant of c-Jun lacking the δ domain (c-Jun δ) and v-Jun. The double-labeling experiments depicted in Figure 9, D–I, show that both c-Jun δ and v-Jun form aggregates in the nucleus that fail to recruit the proteasome. Overexpression of unrelated control proteins produced intranuclear aggregates that failed to recruit proteasomes (our unpublished data).
Figure 9.
Overexpressed c-Jun protein forms aggregates that recruit proteasomes but are distinct from clastosomes. Ultrathin sections of HeLa cells transiently transfected with a plasmid encoding His6-tagged human c-Jun were immunogold-labeled with anti-20S proteasome (A and B) and anti-His6 antibodies (C). The HeLa cells were either mock treated (A) or incubated for 12 h in the presence of MG132 (B) or lactacystin (C). Bar, 0.3 μm. (D–I) HeLa cells were transiently transfected with a plasmid encoding His6-tagged human c-Jun (D and G), human c-Jun harboring a deletion of the δ domain (E and H), and chicken v-Jun (F and I). The cells were double-labeled with anti-His6 (D–F) and anti-20S proteasome antibodies (G–I). All three forms of the Jun protein produce aggregates in the nucleoplasm (D–F arrows); however, only c-Jun aggregates recruit proteasomes (G, arrows). Bar, 10 μm.
In conclusion, we propose the following model for distribution of proteasomes in the mammalian cell nucleus. All cells contain 19S and 20S proteasomes spread throughout the nucleoplasm. When there is a high demand for proteasome-dependent proteolysis, the excessive protein substrates associated with components of the ubiquitin–proteasome pathway queue up for degradation forming a transient and conserved structure, the clastosome. Once formed, clastosomes recruit more proteins targeted for degradation, thus enhancing the availability of substrates for the proteolytic machine. Under certain pathological or nonphysiological conditions (for example, overexpression), abnormal protein aggregates form in the nucleus. In contrast to the conserved doughnut profile of clastosomes, abnormal deposits of aggregated proteins tend to be amorphous and very irregularly shaped. Proteasomes are recruited and possibly trapped within these clumps whenever the aggregates contain proteins recognized by the ubiquitination system.
DISCUSSION
In the nucleus of higher eukaryotes, the majority of short-lived proteins are degraded via the ubiquitin–proteasome pathway (Rock et al., 1994). In contrast with the extensive and detailed knowledge on the structure and enzymatic activity of the proteasome, current understanding of its spatial organization in vivo is lagging behind. Herein, we show that the mammalian cell nucleus contains a specific and conserved domain enriched in proteasomes, which we refer to as the clastosome. The clastosome concentrates 20S and 19S proteasomes, ubiquitin-conjugates and molecular chaperones Hsp70/Hsc70, as well as protein substrates of the proteasome such as c-Fos, c-Jun, PML, and adenoviral E1A proteins. Clastosomes are dynamic structures: they form in response to stimuli that activate proteasome-dependent proteolysis and they disappear when proteasome function is inhibited. This suggests that proteasome-dependent proteolysis is taking place in clastosomes.
When viewed at the electron microscope, clastosomes correspond to nuclear bodies. These structures were first described by De Théet al. (1960), and since then, numerous electron microscopy studies reported the observation of nuclear bodies in a variety of cell types (Bouteille et al., 1967, 1974; Padykula and Clark 1981; Brasch et al., 1989). On the basis of their structure, nuclear bodies have been classified as either simple or complex (Bouteille et al., 1974). The simple nuclear bodies are small (0.2–0.5 μm) and finely fibrillar, whereas the complex nuclear bodies are larger (0.2–1.2 μm) and enveloped by a peripheral capsule, which gives them a doughnut-shaped appearance. Based on the data presented herein we propose that the doughnut-shaped complex nuclear bodies correspond to clastosomes.
Early electron microscopic studies showed that complex nuclear bodies are normally scarce, but their number increases under certain conditions such as viral infection and hormonal stimulation (Padykula and Clark, 1981; Padykula et al., 1981; Brasch et al., 1989; Brasch and Ochs, 1992). Accordingly, we show herein that clastosomes are absent or scarce in normal cells, indicating that clastosomes are not essential for proteasome function. Under normal conditions misfolded, unassembled, or damaged proteins are rapidly and efficiently degraded by cellular proteasomes. This is probably the reason why clastosomes are not normally detected. In the presence of elevated levels of proteins targeted for proteasome-dependent degradation (for example, in response to viral infection, hormonal stimulation, or stress) these may transiently queue up for proteolysis giving rise to clastosomes. Once formed, clastosomes may act by recruiting additional proteins targeted to the proteasome, thus enhancing the local availability of proteolytic machines for substrate degradation.
ACKNOWLEDGMENTS
We gratefully acknowledge Göran Akusjärvi and Catharina Svensson (Uppsala University, Uppsala, Sweden) for the gift of the E1A plasmid, Gideon Dreyfuss (University of Pennsylvania, College Park, PA) for anti-SMN antibody, Angus Lamond (University of Dundee, Dundee, United Kingdom) for anti-coilin antibody, and Stéphane Richard (McGill University, Montreal, Quebec, Canada) for anti-Sam68 antibody. This study was supported by grants from Fondo de Investigaciones Sanitarias (FIS 00/0947) and Fundación Marqués de Valdecilla (00/2), Spain, and Fundação para a Ciência e Tecnologia, Portugal.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–03–0122. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–03–0122.
REFERENCES
- Amsterdam A, Pitzer F, Baumeister W. Changes in intracellular localization of proteasomes in immortalized ovarian granulosa cells during mitosis associated with a role in cell cycle control. Proc Natl Acad Sci USA. 1993;90:99–103. doi: 10.1073/pnas.90.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribas J, Arizti P, Castaño JG. Antibodies against the C2 COOH-terminal region discriminate the active and latent forms of the multicatalytic proteinase complex. J Biol Chem. 1994;269:12858–12864. [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:3673–3680. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1467. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Bercovich B, Stancovski I, Mayer A, Blumenfeld N, Laszlo A, Schwartz AL, Ciechanover A. Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J Biol Chem. 1997;272:9002–9010. doi: 10.1074/jbc.272.14.9002. [DOI] [PubMed] [Google Scholar]
- Bochtler M, Ditzel L, Groll M, Hartmann C, Huber R. The proteasome. Annu Rev Biophys Biomol Struct. 1999;28:295–317. doi: 10.1146/annurev.biophys.28.1.295. [DOI] [PubMed] [Google Scholar]
- Bohmann K, Ferreira JA, Lamond AI. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J Cell Biol. 1995;131:817–831. doi: 10.1083/jcb.131.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondesson M, Mannervik M, Akusjärvi G, Svensson C. An adenovirus E1A transcriptional repressor domain functions as an activator when tethered to a promotor. Nucleic Acids Res. 1994;22:3053–3060. doi: 10.1093/nar/22.15.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Weissman AM. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouteille M, Kalifat SR, Delarue J. Ultrastructural variations of nuclear bodies in human diseases. J Ultrastruct Res. 1967;19:474–486. doi: 10.1016/s0022-5320(67)80074-1. [DOI] [PubMed] [Google Scholar]
- Bouteille M, Laval M, Dupuy-Coin AM. Localization of nuclear functions as revealed by ultrastructural autoradiography and cytochemistry. In: Busch H, editor. The Cell Nucleus. I. New York: Academic Press; 1974. pp. 3–71. [Google Scholar]
- Brasch K, Harrington S, Blake H. Isolation and analysis of nuclear bodies from estrogen-stimulated chick liver. Exp Cell Res. 1989;182:425–435. doi: 10.1016/0014-4827(89)90247-4. [DOI] [PubMed] [Google Scholar]
- Brasch K, Ochs RL. Nuclear bodies (NBs): a newly “rediscovered” organelle. Exp Cell Res. 1992;202:211–223. doi: 10.1016/0014-4827(92)90068-j. [DOI] [PubMed] [Google Scholar]
- Braun BC, Glickman M, Kraft R, Dahlmann B, Kloetzel P-M, Finley D, Schmidt M. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat Cell Biol. 1999;1:221–226. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]
- Bravo R, Burckhardt J, Curran T, Muller R. Expression of c-fos in NIH-3T3 cells is very low but inducible throughout the cell cycle. EMBO J. 1986;5:695–700. doi: 10.1002/j.1460-2075.1986.tb04269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockes JP, Fields KL, Raff MC. Studies on cultured rat Schwann cells: I. Establishment of purified populations from cultures of peripheral nerve. Brain Res. 1979;165:105–118. doi: 10.1016/0006-8993(79)90048-9. [DOI] [PubMed] [Google Scholar]
- Brooks P, Fuertes G, Murray RZ, Bose S, Knecht E, Rechsteiner MC, Hendil KB, Tanaka K, Dyson J, Rivett J. Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem J. 2000;346:155–161. [PMC free article] [PubMed] [Google Scholar]
- Ceccateli S, Villar MJ, Goldstein M, Hokfelt T. Expression of c-Fos immunoreactivity in transmitter-characterized neurons after stress. Proc Natl Acad Sci USA. 1989;86:9569–73. doi: 10.1073/pnas.86.23.9569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Boisvert FM, Bazett-Jones DP, Richard S. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cells. Mol Biol Cell. 1999;10:3015–3033. doi: 10.1091/mbc.10.9.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, DiGiuseppe JA, Bercovich B, Orian A, Richter JD, Schwartz AL, Brodeur GM. Degradation of nuclear oncoproteins by the ubiquitin system in vitro. Proc Natl Acad Sci USA. 1991;88:139–143. doi: 10.1073/pnas.88.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- De Thé G de, Rivière M, Bernhard W. Examen au microscope électronique de la tumeur VX2 du lapin domestique dérivée du papillome de Shope. Bull Cancer. 1960;47:569–584. [Google Scholar]
- Dick LR, Cruikshank AA, Grenier L, Melandri FD, Nunes SL, Stein RL. Mechanistic studies on the inactivation of the proteasome by lactacystin: a central role for clasto-lactacystin β-lactone. J Biol Chem. 1996;271:7273–7236. doi: 10.1074/jbc.271.13.7273. [DOI] [PubMed] [Google Scholar]
- Ding JM, Carver WC, Terracio L, And Buggy J. Proto-oncogene c-fos and the regulation of vasopressin gene expression during dehydration. Mol Brain Research. 1994;21:247–55. doi: 10.1016/0169-328x(94)90255-0. [DOI] [PubMed] [Google Scholar]
- Enenkel C, Lehmann A, Kloetzel P-M. Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J. 1998;17:6144–6154. doi: 10.1093/emboj/17.21.6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- Fischer EA, Zhou M, Mitchell DM, Wu X, Omura S, Wang H, Goldberg AL, Ginsberg HN. The degradation of apolipoprotein B100 is mediated by the ubiquitin-proteasome pathway and involves heat shock protein 70. J Biol Chem. 1997;272:20427–20434. doi: 10.1074/jbc.272.33.20427. [DOI] [PubMed] [Google Scholar]
- Gall JC. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Galvão R, Mendes-Soares L, Camara J, Jaco I, Carmo-Fonseca M. Triplet repeats, RNA secondary structure and toxic gain-of-function models for pathogenesis. Brain Res Bull. 2001;56:191–201. doi: 10.1016/s0361-9230(01)00651-7. [DOI] [PubMed] [Google Scholar]
- Grand RJ, Turnell AS, Mason GG, Wang W, Milner AE, Mymryk JS, Rookes SM, Rivett AJ, Gallimore PH. Adenovirus early region 1A protein binds to mammalian SUG1-a regulatory component of the proteasome. Oncogene. 1999;18:449–458. doi: 10.1038/sj.onc.1202304. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of the 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Habets WJ, Hoet MH, de Jong BA, van der Kemp A, van Venrooij WJ. Mapping of B cell epitopes on small nuclear ribonucleoproteins that react with human autoantibodies as well as with experimentally-induced mouse monoclonal antibodies. J Immunol. 1989;143:2560–2566. [PubMed] [Google Scholar]
- Harlow E, Franza BJ, Schley C. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J Virol. 1985;55:533–546. doi: 10.1128/jvi.55.3.533-546.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hateboer G, Kerhoven RM, Shvarts A, Bernards R, Beijersbergen RL. Degradation of E2F by the ubiquitin-proteasome pathway: regulation by retinoblastoma family of proteins and adenovirus transforming proteins. Genes Dev. 1996;10:2960–2970. doi: 10.1101/gad.10.23.2960. [DOI] [PubMed] [Google Scholar]
- Herman JP, Schafer MK-H, Watson SJ, Sherman T. In situ hybridization analysis of arginine vasopressin gene transcription using intron specific probes. Mol Endocrinol. 1991;5:1447–1456. doi: 10.1210/mend-5-10-1447. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Huang J-Y, Raff JW. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 1999;18:2184–2195. doi: 10.1093/emboj/18.8.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Maniatis T. Regulation of interferon-gamma-activated STAT1 by the ubiquitin-proteasome pathway. Science. 1996;273:1717–1719. doi: 10.1126/science.273.5282.1717. [DOI] [PubMed] [Google Scholar]
- Lafarga M, Berciano MT, Garcia-Segura LM, Andres MA, Carmo-Fonseca M. Acute osmotic/stress stimuli induce a transient decrease of transcriptional activity in the neurosecretory neurons of supraoptic nuclei. J Neurocytol. 1998;27:205–217. doi: 10.1023/a:1006937032068. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, et al. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced P.M.L. or P.M.L/retinoic acid receptor α degradation. J Exp Med. 2001;193:1361–1371. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- Luders J, Demand J, Hohfeld J. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J Biol Chem. 2000;275:4613–4617. doi: 10.1074/jbc.275.7.4613. [DOI] [PubMed] [Google Scholar]
- Mengual E, Arizti P, Rodrigo J, Gimenez-Amaya JM, Castano JG. Immunohistochemical distribution and electron microscopic subcellular localization of the proteasome in the rat CNS. J Neurosci. 1996;16:6331–6341. doi: 10.1523/JNEUROSCI.16-20-06331.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. J Cell Sci. 2000;113:1841–1849. doi: 10.1242/jcs.113.11.1841. [DOI] [PubMed] [Google Scholar]
- Moneron A, Bernhard W. Fine structural organization of the interphase nucleus in some mammalian cells. J Ultrastruct Res. 1969;27:266–288. doi: 10.1016/s0022-5320(69)80017-1. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Morita K, Tsunoda H, Imajoh-Ohmi S, Tanaka H, Yasuda H, Oda K. Stabilization of p53 by adenovirus E1A occurs through its amino-terminal region by modification of the ubiquitin-proteasome pathway. J Biol Chem. 1998;273:20036–20045. doi: 10.1074/jbc.273.32.20036. [DOI] [PubMed] [Google Scholar]
- Omura S, Matsuzake K, Fujimoto T, Kosuge K, Furuya T, Fujita S, Nakagawa A. Structure of lactacystin, a new microbial metabolite which induces differentiation of neuroblastoma cells. J Antibiot (Tokyo) 1991;44:117–118. doi: 10.7164/antibiotics.44.117. [DOI] [PubMed] [Google Scholar]
- Orr HT. Beyond the Qs in the polyglutamine diseases. Genes and Development. 2001;15:925–932. doi: 10.1101/gad.888401. [DOI] [PubMed] [Google Scholar]
- Padykula HA, Clark JH. Nuclear bodies as functional indicators in the target cells of sex steroid hormones. In: Busch H, editor. The Cell Nucleus, vol. IX. New York: Academic Press; 1981. pp. 309–339. [Google Scholar]
- Padykula HA, Fitzgerald M, Clark JH, Hardin J. Nuclear bodies as structural indicators of estrogenic stimulation in uterine luminal epithelial cells. Anat Rec. 1981;201:679–696. doi: 10.1002/ar.1092010412. [DOI] [PubMed] [Google Scholar]
- Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell. 1994;78:78–85. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- Reits EAJ, Benham AM, Plougastel B, Neefjes J, Trowsdale J. Dynamics of proteasome distribution in living cells. EMBO J. 1997;16:6087–6094. doi: 10.1093/emboj/16.20.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clask K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Seemüller E, Lupas A, Stock D, Löwe J, Huber R, Baumeister W. Proteasome from Thermoplasma acidophilum: a threonine protease. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- Sharp FR, Sagar SM, Hicks K, Lowenstwein D, Hisinaga K. c-fos mRNA, Fos, and Fos-related antigen induction by hypertonic saline and stress. J Neurosci. 1991;11:2321–31. doi: 10.1523/JNEUROSCI.11-08-02321.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman TG, McKelvy JF, Watson SJ. Vasopressin mRNA regulation in individual hypothalamic nuclei: a northern and in situ hybridization analysis. J Neurosci. 1986;6:1685–1694. doi: 10.1523/JNEUROSCI.06-06-01685.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H, Yanagida M. Cut8, essential for anaphase, controls localization of 26S proteasome, facilitating destruction of cyclin and Cut2. Curr Biol. 2000;10:1329–1338. doi: 10.1016/s0960-9822(00)00773-9. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–98. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- Turnell AS, Grand RJ, Gorbea C, Zhang X, Wang W, Mymryk JS, Gallimore PH. Regulation of the 26S proteasome by adenovirus E1A. EMBO J. 2000;19:4759–4773. doi: 10.1093/emboj/19.17.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Deshaies RJ. A proteasome howdunit: the case of the missing signal. Cell. 2000;101:341–344. doi: 10.1016/s0092-8674(00)80843-0. [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Wang K, Guldenaar SEF, McCabe JT. Fos and Jun expression in rat supraoptic nucleus neurons after acute vs. repeated osmotic stimulation. Brain Res. 1997;746:117–125. doi: 10.1016/s0006-8993(96)01216-4. [DOI] [PubMed] [Google Scholar]
- Wilkinson CRM, Wallace M, Morphew M, Perry P, Allshire R, Javerzat J-P, McIntosh JR, Gordon C. Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J. 1998;17:6465–6476. doi: 10.1093/emboj/17.22.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J-J, Hatton GI. Differential responses of oxytocin and vasopressin neurons to the osmotic and stressful components of hypertonic saline injections: a Fos protein double-labeling study. Brain Res. 1996;719:143–153. doi: 10.1016/0006-8993(95)01466-7. [DOI] [PubMed] [Google Scholar]
- Zhong S, Salomoni P, Pandolfi PP. The transcriptional role of PML and the nuclear body. Nat Cell Biol. 2000;2:E85–E90. doi: 10.1038/35010583. [DOI] [PubMed] [Google Scholar]
- Zhu J, Lallemand-Breitenbach V, de Thé H. Pathways of retinoic acid-or arsenic trioxide-induced PML/RARα catabolism, role of oncogene degradation in disease remission. Oncogene. 2001;20:7257–7265. doi: 10.1038/sj.onc.1204852. [DOI] [PubMed] [Google Scholar]
- Zwickl P, Baumeister W, Steven A. Dis-assembly lines: the proteasome and related ATPase-assisted proteases. Curr Opin Struct Biol. 2000;10:242–250. doi: 10.1016/s0959-440x(00)00075-0. [DOI] [PubMed] [Google Scholar]