Abstract
Sorting nexins (Snxs) are a recently discovered family of conserved hydrophilic cytoplasmic proteins that have been found associated with membranes of the endocytic system and that are implicated in the trafficking of many endosomal membrane proteins, including the epidermal growth factor receptor and transferrin receptor. Snx proteins are partly defined by the presence of a p40 phox homology domain that has recently been shown to bind phosphatidylinositol 3-phosphate. Most Snx proteins also contain a predicted coiled-coils domain in the carboxyl-terminal half of the protein and have been shown to form dimers with other members of the Snx family. The yeast sorting nexins Vps5p and Vps17p form a dimer and are also components of the retromer complex that mediates endosome-to-Golgi transport of the carboxypeptidase Y receptor Vps10p. To functionally define the different domains of the yeast sorting nexins Vps5p and Vps17p, we have generated various truncations to examine the role that the different domains of Vps5p/Vps17p play in their respective functions. Herein, we show that the C-terminal halves of Vps5p and Vps17p, which contain the coiled-coils domains, are necessary and sufficient for their interaction. We have also mapped the retromer assembly domain to the N-terminal half of Vps5p and found that binding of Vps5p by Vps17p synergizes the interaction between Vps5p and other retromer components. Additionally, we have examined which domain(s) of Vps5p is necessary for membrane association.
INTRODUCTION
The various compartments that make up the secretory and endocytic pathways of eukaryotic cells each have a discrete complement of resident proteins that define the function of the particular organelle. Vesicle-mediated transport between the organelles is necessary to deliver and maintain each organelle's set of resident proteins (Rothman and Orci, 1992; Schekman and Orci, 1996). The complexity of the endocytic and secretory pathways dictates that each protein must be subject to multiple sorting events as they traverse these pathways to their final destination. Cytoplasmic vesicle coat proteins are key components of this sorting machinery and have been shown to provide the selectivity in vesicle-mediated protein sorting (Kirchhausen et al., 1997; Robinson, 1997).
Recently, a new class of vesicle-associated proteins has been shown to play a key role in trafficking within the endocytic system. The family of sorting nexin (Snx) proteins is conserved from yeast to humans (Kurten et al., 1996; Renfrew-Haft et al., 1998). Generally, Snx proteins are between 500 and 700 amino acids in length and are charged hydrophilic proteins. One of the defining features of the sorting nexin family is a conserved domain, the p40 phox homology (PX) domain (Ponting, 1996; Renfrew-Haft et al., 1998). The PX domain is ∼100–120 amino acids in size and has recently been shown to bind the lipid phosphatidylinositol 3-phosphate (PtdIns 3-P) (Cheever et al., 2001; Song et al., 2001; Xu et al., 2001; Yu and Lemmon, 2001). Most Snx proteins share an overall domain arrangement in which the PX domain is located in the middle or N-terminal half of the Snx protein, whereas the C-terminal half usually has one or two regions of predicted coiled coils.
The first Snx protein to be characterized was SNX1, which was identified through the yeast two-hybrid system by using the tail of the epidermal growth factor (EGF) receptor as bait (Kurten et al., 1996). Subsequent studies have identified more than a dozen members of the Snx family in mammals and they have been found associated with several membrane proteins that traffic throughout the endocytic system, including the insulin receptor, the transferrin receptor and the PDGF receptor (Renfrew-Haft et al., 1998; Otsuki et al., 1999; Parks et al., 2001). SNX1 has been implicated in the down-regulation of activated EGF receptors in lysosomes and overexpression of SNX1 can lead to increased EGF receptor degradation (Kurten et al., 1996). However, the association of SNX1 with several receptors, including the transferrin receptor, also argues for SNX1 having a more general role in trafficking within the endocytic system.
The yeast homolog of SNX1 is the product of the vacuolar protein sorting gene VPS5. Mutants of vps5 fail to deliver vacuolar proteases such as carboxypeptidase Y (CPY) to the vacuole resulting in the secretion of CPY from vps5 mutants (Horazdovsky et al., 1997; Nothwehr and Hindes, 1997). This defect is actually a result of the mislocalization of the CPY receptor Vps10p. Vps5p forms a dimer with Vps17p (Horazdovsky et al., 1997). Vps17p is also an Snx protein with a PX domain in its N-terminal half and predicted coiled coils in the C-terminal half. The Vps5p/Vps17p dimer is a subcomplex of the heteropentameric retromer complex, which has been shown to mediate the endosome-to-Golgi retrieval of Vps10p (Seaman et al., 1998). The other members of the yeast retromer are Vps35p, Vps29p, and Vps26p. Mammalian SNX1 forms a dimer with SNX2 and has been shown to associate with the mammalian orthologues of Vps35p, Vps29p, and Vps26p (Renfrew-Haft et al., 2000). SNX1 therefore is part of mammalian retromer and may well function in an analogous role transporting membrane proteins between the endosome and the Golgi. A general property of Snx proteins therefore may be to form dimers, which may in turn bind other proteins that function in vesicle trafficking at various sites in the cell. One of these Snx binding proteins seems to be Hrs1 (Chin et al., 2001), a homolog of the yeast VPS27 gene and a FYVE domain containing protein that can bind PtdIns 3-P (Burd and Emr, 1998).
Along with assembling into the retromer complex, Vps5p has been shown to have intrinsic self-assembly activity in vitro. Recombinant Vps5p is able to form large (15–20-nm) particles, which appear by electron microscopy to be uniform in size and shape (Seaman et al., 1998). SNX1 has also been shown to self-assemble and this property may play an important role in Snx function (Renfrew-Haft et al., 2000; Kurten et al., 2001). Very little is known about the functional importance of the respective domains of the Snx proteins. To date, various studies conducted in mammalian cells have attempted to address the questions of which Snx domain mediates membrane binding and/or membrane targeting (Teasdale et al., 2001). This approach has the caveat that the endogenous Snx protein will always be present making the study of the localization of Snx domains problematic due to potential interactions between the transfected Snx protein and its Snx dimer partner or due to self-assembly interactions between the transfected Snx and the endogenous protein.
Therefore, we have conducted an extensive study of the functional domains of the yeast Snx protein Vps5p. Yeast provides an excellent model system for these studies because the use of deletion mutants solves many of the problems associated with performing these studies in systems where the endogenous proteins are present. We can also explore the functional importance of the domains of Vps5p by using functional assays for trafficking to the vacuole, retromer assembly, and membrane association. Using this approach, we have mapped the domain of Vps5p that interacts with the other retromer components, identified the region of Vps5p required to interact with its Snx dimer partner Vps17p, and investigated the domain required for membrane association.
MATERIALS AND METHODS
Reagents, Yeast, and Bacterial Cell Culture
Most laboratory reagents were purchased from Sigma-Aldrich (Poole, Dorset, United Kingdom). Restriction enzymes were obtained from New England Biolabs (Hitchen, Herts, United Kingdom). Protein A-Sepharose, 125I-protein A, and [35S]methionine/cysteine (Promix) were obtained from Amersham Biosciences (St. Albans, Herts, United Kingdom). Antisera against Vps5p, Vps35p, Vps29p, and Vps17p were provided by Scott Emr (University of California, San Diego, San Diego, CA). Antisera against CPY was also obtained from Scott Emr. Antisera against Vps17p was also raised during the course of this study (see below). Antisera against Vps26p was generated in our laboratory as described in Reddy and Seaman (2001). Escherichia coli were grown in LB media (supplemented with appropriate antibiotics). Bacterial transformations were performed according to Hanahan (1983). Yeast strains (Table 1) were grown on/in either rich media (yeast extract, peptone, dextrose-YPD) or minimal media (yeast nitrogen base, dextrose; YNB) supplemented with appropriate amino acids. Yeast transformations were performed according to Elble (1992).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| SEY6210 | MATα leu2-3,112 ura3-52 his3Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | Robinson et al., 1988 |
| TVY614 | SEY6210 praΔ::LEU2 prbΔ::HISG prcΔ::HIS3 | Wurmser and Emr, 1998 |
| MSY2600 | SEY6210 vps26Δ::LEU2 | Seaman et al., 1998 |
| EMY18 | SEY6210 vps35Δ::HIS3 | Seaman et al., 1997 |
| PSY1-29 | SEY6210 vps29Δ::HIS3 | Seaman et al., 1997 |
| BHY152 | SEY6210 vps5Δ::HIS3 | Horazdovsky et al., 1997 |
| PHY102 | SEY6210 vps34Δ::TRP1 | Herman and Emr, 1990 |
| YES102 | pik1-83::TRP1 | Hama et al., 1999 |
| MSY1700 | SEY6210 vps17Δ::HIS3 | Reddy and Seaman, 2001 |
| MSY5211 | SEY6211 vps5Δ::HIS3 | Reddy and Seaman, 2001 |
| MSY3426 | PHY102 vps26Δ::LEU2 | This study |
| MSY3435 | PHY102 vps35Δ::LEU2 | This study |
| MSY0517 | vps5Δ::HIS3 vps17Δ::HIS3 | This study |
| MSY2700 | SEY6210 vps27Δ::LEU2 | This study |
The vps5Δ vps17Δ strain (MSY0517) was made by mating the strains MSY1700 and MSY5211. Diploids were sporulated and the tetrads were disrupted by repeated extractions with ether. Spores were germinated on rich media and then replica plated onto −HIS minimal media. HIS+ strains were tested by polymerase chain reaction (PCR) to identify the double mutants. The vps27Δ strain (MSY2700) was made by disruption of the VPS27 gene by insertion with the LEU2 gene. VPS27 was amplified by PCR from genomic DNA. The product was cloned using the pCR blunt vector (Invitrogen, Paisley, Scotland). The VPS27 gene was digested with ClaI, blunted, and then digested with BglII to excise a 1.4-kbp fragment. This was replaced with the LEU2 gene, which had been excised from pBluescript by using HindIII and BamHI (the HindIII site was blunted before BamHI digestion). The vps27Δ::LEU2 construct was amplified by PCR, and the PCR product was transformed into SEY6210. LEU+ transformants were tested by PCR to confirm the disruption of the VPS27 gene. The vps34Δvps35Δ and vps34Δvps26Δ strains were generated by respective disruption of the VPS35 and VPS26 genes in the PHY102 strain. The vps26::LEU2 construct used is described in Seaman et al. (1998). The vps35::LEU2 knockout construct was created by replacing ∼90% of the VPS35 coding region contained in a HindII fragment with the LEU2 gene. The vps35::LEU2 construct was amplified by PCR by using flanking primers that anneal ∼200 base pairs outside of the coding region and transformed into the PHY102 strain. Transformants were selected for on YNB-LEU plates. Disruption of the VPS35 and VPS26 genes was confirmed by PCR.
Western Blotting
Western blotting was performed as described in Reddy and Seaman (2001).
DNA Manipulation/Generation of Vps5p/Vps17p Truncations
Vps5p C-Terminal Truncations.
To aid in the manipulation of VPS5, the ∼3-kbp SmaI-XhoI fragment containing full-length VPS5 was subcloned into pBluescript (which had been digested with SmaI and XhoI) to create pVPS5-pBS. To generate the C-terminal truncations, oligonucleotide primers containing a stop codon were used with the primer VPS55 (Table 2) to amplify a specific region of VPS5. The PCR reaction was performed using either Pfu or Vent polymerase and the product was cloned using the pCR blunt cloning vector. The PCR product was then excised from pCR blunt (Invitrogen) by digestion with SnaBI and NsiI and was subcloned into SnaBI/NsiI digested pVPS5-pBS. The C-terminal truncation was then excised with SmaI and KpnI and subcloned into pRS316 cut with SmaI-KpnI. This method ensured that the NsiI-XhoI region of VPS5 that contains the 3′ untranslated region was retained.
Table 2.
Oligonucleotide primers used in construction of Vps5p truncation mutants
| Construct | Forward primer | Reverse primer |
|---|---|---|
| ΔC130 | CGGTCGCGGCAGTCTTCAAAA (VPS55) | GACTCTTAACTAAAGTCATC |
| ΔC280 | VPS55 | CCATCGTCTAAGTCTCTTGC |
| ΔC392 | VPS55 | CAACTTAGAAAGCCACAGC |
| Δ441-491 | VPS55 | GATTCCGCGTCGACACCCCC |
| ΔN130 | CTCTCCATGGTTTTTGACGATAC | GATGCTACGTCGCACTGGCTG (VPS53) |
| ΔN160 | CCATGGATTCCGCCAGAGCTCAAAGA | VPS53 |
| ΔN190 | CCATGGATCCCTTAAAGAAAGCTGAA | VPS53 |
| ΔN220 | CCATGGAGGGCAAGTTCACAGCTTCT | VPS53 |
| ΔN280 | GGCCATGGCTTTCAAAGTTGAAG | VPS53 |
| ΔN390 | GCCATGGATGACTTTAGTTCAGAGTCT | VPS53 |
| 5ΔPX (N) | VPS55 | GAAAGCTTCAGCCTTTTCGG |
| 5ΔPX (C) | AAAGCTTACTTTGTTCAGAGTCT | VPS53 |
The Δ441–491 truncation required a slightly different strategy. In this case, a reverse primer that incorporated a SalI site was used with the ΔN130 forward primer. The PCR product was cloned into the pCR blunt vector. The insert was digested with StuI and SalI, and the fragment was gel purified and then cloned into pVPS5-pBS, which had been digested with StuI and SalI. The construct was then further subcloned into pRS316 as described above.
Vps5p N-Terminal Truncations.
To aid in the manipulation of VPS5, the ∼3-kbp SmaI-XhoI fragment containing full-length VPS5 was subcloned into pRS424 (which had been digested with SmaI and XhoI) to create pVPS5–424. PCR was used to amplify specific regions of VPS5. The forward primer (Table 2) was designed with an NcoI site (at the 5′ end) in frame with the endogenous NcoI site at the start codon of VPS5. The VPS53 reverse primer was used for all N-terminal deletions. PCR products were cloned using pCR blunt. After digestion with NcoI and SalI, the insert was gel purified and subcloned into pVPS5–424, which had been digested with NcoI and SalI. The N-terminal truncations were then excised by digestion with SmaI and XhoI subcloned into the CEN vector pRS316. The 5ΔPX construct was generated by amplifying the N- and C-terminal halves of VPS5 either side of the PX domain. The primers used were designed to create a HindIII site, which was used to join the to halves together. PCR products were cloned using the pCRblunt vector (Invitrogen). The N-terminal half was digested with NcoI and HindIII, whereas the C-terminal half was excised with HindIII and SalI. The two halves were simultaneously ligated into NcoI-SalI digested VPS5 in pRS424. The 5ΔPX construct was then excised from pRS424 by SmaI-XhoI digestion and subcloned into pRS316 for expression at CEN levels. All N- and C-terminal truncations were sequenced to confirm the truncation and ensure that no other mutation had occurred.
Vps17p Truncations.
To remove the N terminus (and PX domain) of Vps17p, VPS17 (∼2.5-kbp genomic DNA fragment) in pRS414 was digested with NdeI. The two NdeI sites are in frame with each other and so the cut pVPS17-414 was simply religated. To truncate the C terminus, VPS17-414 was cut with NcoI and AflIII, treated with T4 polymerase and religated. This method preserved the 3′ UTR of VPS17 and resulted in a stop codon being created and therefore truncation of Vps17p. The N- and C-terminal Vps17p truncations were subcloned into pRS424 and pRS425, respectively. The 17ΔPX construct was generated by digesting the NdeI dropout construct (Δ8–250) with NdeI followed by blunting with Mung bean nuclease. The construct was then further digested with MluI. Wild-type VPS17 was digested with AgeI, blunted with T4 polymerase and then further digested with MluI. The MluI-AgeI fragment was ligated into the MluI-NdeI digested VPS17NdeI dropout in pRS414. This construct effectively removed the Vps17p PX domain.
CPY Sorting Assay
Whole cell CPY sorting assays were performed as explained in Reddy and Seaman (2001) and described briefly as follows. Cells grown in selective minimal media were harvested by centrifugation. Approximately 4 OD600 nm equivalents of each strain were resuspended into fresh media, labeled for 10 min with [35S]methionine (∼60 μCi/sample), and then chased for 30 min in the presence of excess cold methionine/cysteine. The cells were then transferred to ice and all labeled proteins were precipitated with trichloroacetic acid (TCA). After centrifugation, the pellet was washed twice with acetone, dried, and then resuspended into 100 μl of urea cracking buffer (50 mM Tris-HCl pH 7.4, 6 M urea, and 1% wt/vol SDS). The cells were lysed by vortexing with glass beads and then heated to 70°C for 5 min. Then 1 ml of immunoprecipitation buffer (50 ml Tris-HCl pH 7.4, 150 mM NaCl, 0.5 mM EDTA, and 0.5% [vol/vol] Tween 20) was added and the samples were cleared by centrifugation for 10 min. After transferring the supernatant to a fresh tube, antisera against CPY was added. Protein A-Sepharose was used to capture immune complexes; samples were then washed, dried, and analyzed by SDS-PAGE/fluorography.
Vps5p Localization Assay
Vps5p localization was performed using the differential centrifugation assay described in Reddy and Seaman (2001) and also Seaman et al. (1997, 1998). Because Vps5p does not localize to P13 (13,000 × g membrane pellet) membranes, only P100 (pellet fraction) and S100 (cytosolic fraction) were analyzed for Vps5p. Autoradiograms produced from these experiments were scanned and quantified using NIH Image software.
Vps17p Stability Assay
The Vps17p stability assay was performed essentially the same as the Vps10p and Vps35p stability assays in Reddy and Seaman (2001). Briefly, cells were grown in YNB media to an OD of ∼0.7 OD600 nm/ml. Cells were then harvested and resuspended into 3 ml of fresh media at a concentration of ∼2–3 OD600 nm/ml. Cells were pulse labeled with ∼150 μCi of [35S]methionine (Promix) for 15 min. Then 10× chase (50 mM methionine, 10 mM cysteine, 5% yeast extract, and 10% glucose) was added. Aliquots (1 ml) were removed at 0, 45, and 90 min of chase and precipitated on ice with 10% TCA. The cells were collected by centrifugation, washed twice with ice cold acetone, and then dried in a Speed Vac. Cells were lysed by vortexing with glass beads in urea cracking buffer (50 mM Tris-HCl pH 7.4, 6 M urea, and 1% [wt/vol] SDS). After heating to 75°C for 5 min, the lysate was diluted with the addition of 1 ml of immunoprecipitation buffer (50 ml Tris-HCl pH 7.4, 150 mM NaCl, 0.5 mM EDTA, and 0.5% [vol/vol] Tween 20). The lysate was then cleared by centrifugation at 13,000 × g for 10 min, transferred to a fresh tube, and affinity-purified anti-Vps17p antisera was added (1 μl/OD). After incubation on a rotating wheel at 4°C overnight, protein A-Sepharose was added and the mix was incubated at 4°C for a further 2 h. The immune complexes were then washed with immunoprecipitation buffer, followed by urea buffer (50 ml Tris-HCl pH 7.4, 200 mM NaCl, 2 M urea, 1 mM EDTA, and 0.5% Tween 20), 0.1% (wt/vol) SDS, and finally with phosphate-buffered saline. After drying the immunoprecipitates in the Speed Vac, the samples were subjected to SDS-PAGE and fluorography.
Native Immunoprecipitation Assay
Native immunoprecipitations to examine retromer assembly were performed essentially as described in Reddy and Seaman (2001). This involved labeling spheroplasted yeast with [35S]methionine for 15 min, chasing for 45 min, and then lysing the yeast in cytosol buffer (0.2 M sortbitol, 20 mM HEPES-KOH pH 7.0, 50 mM potassium acetate, and 2 mM EDTA) containing 0.5% (vol/vol) Triton X-100 and protease inhibitors. The lysate was cleared by centrifugation at 13,000 × g for 5 min. Antisera was added and the samples were allowed to incubate at 4°C for 90 min on a rotating wheel. Protein A-Sepharose was added and the incubated with the samples for 45 min. The immunoprecipitates were washed with cytosol buffer plus triton four times and then dried in a Speed Vac. After resuspending the protein A-Sepharose into urea cracking buffer, the sample were diluted with 1 ml of immunoprecipitation buffer and reimmunoprecipitated with antisera against retromer components.
Vps17p Antisera Production
Antisera against the N-terminal half of Vps17p was generated using a glutathione S-transferase (GST) fusion protein of amino acids 1–264 of Vps17p. Oligonucleotide primers were used to amplify the N-terminal half of Vps17p, adding a BamHI site at the 5′ end in the process. This was cloned using the pCR blunt cloning vector (Invitrogen). The Vps17p N-terminal half was excised by digestion with BamHI and XhoI and subcloned into pGEX 4T-2 (Amersham Biosciences). The same fragment was also subcloned into pQE30.1 (QIAGEN, Crawley, West Susssex, United Kingdom) (cut with BamHI and SalI) for expression as a His6-tagged protein in bacteria. GST-Vps17p (N-terminal half) was expressed in XL1blue cells and purified using glutathione-Sepharose following manufacturer's instructions. A New Zealand White rabbit was immunized with ∼500 μg of fusion protein on three occasions following a standard immunization protocol. The antiserum was affinity purified using antigen coupled to cyanogen bromide-activated Sepharose.
Recombinant Fusion Protein Binding Assay
The Vps5p deletion constructs were subcloned into pQE30.1, which had been cut with SmaI and SalI. The VPS5 truncations were excised from pRS316 by digestion with NcoI, and XhoI (the NcoI site was blunted before XhoI digestion). For the binding assay, wild-type His6-Vps5p, His6-ΔN280 and His6-ΔC280 and His6-Vps17 N-terminal (amino acids 1–264) were expressed in XL1blue cells. Fusion protein was isolated from 500 ml of bacterial culture following manufacturer's instructions. The fusion protein bound to Ni2+-agarose was incubated with 10 ml of yeast extract prepared from the protease deficient strain TVY614. The extract was prepared from 3000 OD 600 nm of cells that were spheroplasted, lysed in 50 ml of cytosol buffer plus 0.5% Triton (vol/vol) and 10 mM imidazole, and then centrifuged for 10 min at 13,000 × g. The lysate was cleared by incubation with just Ni2+-agarose for 15 min at 4°C on a rotating wheel. After removing the Ni2+-agarose by centrifugation, the fusion protein-Ni2+-agarose was added to the extract and incubated for 30 min at 4°C on a rotating wheel. After several washes with lysis buffer, the bound protein was eluted by four 250-μl applications of 250 mM imidazole in 50 mM Tris-HCl pH 7.4. The eluate was precipitated with TCA, and subjected to SDS-PAGE. The proteins were electrophoretically transferred to nitrocellulose and Western blotted with antisera against Vps35p and Vps26p.
Vps5p Self-Assembly Assay
Wild-type Vps5p and the ΔN280 and ΔC280 truncations were expressed in bacteria as His6-tagged fusion proteins (see above). The protein was purified on Ni2+-agarose beads, eluted with imidazole, and then dialyzed against cytosol buffer (20 mM HEPES-KOH pH 7.0, 50 mM potassium acetate, 2 mM EDTA, and 0.2 M sorbitol) overnight at 4°C. The protein was then subjected to gel filtration chromatography by using a Sephacryl S300 column. Fractions were collected, TCA precipitated, and then loaded onto a 10% polyacrylamide gel. The gel was silver stained and then scanned. The band corresponding to the fusion protein was quantified using NIH Image software.
RESULTS
Dissection of Vps5p to Identify Functional Domains
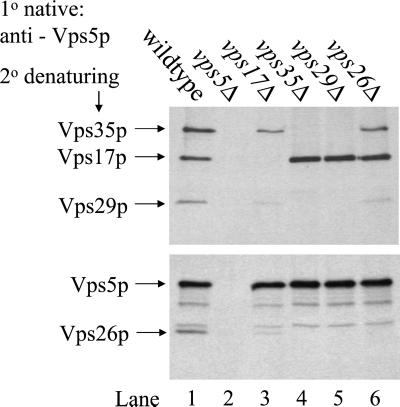
Vps5p is homologous to mammalian SNX1 (26% identity over 495 residues) and therefore understanding the assembly of Vps5p with Vps17p and subsequent interactions with the other retromer components may shed much light upon the function of SNX1 and possible other Sorting Nexins in trafficking within the trans-Golgi network/endocytic system. To better understand the interactions that Vps5p undergoes with the other members of the retromer complex we performed a series of native immunoprecipitation (IP) experiments from wild-type cells and retromer mutants. Vps5p was immunoprecipitated under native conditions from lysates generated from [35S]methionine-labeled cells. After the primary IP, the immunoprecipitates were denatured and the resulting lysates were reimmunoprecipitated with antisera against retromer proteins. In Figure 1, Vps5p coimmunoprecipitated the other four members of retromer from wild-type extracts (lane 1). This is dependent upon Vps5p being present as no additional bands are observed when vps5Δ extracts are immunoprecipitated (lane 2). If Vps35p or Vps29p is absent (lanes 4 and 5) then Vps5p will only coimmunoprecipitate with Vps17p. When Vps26p is absent, Vps5p will maintain only weak interactions with Vps35p/Vps29p, but will still strongly coimmunoprecipitate Vps17p (lane 6). Interestingly, when Vps17p is deleted, Vps5p is still capable of interacting with the remaining components (Vps35p, Vps29p and Vps26p) but at a much reduced level (lane 3). This experiment provides a benchmark for further studies to dissect Vps5p and identify the key domains that mediate its interactions with Vps17p and other retromer components.
Figure 1.
Examination of the interactions between Vps5p and the other retromer components. Approximately 6 OD600 nm equivalents of spheroplasted cells were labeled with [35S]methionine for 20 min and then chased for 40 min. The cells were lysed in cytosol buffer containing 0.5% Triton X-100. The lysate was cleared by centrifugation for 5 min at 13,000 × g and transferred to a fresh tube. Vps5p was immunoprecipitated under native conditions from the lysate by using an affinity purified polyclonal antisera (Horazdovsky et al., 1997). After several washes the samples were boiled in urea cracking buffer and a fresh lysate was generated. Vps35p, Vps17p, and Vps29p were then simultaneously immunoprecipitated after which the lysate was reimmunoprecipitated with antibodies against both Vps5p and Vps26p. This experiment provides a benchmark for further experiments to dissect Vps5p and identify its functional domains.
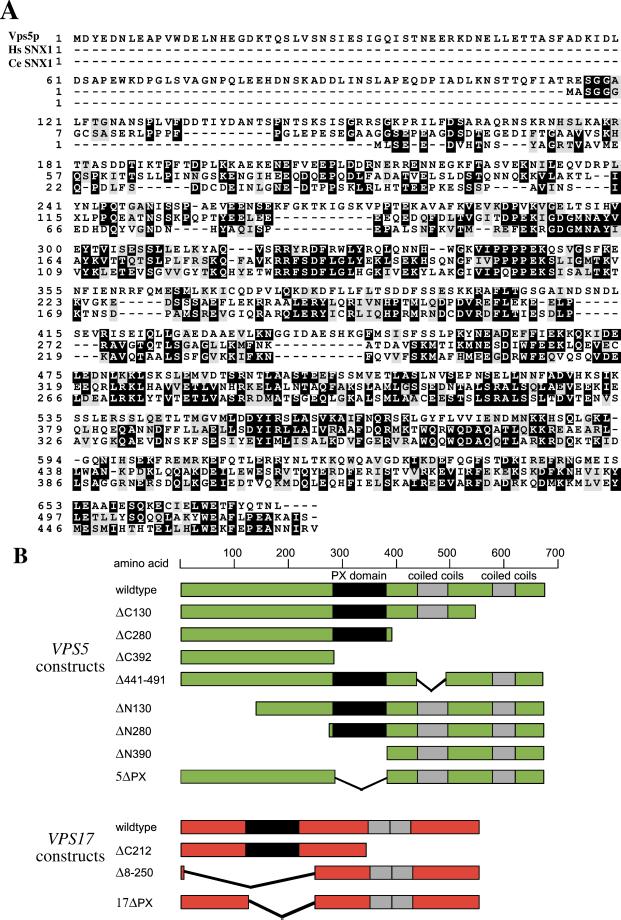
To aid in the dissection of Vps5p we have aligned the Vps5p protein sequence with that of human SNX1 and the SNX1 homolog in nematode (Caenothabditis elegans) (Figure 2a). It is immediately apparent that there is significant sequence conservation between the different species but clearly Vps5p also contains extra sequences within the N-terminal half that are not conserved. To examine the functional domains of Vps5p we have performed a systematic deletion analysis of Vps5p by using the PX domain and the two regions of predicted coiled coils as boundaries for the various truncation mutants. A schematic diagram of the Vps5p deletion mutants is shown in Figure 2b. The N-terminal half of Vps5p does not contain any predicted structural domains; therefore, truncations were designed that would break up the N-terminal half into two regions encompassing amino acids 1–130 and 131–280, respectively. This mirrors the division of the C-terminal half by the two regions of predicted coiled coils. Although Vps17p is less well conserved among the family of sorting nexins, we have also generated simple truncations of Vps17p to effectively cleave the protein into two halves, the N-terminal half that contains the PX domain and the C-terminal half that has the predicted coiled-coils domain (Figure 2b). Because PX domains have recently been shown to bind to PtdIns 3-P (Cheever et al., 2001; Song et al., 2001; Xu et al., 2001; Yu and Lemmon, 2001), we have also generated constructs in which the PX domains of Vps5p and Vps17p have been removed.
Figure 2.
(A) Alignment of Vps5p with human (Hs) and nematode (Ce) SNX1. Sequences were aligned using the CLUSTAL program available at the embnet server (http://www.ch.embnet.org/index.html). There is extensive sequence conservation particularly over the PX domain and in the C-terminal half. Vps5p is larger than human or nematode SNX1 with unconserved sequences at the N-terminal end of the protein. (B) Schematic diagram of the truncation/deletion mutants of Vps5p and Vps17p. Vps5p was dissected using the PX domain, coiled-coil domain, and sequence homology as boundaries for making truncations.
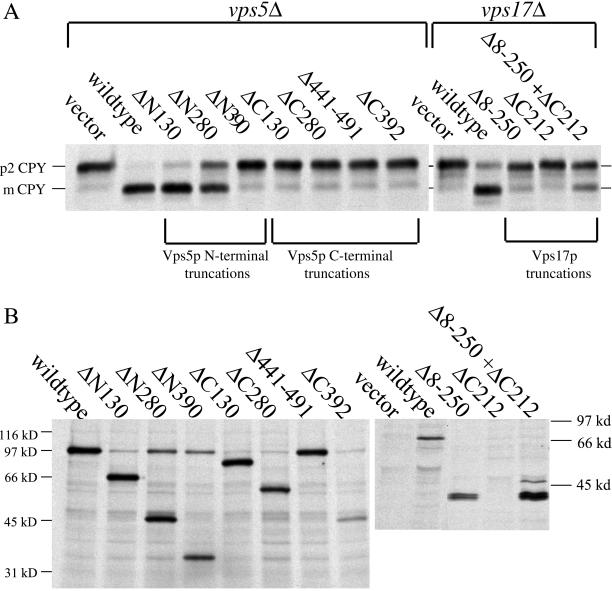
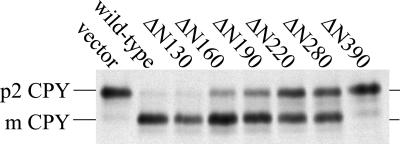
To test the Vps5p and Vps17p truncation mutants for function we have examined the ability of the various truncations to complement the CPY sorting defect in either vps5Δ or vps17Δ cells. A whole cell CPY sorting assay (see MATERIALS AND METHODS) was used in which both cell-associated and secreted CPY can be analyzed simultaneously. The Vps5p truncations were expressed from centromeric (CEN) plasmids to ensure expression levels comparable to wild-type levels but the Vps17 proteins were expressed from multicopy vectors. As shown in Figure 3a, deletion of any part of the C terminus of Vps5p results in the protein becoming nonfunctional and unable to complement the vps5Δ deletion mutant. This is in stark contrast to deletions of the N-terminal half. The ΔN130 construct retains almost 100% complementation activity, whereas the ΔN280 has ∼50% activity. The ΔN390 truncation that has lost the PX domain has no ability to complement the CPY sorting defect in vps5Δ cells. Truncation of Vps17p also results in Vps17p becoming nonfunctional. Interestingly, however, when both halves of the protein are expressed from different plasmids, there is a small degree of complementation observed.
Figure 3.
(A) CPY sorting of cells expressing Vps5p or Vps17p truncations. vps5Δ cells or vps17Δ cells were transformed with plasmids to express the various truncations in the respective null mutants. All Vps5p truncations were expressed from CEN plasmids, whereas Vps17p truncations were expressed from multicopy plasmids. Cells were pulse labeled with [35S]methionine for 10 min and then chased for 30 min. Lysates were generated by glass bead lysis and were then immunoprecipitated with antibodies against CPY. The immunoprecipitates were analyzed by SDS-PAGE/fluorography. Truncation of Vps5p from the C terminus renders the protein nonfunctional, whereas truncation from the N-terminus can be partially tolerated. Truncation of Vps17p renders the protein nonfunctional although expression of both halves of the protein separately restores some CPY sorting. (B) Vps5p truncations were expressed in wild-type cells from multicopy plasmids. Cells were pulse labeled with [35S]methionine for 10 min and chased for 30 min with excess unlabeled methionine. Vps5p was immunoprecipiated from lysates and subjected to SDS-PAGE. Vps17p truncations were expressed in vps17Δ cells. Vps17p was immunoprecipitated from lysates and subjected to SDS-PAGE. All Vps5p truncations seem to be expressed and stable. The Vps17p ΔC212 truncation was only detectable when coexpressed with the Δ8–250 truncation.
To assess the expression and stability of the various truncation mutants, multicopy plasmids containing the respective Vps5p truncation mutants were introduced into wild-type cells, and the Vps17p truncations were introduced into vps17Δ cells. In Figure 3b, cells have been pulse labeled with [35S]methionine and then chased for 30 min. Cells lysates were then immmunoprecipitated with antisera against Vps5p. All the Vps5p truncation mutants seem to be expressed and are stable over the 30-min chase period. This contrasts with the expression of the Vps17p truncation mutants. The Vps17pΔC212 mutant was undetectable unless expressed along with the Vps17pΔ8-250 truncation.
C-Terminal Half of Vps5p Interacts with C-Terminal Half of Vps17p
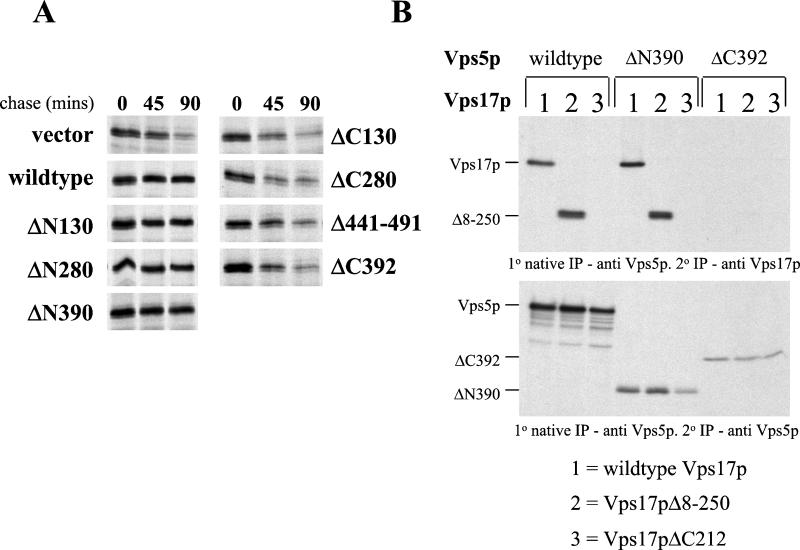
The truncations to the C-terminal half of Vps5p resulted in the protein becoming nonfunctional most likely because the coiled coils in the C-terminal half are responsible for mediating the interaction between Vps5p and either Vps17p or the other retromer components. Because Vps17p also has a region of coiled coils in its C-terminal half we examined the possibility that the C-terminal half of Vps5p interacts with Vps17p. It has been previously reported that Vps17p is unstable in a vps5Δ but not in any other retromer mutants (Seaman et al., 1998). Interaction with Vps5p is therefore necessary for Vps17p stability and hence Vps17p stability can be used to determine whether the various truncations of Vps5p are competent to interact with Vps17p. In Figure 4a we have used a pulse-chase assay to score the stability of Vps17p. In vps5Δ cells Vps17p is degraded with a half-time of ∼45 min. Wild-type Vps5p restores the stability of Vps17p as do the three N-terminal truncations. However, all of the C-terminal truncations result in Vps17p becoming unstable and degraded. This indicates that the two coiled-coils regions in Vps5p are both required to form a stable interaction with Vps17p.
Figure 4.
(A) C-terminal half of Vps5p is required for Vps17p stability. vps5Δ cells were transformed with CEN plasmids to express the various Vps5p truncations. Cells were labeled for 15 min with [35S]methionine and then chased for up to 90 min. Aliquots of cells were removed after 0 and 45 min. Lysates were generated using glass bead lysis and Vps17p was immunoprecipitated and subjected to SDS-PAGE/fluorography. In the absence of Vps5p, Vps17p is unstable and is degraded with a half-life of ∼45 min. Truncation of the C-terminus results in Vps5p being unable to rescue Vps17p stability, whereas truncation of the N-terminus has no effect. (B) C-terminal half of Vps5p interacts with the C-terminal half of Vps17p. vps5Δ vps17Δ double knockout cells were transformed with CEN plasmids to express the various N- and C-terminal truncations of Vps5p and Vps17p. Using the native immunoprecipitation assay (Figure 1), the interaction between Vps5p and Vps17p was examined and found to be mediated by the C-terminal halves of Vps5p and Vps17p.
Does the coiled-coils domain of Vps5p interact with the coiled-coils of Vps17p? To test this directly, we have generated vps5Δvps17Δ double mutant in which we can express the truncations of Vps5p and Vps17p in various combinations. vps5Δvps17Δ cells were transformed with CEN plasmids to express the wild-type genes and the N- and C-terminal truncation mutants of Vps5p and Vps17p in all the combinations possible. Spheroplasted cells were pulse labeled, chased, and then lysed under native conditions. The various truncations of Vps5p were immunoprecipitated. After washes, the immunoprecipitates were denatured and Vps17p antisera was added. The presence of Vps17p was determined by SDS-PAGE and fluorography. In Figure 4b (top), full-length Vps5p will coimmunoprecipitate with both full-length Vps17p and also the Vps17pΔ8–250 mutant but not the Vps17pΔC212 mutant. Similarly, the Vps5pΔN390 mutant was able to coimmunoprecipitate both full-length Vps17p and the Vps17pΔ8–250 mutant. In contrast, however, the Vps5pΔC392 mutant was unable to coimmunoprecipitate any of the Vps17p truncation mutants. These data are consistent with the stability experiment shown in Figure 4a and indicate that the Vps5p/Vps17p dimer is formed from the interactions between the respective coiled-coils domains.
N-Terminal Half of Vps5p Mediates Interaction with Other Retromer Components and Self-Assembly
The respective coiled-coils domains in the C-terminal halves of Vps5p and Vps17p seem to be both necessary and sufficient to mediate the interactions between the two proteins. What role then do the N-terminal halves play in Vps5p/Vps17p function? To address this question we made a more comprehensive series of truncations of Vps5p in which the region between residues 130–280 was dissected generating three additional mutants: ΔN160, ΔN190, and ΔN220. These new mutants were transformed into vps5Δ cells along with the other N-terminal vps5 mutants and their respective abilities to complement the CPY sorting defect present were determined by pulse-chase assay. In Figure 5 we find that truncation of Vps5p between residues 130–280 results in a progressively worse sorting defect such that the ΔN160 and ΔN190 mutants exhibit a 30–35% CPY sorting defect, whereas the ΔN220 and ΔN280 mutants have a 50% sorting defect. Immunoprecipitation of the Vps5p N-terminal truncation mutants from [35S]methionine labeled lysates revealed that all the truncation mutants are equally expressed and stable and that the progressive CPY sorting defect is not due to progressively worse stability of the N-terminal truncation mutants (our unpublished data).
Figure 5.
Truncation of the N terminus of Vps5p results in a progressively worse CPY sorting defect. Additional truncations of the N-terminal half of Vps5p were generated. CEN plasmids to express the truncations were transformed into vps5Δ cells and CPY sorting was tested. Truncation of Vps5p beyond amino acid 130 results in a defect in CPY sorting, which becomes worse as more of the N-terminus is truncated.
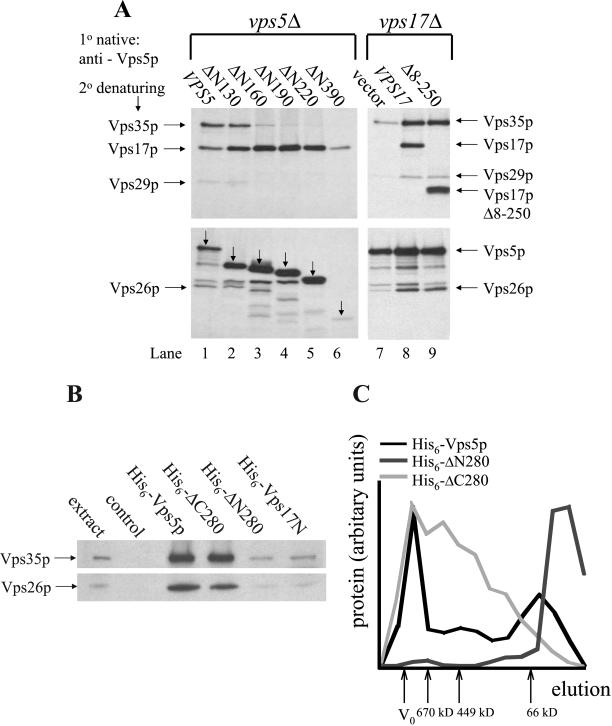
One possibility to explain the progressive CPY sorting defect displayed by the N-terminal Vps5p truncations is that the N-terminal half of Vps5p mediates interactions with the other components of retromer, namely, Vps35p, Vps29p, and Vps26p. We have tested this hypothesis by performing native immunoprecipitations from vps5Δ cells expressing the N-terminal truncations. In Figure 6a we find that the ΔN130 mutant is able to coimmunoprecipitate the other members of retromer in a manner similar to wild-type Vps5p (compare lanes 1 and 2). This is consistent with the observation that the ΔN130 mutant is fully able to complement the CPY sorting defect in vps5 mutants. Further truncation of the N-terminal half of Vps5 results in a weakening of the interactions with the other components of retromer such that Vps35p cannot be detected in the native immunoprecipitates from cells expressing the ΔN220 mutant (lane 5). Vps17p continues to coimmunoprecipitate with the N-terminal truncations of Vps5p in the absence of interactions with Vps35p/Vps29p/Vps26p. Fluctuations in the amount of Vps17p present correlate with the amount of Vps5p that is immunoprecipitated consistent with the observations in Figure 4 that demonstrate the role of the C-terminal domains of Vps5p and Vps17p in mediating the interaction of these two proteins.
Figure 6.
(A) Truncation of Vps5p N-terminal half ablates the interaction with Vps35p, Vps29p, and Vps26p. vps5Δ or vps17Δ cells expressing the respective truncations from CEN plasmids were pulse labeled for 20 min with [35S]methionine and chased for 40 min with excess unlabeled methionine. After lysis, Vps5p was immunoprecipitated under native conditions. Vps35p, Vps17p, Vps29p, Vps26p, and also Vps5p (indicated with the downward pointing arrows) were subsequently immunoprecipitated under denaturing conditions and then subjected to SDS-PAGE. The N-terminal half of Vps5p seems to be necessary for interactions with other retromer components, whereas binding of Vps17p (full length or truncated) to Vps5p seems to facilitate the interactions with Vps35p, Vps29p, and Vps26p. (B) Recombinant His6-tagged Vps5p can bind to Vps35p and Vps26p. Bacterially expressed His6-fusion proteins purified on Ni2+-agarose were incubated with a yeast lysate. After several washes of the Ni2+-agarose, the bound proteins were eluted with 250 mM imidazole, precipitated, and then subjected to SDS-PAGE. Vps35p and Vps26p were detected by Western blotting. The N-terminal half of Vps5p is sufficient for interactions with Vps35p and Vps26p. (C) Self-assembly activity is mediated by the N-terminal half of Vps5p. Bacterially expressed His6-fusion proteins purified on Ni2+-agarose were dialyzed against cytosol buffer and then size fractionated on a Sephacryl S300 column. Fractions were analyzed for fusion protein by SDS-PAGE. The gels were scanned and the amount of fusion protein present in each fraction quantitated using NIH Image software.
This finding suggests that the N-terminal half of Vps5p provides the binding activity required for interactions with the other retromer complex components. However, as shown in Figure 1 in the absence of Vps17p, Vps5p can only weakly interact with the other retromer proteins. We therefore tested the effect that Vps17p has upon the interaction between Vps5p and Vps35p/Vps29p/Vps26p by using the Vps17pΔ8-250 truncation that we have shown is necessary and sufficient to interact with Vps5p. In Figure 6a (lanes 7–9) Vps5p has been immunoprecipitated under native conditions and the association of the other retromer components examined by denaturing IP. We find that in the absence of Vps17p, Vps5p only weakly interacts with the remaining members of retromer, but in the presence of either wild-type Vps17p or the Vps17pΔ8–250 truncation, Vps5p can interact strongly with the other retromer components.
These data indicate that the N-terminal half of Vps5p is responsible for binding other members of the retromer complex. This was tested directly by production of His6-tagged fusion proteins in bacteria. Three fusion proteins of Vps5p were produced: full-length Vps5p, the N-terminal half corresponding to amino acids 1–395 (ΔC280), and the C-terminal half corresponding to 281–675 (ΔN280). The N-terminal half of Vps17p was also produced in bacteria as a His6-tagged fusion. The fusion proteins were isolated from 500 ml of bacterial culture, full-length Vps5p, and the N-terminal half of Vps5p are both poorly expressed relative to the Vps5p C-terminal half and the Vps17p N-terminal half (our unpublished data). After capturing the fusion proteins on Ni2+-agarose, the proteins were added to a yeast lysate from protease deficient cells. After binding at 4°C, the Ni2+-agarose was washed extensively and the bound proteins eluted with 250 mM imidazole. After precipitation, the proteins were subjected to SDS-PAGE and then Western blotting. In Figure 6b we find that full-length Vps5p and the N-terminal half can both strongly bind to Vps35p and Vps26p. The Vps5p C-terminal half and Vps17p N-terminal halves did not strongly interact with Vps35p or Vps26p, although a small amount of Vps35p and Vps26p was detected by Western blotting. This experiment confirms that the N-terminal half of Vps5p is both necessary and sufficient to bind to the other members of retromer.
Previously, we have reported that Vps5p can self-assemble in vitro (Seaman et al., 1998). This property has been also demonstrated for SNX1 (Renfrew-Haft et al., 2000; Kurten et al., 2001) and therefore may play an important role in sorting nexin function. We therefore investigated which domain of Vps5p was necessary for its self-assembly. Wild-type Vps5p, ΔN280, and ΔC280 were expressed as His6-tagged fusion proteins in bacteria, purified, and dialyzed against cytosol buffer. The protein was then size fractionated on a Sephacryl S300 column, and the fractions were analyzed for the presence of the fusion protein. In Figure 6c, the elution profiles of the His-tagged fusion proteins are shown. Vps5p was able to self-assemble and eluted in fractions consistent with it forming a complex >1000 kDa. The ΔN280 protein was unable to self-assemble and eluted in later fractions consistent with the predicted size of the monomeric protein. The ΔC280 protein was able to self-assemble but did not seem to form particles of a uniform size like the full-length Vps5p and hence the elution profile did not correspond to a sharp peak in the early fractions. These data indicate that the N-terminal half of Vps5p is also necessary for self-assembly activity.
Membrane Targeting of Vps5p Requires Interaction with Vps17p and N-Terminal Domain
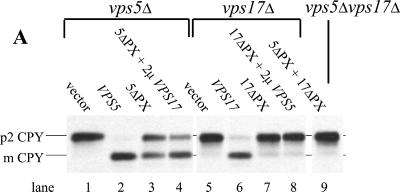
One of the characteristic properties of the family of sorting nexin proteins is that they are peripherally associated with membranes and are often found in both membrane and cytosolic fractions. Indeed, they have been proposed to cycle on and off the membrane as part of their function (Kurten et al., 2001). How this is achieved is not yet known. We have examined the membrane association of Vps5p by localizing Vps5p in various mutants and by investigating which domain of Vps5p is required for membrane association. Because PX domains have been shown to bind PtdIns 3-P (Cheever et al., 2001; Song et al., 2001; Xu et al., 2001; Yu and Lemmon, 2001), the PX domain is the most likely candidate for mediating the membrane association of Vps5p/Vps17p. The PX domains of Vps5p and Vps17p were deleted (Figure 2b) and the function of the respective truncations examined by CPY sorting. Deletion of the Vps5p PX domain resulted in a protein that could complement the CPY sorting defect present in the vps5Δ strain by ∼50% (Figure 7a, lane 3). Overexpression of Vps17p slightly improved the CPY sorting of the 5ΔPX mutant (compare lanes 3 and 4). Deletion of the Vps17p PX domain had a more pronounced effect upon the ability of the truncated protein to complement the CPY sorting defect in the vps17Δ strain (Figure 7a, lane 7). The 17ΔPX construct could not support CPY sorting any better than empty vector. Overexpression of Vps5p with the 17ΔPX construct also did not result in correct CPY sorting. Both the 5ΔPX and 17ΔPX constructs were properly expressed and were found to be stable (our unpublished data). The 5ΔPX could also stabilize Vps17p in a vps5Δ background and therefore could associate with Vps17p (our unpublished data).
Figure 7.
(A) Deletion of the Vps5p and Vps17p PX domains exhibit different CPY sorting activity. Cells expressing the PX domain deletions with subjected to the whole cell CPY sorting assay. The 5ΔPX construct retained 50% CPY sorting activity, whereas the 17ΔPX mutant was unable to complement the CPY sorting defect in vps17Δ cells. (B) Membrane association of Vps5p seems to require binding by Vps17p and the N-terminal half of the protein. Approximately 5 OD600 nm equivalents of spheroplasted cells were labeled with [35S]methionine, chased, and then lysed. The lysates were spun at 13,000 × g to remove larger membranes such as vacuoles, endoplasmic reticulum, and plasma membrane. The supernatant was then further centrifuged at 100,000 × g for 1 h to pellet small membranes such as endosomes, vesicles, and Golgi membranes. Proteins from the supernatant (S) and pellet (P) fractions were precipitated and a lysate generated. Vps5p was recovered from the lysate by immunoprecipitation and subjected to SDS-PAGE and fluorography. The resulting autoradiogram was scanned and quantitated using NIH Image software.
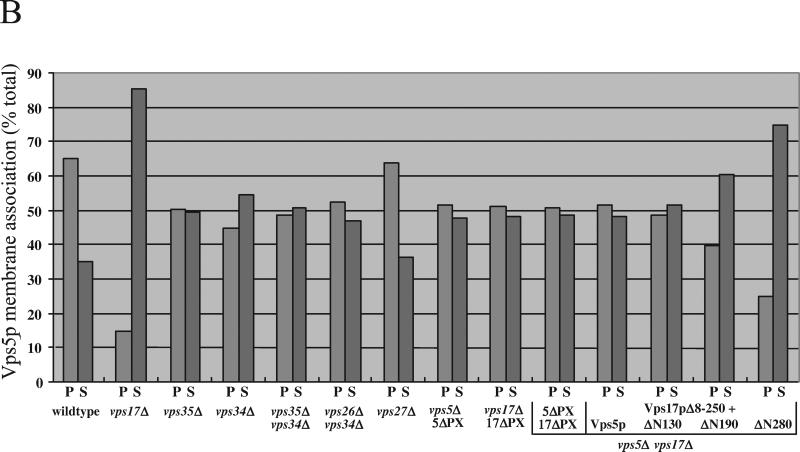
Vps5p has been previously localized to a 100,000 × g membrane pellet and also the cytosolic fraction by using a differential centrifugation assay (Horazdovsky et al., 1997; Nothwehr and Hindes, 1997). The membranes in this P100 are predominately Golgi, endosomal, and small vesicles. In Figure 7b, we find that in wild-type cells, Vps5p is indeed predominately localized to the P100 fraction with ∼65% being present in the P100. However, this membrane association is dependent upon Vps17p as was originally reported by Horazdovsky et al. (1997). We next examined the membrane association of Vps5p in various deletion strains. Vps5p was found to be equally divided between the P100 and S100 fractions in vps35Δ mutants. A similar effect was upon the Vps5p membrane association was observed in vps34Δ mutants in which there will be no PtdIns 3-P present (Herman and Emr, 1990; Stack et al., 1995). Deletion of both VPS35 and VPS34 together or VPS26 and VPS34 did not result in more Vps5p becoming soluble. Similarly, the role for PtdIns 4-P was tested using a temperature conditional PtdIns 4-kinase (PIK1) mutant (Hama et al., 1999) but was found to have no apparent effect (our unpublished data). Additionally, deletion of VPS27, the yeast equivalent of the mammalian protein Hrs1 resulted in Vps5p membrane association no different from wild-type cells.
Next, we examined the membrane association of the Vps5pΔPX protein and Vps5p in cells expressing the 17ΔPX construct. The 5ΔPX protein was found equally in both the P100 and S100 fractions. Similarly, the expression of the 17ΔPX protein in vps17Δ cells resulted in Vps5p being present both in pellet and soluble fractions. Even when both the 5ΔPX and 17ΔPX constructs were expressed together in vps5Δvps17Δ double mutants, the membrane association of the 5ΔPX protein was no worse than in cells expressing the single PX domain deletions. The effect of deletion of Vps17p upon Vps5p membrane association suggested that interaction with Vps17p may be the key to Vps5p membrane association. Therefore, we tested whether just the C-terminal half of Vps17p (Vps17pΔ8-250) was sufficient to promote the membrane association of Vps5p. This domain of Vps17p is sufficient to interact with Vps5p (Figure 4). vps5Δvps17Δ cells were transformed with the Vps17pΔ8-250 construct along with either full-length Vps5p or various N-terminal truncations. As shown in Figure 7b, full-length Vps5p with the Vps17pΔ8-250 is split equally between the P100 and the S100, as is the Vps5pΔN130 mutant. However, the Vps5pΔN190 and Vps5pΔN280 are progressively less P100 associated. These mutants are also unable to interact with the other members of the retromer complex (Figure 6a). However, deletion of VPS35 did not have a strong effect upon Vps5p membrane association and therefore lack of interaction between Vps5p and Vps35p/Vps29p/Vps26p is unlikely to be the cause of the loss of membrane association of the Vps5pΔN190 and Vps5pΔN280 constructs. In summary, only deletion of Vps17p, or truncation of Vps5p from the N terminus was able to significantly shift Vps5p into the cytosolic fraction.
DISCUSSION
Sorting nexins are a recently discovered large family of conserved proteins that are believed to be important components of the molecular machinery responsible for sorting membrane proteins in the trans-Golgi network/endocytic system (Kurten et al., 1996; Renfrew-Haft et al., 1998). We have attempted to functionally define the different domains of the yeast sorting nexins, Vps5p and Vps17p. By dissecting Vps5p and Vps17p and expressing the various truncations in a null background, we have identified the respective domains of Vps5p and Vps17p that are responsible for their interaction with each other, and the Vps5p domain that mediates the interaction with the other members of the retromer complex. We have also examined the role of the respective domains of Vps5p in targeting Vps5p to the membrane.
Vps5p is homolog to SNX1 (Figure 2a), the first sorting nexin identified (Kurten et al., 1996). Truncations of Vps5p were designed by comparing the sequences of the two proteins and by using the PX domain and the predicted coiled-coils domain of Vps5p as boundaries. Vps17p is less well conserved but does conform to the sorting nexin prototype of having an N-terminal PX domain and predicted coiled-coils in the C-terminal half. Relatively simple truncations of Vps17p were generated so that the protein could be bisected to separate the N-terminal PX domain-containing portion from the C-terminal coiled-coiled domain.
The respective truncations were expressed in vps5Δ or vps17Δ cells, and their ability to complement the CPY sorting defect was examined (Figure 3a). All of the Vps5p C-terminal truncations were completely nonfunctional and resulted in CPY sorting no different from the null mutant. Truncations of the Vps5p N-terminal half, on the other hand, retained a significant amount of function. Deletion of the N-terminal 130 amino acids seemed to have virtually no effect upon the Vps5 protein, whereas deletion of the N-terminal 280 amino acids produced a truncation that retained ∼50% CPY sorting ability. Vps17p truncations were both nonfunctional, although interestingly when both were expressed together from different plasmids, the two domains could partially rescue the CPY sorting defect. All of the Vps5p truncations seemed to be expressed, stable, and detectable by immunoprecipitation after a 30-min chase period (Figure 3b). Differences in the apparent expression are most likely due to differences in the numbers of methionine and cysteine residues present in the truncated protein (most Met and Cys residues are in the C-terminal half) and also due to the preference of the antiserum for certain epitopes. The Vps17p truncations on the other hand displayed very different stability. The removal of the N-terminal domain in the Δ8–250 mutant did not significantly affect the stability of the resulting Vps17p truncation, but removal of the C-terminal domain, leaving only the N-terminal half (ΔC212), caused the protein to be completely undetectable when expressed in the absence of any other Vps17 protein. The N-terminal half was marginally stabilized and detectable when coexpressed in trans with the C-terminal half of Vps17p, which correlated with partial rescue of the CPY sorting defect.
The C-terminal truncations of Vps5p were found to be nonfunctional in the CPY sorting assay. Given that this region of Vps5p contains predicted coiled-coils domains, we tested the possibility that these domains were responsible for interactions with Vps17p. This indeed was the case. Using the stability of Vps17p in a vps5Δ mutant as an assay we found that the C-terminal truncations could not rescue Vps17p stability, whereas the N-terminal truncations were like wild-type Vps5p in this respect (Figure 4a). Furthermore, using the native immunoprecipitation assay, we found that the C-terminal half of Vps5p was sufficient to interact with both full-length Vps17p and also a truncation of Vps17p (Δ8–250) that lacked the N-terminal half and PX domain but retained the coiled-coils domain (Figure 4b). Thus, in the case of the yeast sorting nexins Vps5p and Vps17p, their interaction with each other seems to be mediated solely by the coiled-coils domains. This may in fact be generally true for all sorting nexins because most proteins that have been classified as Snxs have a region of predicted coiled-coils in the C-terminal half of the protein.
When comparing the sequences of Vps5p and its homolog SNX1, it is interesting to note that the homology between Vps5p and SNX1 is not apparent in the first ∼100 amino acids of Vps5p. It is therefore not surprising that the N-terminal 130 amino acids seem to be dispensable for Vps5p function. The role that this piece of Vps5p plays in its function as a sorting nexin and retromer component remains to be determined. Perhaps it provides a binding site for an accessory protein that has yet to be identified and is unique to yeast, or alternatively it may play a subtle role in binding of the other components of retromer but is not essential for this function. Further truncation of the N-terminal half of Vps5p resulted in a progressively worse CPY sorting defect, which was found to correlate with a weakening of the interaction between Vps5p and the Vps35p/Vps29p/Vps26p components of retromer (Figures 5 and 6a). None of the vps5 N-terminal truncation mutants demonstrated any preference toward binding Vps35p/Vps29p/Vps26p and when interactions between Vps5p and Vps35p/Vps29p/Vps26p are affected, all interactions are affected equally, indicating that Vps35p/Vps29p/Vps26p binds to Vps5p/Vps17p as a single entity. Our data are also consistent with the hypothesis that Vps5p binds directly to Vps35p/Vps29p/Vps26p but we cannot formally rule out the possibility that another protein is also involved in this interaction. This seems unlikely, however, because previous cross-linking studies (Seaman et al., 1998) did not indicate the presence of any additional proteins interacting with the retromer complex. These observations extend the studies of Renfrew-Haft et al., (2000) in which they mapped binding sites for SNX1, mammalian VPS26, and VPS29 onto mammalian VPS35.
Interestingly, although Vps17p seems not to interact directly with Vps35p/Vps29p/Vps26p, it clearly facilitates the interaction between Vps5p and Vps35p/Vps29p/Vps26p. Truncation of Vps17p to remove the N-terminal half and PX domain resulted in a protein that could interact with Vps5p and apparently synergize the interaction between Vps5p and Vps35p/Vps29p/Vps26p (Figure 6a). We cannot formally rule out the possibility that the C-terminal half of Vps17p contributes to the binding of Vps35p/Vps29p/Vps26p but we presently have no evidence to suggest this.
The N-terminal half of Vps5p was subsequently found to be both necessary and sufficient for interactions with Vps35p/Vps29p/Vps26p when His6-tagged Vps5p fusion proteins were used to fish for interactions with Vps35p and Vps26p. However, it is somewhat surprising that even though the ΔN220 Vps5p mutant was unable to interact with Vps35p/Vps29p/Vps26p (Figure 6a) it still retained ∼50% CPY sorting activity (Figure 5). This suggests that perhaps the retromer complex need not be a stable single entity for its function. The Vps35p/Vps29p/Vps26p subcomplex may be able to perform its function of cargo selection without interactions with the Vps5p/Vps17p subcomplex. Vps5p/Vps17p could then promote vesicle formation through the intrinsic self-assembly activity of Vps5p. In this scenario, endosome-to-Golgi retrieval could still occur, albeit in a somewhat inefficient manner, resulting in a CPY sorting defect. It is worth remembering that the class B vps5 and vps17 mutants have a more severe vacuole protein sorting phenotype than the class A vps35 and vps29 mutants (Raymond et al., 1992). These and other phenotypic differences led to the original hypothesis that Vps35p/Vps29p/Vps26p perform cargo selection, whereas Vps5p/Vps17p provide the mechanical force to bud a vesicle (Seaman et al., 1998).
The N-terminal half of Vps5p seems to also be important for the self-assembly of the protein. Gel filtration experiments revealed that the C-terminal half of the protein (ΔN280) could not self-assemble, whereas wild-type Vps5p and the N-terminal half (ΔC280) were able to assemble into larger particles, although the particles produced by the assembly of the N-terminal half were not of a uniform size possibly due to folding problems of the fusion protein. Both the full-length Vps5p and the ΔC280 were generally poorly expressed and significantly proteolyzed during expression.
Sorting nexins have been reported to be peripherally associated with endosomal membranes (Teasdale et al., 2001) and can rapidly cycle on and off the membranes (Kurten et al., 2001). We examined membrane association of Vps5p in various deletion mutants and by using the Vps5p truncation mutants. Consistent with previous observations (Horazdovsky et al., 1997) we have found that binding of Vps17p by Vps5p is essential for its membrane association. Deletion of VPS17 shifts almost all of the Vps5p into the cytosolic fraction. Deletion of Vps35p resulted in Vps5p becoming equally divided between the membrane and cytosolic fractions. The native immunoprecipitation assay in Figure 1 showed that for Vps5p to interact with other members of retromer, Vps35p is necessary. These data therefore suggest that the Vps5p/Vps17p interaction is more important for membrane association than interaction between Vps5p and Vps35p/Vps29p/Vps26p. This assertion is supported by the observation that expression of the Δ8-250 Vps17 protein, which lacks the N-terminal and PX domain to Vps17p is sufficient to restore Vps5p membrane association to close to wild-type levels.
The role of PtdIns 3-P in regulating the membrane association of Vps5p remains unclear. There is compelling evidence that PX domains bind PtdIns 3-P (Cheever et al., 2001; Song et al., 2001; Xu et al., 2001; Yu and Lemmon, 2001) and yet deletion of the only yeast PtdIns 3-kinase, Vps34p (Stack et al., 1995), singularly or in combination with deletion of Vps35p or Vps26p, seemed to have only a modest effect upon membrane association of Vps5p (Figure 7b). Likewise, the deletion of the PX domains of either Vps5p or Vps17p did not cause Vps5p to become soluble. Deletion of the Vps5p PX domain resulted in a 50% CPY sorting defect (Figure 7a), suggesting that the 5ΔPX mutant can retain some ability to complement the vps5Δ mutant. Deletion of the Vps17p PX domain had a more pronounced effect upon the function of the Vps17 protein but surprisingly did not cause Vps5p to become soluble when the 17ΔPX protein was expressed in vps17Δ cells. In fact, deletion of both the Vps5p and Vps17p PX domains did not result in Vps5p becoming soluble. These data therefore argue that either PtdIns 3-P binding by Vps5p/Vps17p may have more subtle effects than simply recruitment of the complex onto an endosomal membrane or that other regions of Vps5p/Vps17p outside of the PX domains can contribute to membrane association. Binding of PtdIns 3-P by the Vps5p and/or Vps17p may in fact be comparatively weak. Indeed, a study by Song et al. (2001) has shown that the Vps5p PX domain has no affinity for PtdIns 3-P in vitro, whereas the Vam7p PX domain binds strongly to PtdIns 3-P. Another study by Lemmon and colleagues did report an interaction between the Vps5p PX domain and PtdIns 3-P, but this interaction was also classified as being weak (Yu and Lemmon, 2001).
Studies on mammalian SNX1 have revealed an interaction between SNX1 and Hrs1, the mammalian ortholog of Vps27p (Chin et al., 2001). Vps27p function has also been shown to be important for endosomal morphology and has been implicated in trafficking to and from endosomes (Piper et al., 1995). We, however, found that deletion of VPS27 had no effect on Vps5p membrane association (Figure 7b).
Although deletion of the Vps5p and/or the Vps17p PX domain did not seem to result in Vps5p becoming cytosolic, truncation of the N-terminal half of Vps5p did result in less Vps5p binding to the membrane. When Vps5p membrane association in vps5Δvps17Δ cells expressing N-terminally truncated Vps5p and the Δ8-250 Vps17p truncation was examined it was found that the Vps5p ΔN190 and ΔN280 constructs were progressively less associated with the membrane (Figure 7b). These data therefore suggest that the interaction between Vps5p and its partner Vps17p is most important for membrane association and that sequences in the N-terminal half of Vps5p may also contribute to membrane association.
It is interesting that the region of Vps5p that seems to be important for membrane binding in our experiments is also the region that may drive self-assembly of Vps5p. The self-assembly of Vps5p might therefore facilitate the membrane targeting of Vps5p by creating Vps5p oligomers that collectively could have a high affinity for PtdIns 3-P, and therefore Vps5p could be recruited/stabilized on the membrane. Given that deletion of Vps34p (and therefore abolition of all PtdIns 3-kinase activity) did not dramatically shift Vps5p into the cytosolic fraction, the role of PtdIns 3-P in directing the membrane association of Vps5p is likely to be more subtle than simply providing a binding site at the endosomal membrane. These subtle roles for PtdIns 3-P binding by Vps5p/Vps17p PX domains include directing interactions between Vps5p/Vps17p and other proteins and/or influencing the kinetics of vesicle budding. This may however be a rather “protein-centric” view to take. One intriguing possibility is that the binding of PtdIns 3-P by the Vps5p/Vps17p PX domains is actually required to cluster the lipid in the membrane and not to recruit the protein. The lipid may have an important role in the membrane dynamics of budding a vesicle, or might function downstream in subsequent vesicle docking/fusion.
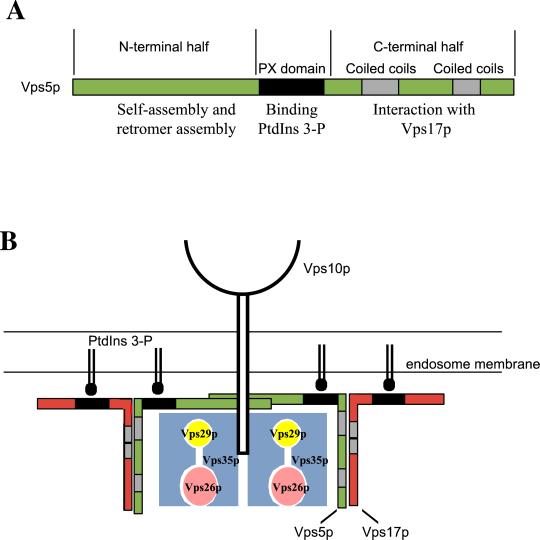
The interactions that Vps5p undergoes with its sorting nexin partner Vps17p and other members of the retromer complex are summarized schematically in Figure 8a. The C-terminal half of Vps5p that contains the coiled-coils domains mediates the interaction with Vps17p. The N-terminal half interacts with Vps35p/Vps29p/Vps26p and also drives the self-assembly of Vps5p. Membrane targeting and assembly of Vps5p into retromer complexes are likely to be the sum of many interactions with both proteins and lipid. Our data are consistent with a model shown in Figure 8b in which self-assembly interactions of Vps5p facilitate or promote the stable interaction with PtdIns 3-P and may also be important for subsequent interactions with other retromer components. The assembly of Vps5p could create multiple binding sites for PtdIns 3-P and thereby stabilize what might be an otherwise weak interaction. One of the key conclusions we can make from our studies is the importance of the interaction between Vps5p and Vps17p for sorting nexin function. Although Vps5p alone can bind the other members of retromer, this interaction is greatly enhanced when Vps5p is able to interact with Vps17p. Similarly, the Vps5p/Vps17p dimer is stabilized on the membrane. Therefore, our view on sorting nexin function should take into account the requirement for these proteins to function as dimers.
Figure 8.
Schematic model of the interactions of Vps5p with retromer and the role that Vps5p/Vps17p plays in retromer-mediated endosome to Golgi retrieval. (A) Interactions between Vps5p and its Snx partner Vps17p are mediated by the C-terminal halves of the respective proteins. The N-terminal half of Vps5p binds to the Vps35p/Vps29p Vps26p retromer subcomplex and also seems to be important for self-assembly activity. (B) Multiple interactions between oligomeric Vps5p/Vps17p and the membrane (possibly via interactions with PtdIns 3-P) facilitate the membrane binding of the Snx dimer and also stabilize the interactions between Vps5p and Vps35p/Vps29p/Vps26p.
What role might the C-terminal half of Vps5p play in membrane association? This question is prompted by the studies by Teasdale et al., (2001), which indicated that the C-terminal half of SNX1 targets the protein to the membrane. Because these studies were conducted by transfection of GFP-Snx chimaeras into mammalian cells containing the endogenous sorting nexins, we believe that these results were due to the C-terminal half of SNX mediating the interaction with its Snx partner. Expression of the C-terminal half alone would therefore target the GFP chimaera to the membrane through coiled-coils interactions of the respective Snx proteins.
The function of the N-terminal half of Vps17p also remains to be determined. Given that truncation of the N-terminal half results in a Vps17 protein that can bind to Vps5p and promote the interactions between Vps5p and the Vps35p/Vps29p/Vps26p and can also promote the membrane association of Vps5p, the N-terminal deletion of Vps17p (Vps17pΔ8-250) is in these respects just like wild-type Vps17p. Clearly, however, the N-terminal half of Vps17p is necessary for function as the Δ8-250 truncation cannot rescue the CPY sorting defect of a vps17Δ mutant but its precise role remains elusive. An interaction between the N-terminal half of Vps17p and an unidentified accessory protein is one possibility or perhaps this domain of Vps17p performs an essential regulatory role that may effect vesicle budding or shedding of the coat.
In these studies we have identified the functional domains of the yeast SNX1 homolog Vps5p. There is much yet to learn about the function of the sorting nexin family of proteins and how they direct the sorting and membrane trafficking of membrane proteins in the endocytic system. The data presented will we believe, provide a template for further studies to refine our knowledge of the precise role that sorting nexins play in membrane trafficking.
ACKNOWLEDGMENTS
We thank Scottie Robinson and Paul Luzio for critical reading of this manuscript and for helpful discussions during the course of this study. We are also indebted to Scott Emr for providing many reagents essential to this study. This work was funded by the Wellcome Trust.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.02–05–0064. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.02–05–0064.
REFERENCES
- Burd CG, Emr SD. Phosphotidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- Cheever ML, Sato TK, de Beer T, Kutateladze TG, Emr SD, Overduin M. Phox domain interaction with PtdIns(3) P targets the Vam7 t-SNARE to vacuole membranes. Nat Cell Biol. 2001;3:613–618. doi: 10.1038/35083000. [DOI] [PubMed] [Google Scholar]
- Chin L-S, Raynor MC, Wei X, Chen H-Q, Li L. Hrs interacts with sorting nexin 1 and regulates degradation of epidermal growth factor receptor. J Biol Chem. 2001;276:7069–7078. doi: 10.1074/jbc.M004129200. [DOI] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Hama H, Schnieders EA, Thorner J, Takemoto JY, DeWald DB. Direct Involvement of Phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J Biol Chem. 1999;274:34294–34300. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherchia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Herman PK, Emr SD. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6742–6754. doi: 10.1128/mcb.10.12.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky BF, Davies BA, Seaman MNJ, McLauglin SA, Yoon S-H, Emr SD. A sorting nexin-1 homologue, Vps5p, forms a complex with Vps17p and is required for recycling the vacuolar protein-sorting receptor. Mol Biol Cell. 1997;8:1529–1541. doi: 10.1091/mbc.8.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T, Bonifacino JS, Riezman H. Linking cargo to vesicle formation: receptor tail interactions with coat proteins. Curr Opin Cell Biol. 1997;9:488–495. doi: 10.1016/s0955-0674(97)80024-5. [DOI] [PubMed] [Google Scholar]
- Kurten RC, Cadena DL, Gill GN. Enhanced degradation of EGF receptors by a sorting nexin, SNX1. Science. 1996;272:1008–1010. doi: 10.1126/science.272.5264.1008. [DOI] [PubMed] [Google Scholar]
- Kurten RC, Eddington AD, Chowdhury P, Smith RD, Davidson AD, Shank BB. Self-assembly and binding of a sorting nexin to sorting endosomes. J Cell Sci. 2001;114:1743–1756. doi: 10.1242/jcs.114.9.1743. [DOI] [PubMed] [Google Scholar]
- Nothwehr SF, Hindes AH. The yeast VPS5/GRD2 gene encodes a sorting nexin-1-like protein required for localizing membrane proteins to the late Golgi. J Cell Sci. 1997;110:1063–1072. doi: 10.1242/jcs.110.9.1063. [DOI] [PubMed] [Google Scholar]
- Otsuki T, Kajigaya S, Ozawa K, Liu JM. SNX5, a new member of the sorting nexin family, binds to the fanconi anemia complementation group A protein. Biochem Biophys Res Comm. 1999;265:630–635. doi: 10.1006/bbrc.1999.1731. [DOI] [PubMed] [Google Scholar]
- Parks WT, et al. Sorting Nexin 6, a novel SNX interacts with the TGF-b family of receptor serine threonine kinases. J Biol Chem. 2001;276:19332–19339. doi: 10.1074/jbc.M100606200. [DOI] [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Yang H, Stevens TH. J. Cell Biol. 131, 603–617. 1995. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP. Novel domains in NADPH oxidase subunits, sorting nexins, and PtdIns 3-kinases: binding partners of SH3 domains? Protein Sci. 1996;5:2353–2357. doi: 10.1002/pro.5560051122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E Vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy JV, Seaman MNJ. Vps26p, a component of retromer, directs the interactions of Vps35p in Endosome-to-Golgi retrieval. Mol Biol Cell. 2001;12:3242–3256. doi: 10.1091/mbc.12.10.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfrew-Haft C, Sierra M, Bafford R, Lesniak MA, Barr VA, Taylor SI. Human orthologs of yeast vacuolar protein sorting proteins Vps26, 29, and 35. Assembly into multimeric complexes. Mol Biol Cell. 2000;11:4105–4116. doi: 10.1091/mbc.11.12.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfrew-Haft C, Sierra M de la luz, Barr VA, Haft DH, Taylor SI. Indentification of a family of sorting nexin molecules and characterization of their association with receptors. Mol Cell Biol. 1998;18:7278–7287. doi: 10.1128/mcb.18.12.7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS. Coats and vesicle budding. Trends Cell Biol. 1997;7:99–102. doi: 10.1016/S0962-8924(96)10048-9. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE, Orci L. Molecular dissection of the secretory pathway. Nature. 1992;355:409–15. doi: 10.1038/355409a0. [DOI] [PubMed] [Google Scholar]
- Schekman R, Orci L. Coat proteins and vesicle budding. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- Seaman MNJ, Marcusson EG, Cereghino J-L, Emr SD. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of VPS29, VPS30 and VPS35 gene products. J Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MNJ, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Xu W, Zhang A, Huang G, Liang X, Virbasius JV, Czech MP, Zhou GW. Phox homology domains specifically bind phosphatidylinositol phosphates. Biochemistry. 2001;40:8940–8944. doi: 10.1021/bi0155100. [DOI] [PubMed] [Google Scholar]
- Stack JH, De Wald DB, Takegawa K, Emr SD. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J Cell Biol. 1995;129:321–334. doi: 10.1083/jcb.129.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale RD, Loci D, Houghton F, Karlsson L, Gleeson PA. A large family of endosome-localized proteins related to sorting nexin 1. Biochem J. 2001;358:7–16. doi: 10.1042/0264-6021:3580007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser AE, Emr SD. Phosphoinositide signaling and turnover: Ptdlns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J. 1998;17:4930–4942. doi: 10.1093/emboj/17.17.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Hortsman H, Seet L, Wong SH, Hong W. SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3) P. Nat Cell Biol. 2001;3:658–666. doi: 10.1038/35083051. [DOI] [PubMed] [Google Scholar]
- Yu JW, Lemmon MA. All Phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphotidylinositol 3-phosphate. J Biol Chem. 2001;276:44179–44184. doi: 10.1074/jbc.M108811200. [DOI] [PubMed] [Google Scholar]