Abstract
Protein import into chloroplasts is mediated by a protein import apparatus located in the chloroplast envelope. Previous results indicate that there may be multiple import complexes in Arabidopsis. To gain further insight into the nature of this multiplicity, we analyzed the Arabidopsis ppi1 and ppi2 mutants, which are null mutants of the atToc33 and atToc159 translocon proteins, respectively. In the ppi2 mutant, in contrast to the extremely defective plastids in mesophyll cells, chloroplasts in guard cells still contained starch granules and thylakoid membranes. The morphology of root plastids in both mutants was similar to that in wild type. After prolonged light treatments, root plastids of both mutants and the wild type differentiated into chloroplasts. Enzymatic assays indicated that the activity of a plastid enzyme was reduced only in leaves but not in roots. These results indicated that both the ppi1 and ppi2 mutants had functional root and guard cell plastids. Therefore, we propose that import complexes are cell type specific rather than substrate or plastid specific.
Most proteins in chloroplasts are nuclear encoded and imported from the cytosol. The chloroplast protein import process is initiated by specific interactions between transit peptides of precursor proteins and the chloroplast protein import machinery in the envelope, followed by translocation of precursor proteins across the envelope. Several components in the machinery have been identified. They are collectively named as Tic (translocon at the inner envelope membrane of chloroplasts) and Toc (translocon at the outer envelope membrane of chloroplasts) proteins (Schnell et al., 1997). Three major Toc proteins, Toc34, Toc75, and Toc159, have been identified from pea (Pisum sativum) chloroplasts by cross-linking with precursor proteins (Kessler et al., 1994; Perry and Keegstra, 1994; Schnell et al., 1994). Antibodies against Toc159 inhibit protein import (Hirsch et al., 1994). Furthermore, Toc159 predominantly interacts with preproteins in the binding step, suggesting that Toc159 is the receptor part of the machinery (Ma et al., 1996). The function of Toc34 is not clear. It is in close proximity to Toc75 and to the preprotein during import (Seedorf et al., 1995; Kouranov and Schnell, 1997). It has been shown that, in vitro, the precursor-binding capacity of Toc34 is regulated by phosphorylation (Sveshnikova et al., 2000). Toc75 contains several transmembrane domains and is likely to function as a protein-conducting channel (Hinnah et al., 1997; Reumann et al., 1999).
Orthologues for pea Toc genes have been found in other plant species. It is interesting that Arabidopsis has three orthologues for Toc159 (atToc159, atToc132, and atToc120) and two orthologues for Toc34 (atToc33 and atToc34). Two Arabidopsis mutants, ppi1 and ppi2, which are defective in atToc33 and atToc159, respectively, recently were isolated from T-DNA-tagged mutants (Jarvis et al., 1998; Bauer et al., 2000). The phenotype of ppi1 was pale green in young leaves, but gradually recovering normal pigmentation in later stages (Jarvis et al., 1998). Ectopic expression of atToc34 in ppi1 could complement the mutant phenotype. This result indicates that the functions of atToc33 and atToc34 are similar (Jarvis et al., 1998). However, these two genes are differentially expressed in various organs (Gutensohn et al., 2000). Protein interaction analysis also showed different affinities of these two proteins for chloroplast precursor proteins (Gutensohn et al., 2000).
The ppi2 mutant is seedling lethal on soil and chloroplast development in the mutant is severely defective. Gene expression and plastid import of proteins essential for photosynthesis are repressed in ppi2. However, the import of atToc75 and atTic110 is normal in the mutant (Bauer et al., 2000). Therefore, it was suggested that the atToc159 mutation limited the capacity of plastids to import a set of highly expressed photosynthetic proteins and chloroplast biogenesis was consequently blocked (Bauer et al., 2000).
In higher plants, there are different types of plastids in different tissues. Although these types of plastids are interconvertable, the morphology and protein content among these plastids are quite different (Thompson and Whatley, 1980). When tested by ectopic expression or in vitro import, proteins from one type of plastid are also imported into other types of plastids (Boyle et al., 1986; Schindler and Soll, 1986; Strzalka et al., 1987; Klšsgen et al., 1989), suggesting that there is a “general import apparatus” for all plastids (Soll and Tien, 1998). However, several pieces of evidence against this “general import apparatus” hypothesis have also arisen. The chlorophyll biosynthesis enzymes NADPH: protochlorophyllide oxidoreductase A and protochlorophyllide oxidoreductase B were shown to be imported into chloroplasts by different import machinery (Reinbothe et al., 2000). Several chloroplast proteins were imported into leucoplasts with a much lower efficiency than into chloroplasts, suggesting that there is a substrate preference for each type of plastid import machinery (Wan et al., 1996). Furthermore, the functions of atToc33 and atToc34 are redundant in protein import (Jarvis et al., 1998), suggesting that there may be multiple chloroplast protein import complexes in Arabidopsis (Chen et al., 2000). If there are multiple import complexes, the next question is: What are the specificities for these different complexes? Do different complexes import different proteins? Are different complexes located on different types of plastids, or can they be located on the same type of plastids at the same time?
As a first step toward understanding the nature of different import complexes, we performed further quantitative analysis of the ppi1 and ppi2 mutants. Previous data indicated that ppi2 completely lacked chloroplast development (Bauer et al., 2000). Our data indicated that, although chloroplasts of mesophyll cells were severely defective in ppi2, the root and guard cells could still contain normal and functional chloroplasts. Our results suggest that, if the three atToc159 homologs assemble into distinct complexes, plastids in different cell types may preferentially use alternative complexes for protein import.
RESULTS
Plastids in ppi1 and ppi2 Guard Cells and Root Tip Cells Contained Starch Granules
Enzymes essential for starch synthesis are all nuclear encoded and must be imported into plastids. If plastid protein import is defective, starch granule formation may also be defective due to the lack of starch synthesis enzymes. Therefore, we asked if the ppi2 mutant was defective in starch accumulation. We stained the wild type and the mutant plants for starch with iodine (Caspar et al., 1991). As shown in Figure 1, starch was detected in the entire cotyledons and at the root tips of the wild-type plants (Fig. 1, A and B; Yu et al., 2000). It is surprising that in ppi2, brown spots were also observed on cotyledons (Fig. 1E). Upon closer examination, these brown spots were pairs of kidney-shaped cells, indicating that they were the guard cells of the stomatal complex. In addition, the root tips of ppi2 also stained for starch as in the wild type (Fig. 1F). These results suggested that, despite the severe defects of plastids in mesophyll cells (Bauer et al., 2000), plastids in guard cells and root tip cells of ppi2 still accumulated significant amounts of starch.
Figure 1.
Starch staining of wild type, ppi1, and ppi2. Cotyledons and root tips of 10-d-old seedlings were harvested and stained with iodine. A, Wild-type (WT) cotyledon; B, wild-type root; C, ppi1 cotyledon; D, ppi1 root; E, ppi2 cotyledon; F, ppi2 root.
The accumulation of starch in cotyledons of ppi1 mutant was also examined. Correlative with the pale-green color of the mutant, the brown color of starch staining was lighter than that of the wild type (Fig. 1C), indicating reduced amount of starch. Root tip cells of ppi1 also contained starch as in the wild type (Fig. 1D).
Ultrastructure of Plastids in Leaf Cells
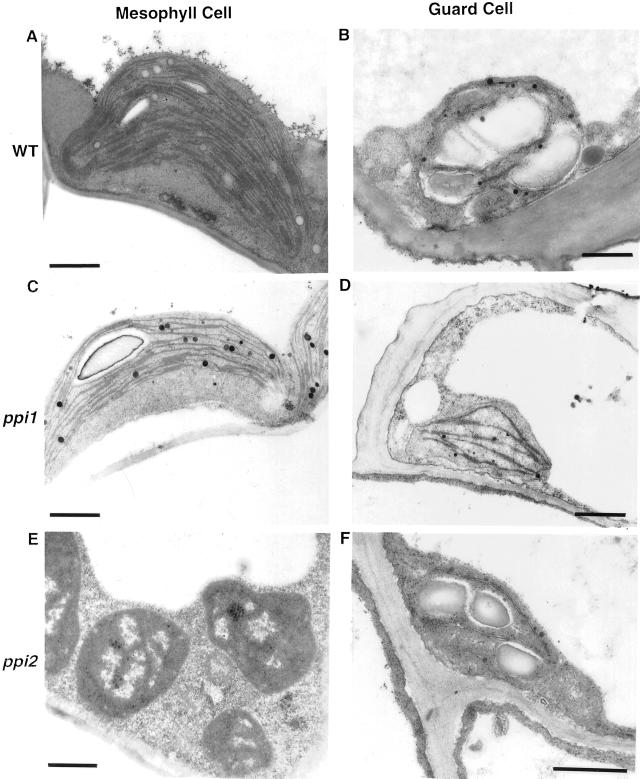
We further inspected the leaf plastids of ppi1 and ppi2 with electron microscopy. In wild type and ppi1, chloroplasts in both the mesophyll cells and the guard cells contained starch granules and thylakoid membranes (Fig. 2, A–D). The amounts of thylakoid membranes in ppi1 chloroplasts were less than that of the wild-type chloroplasts (Jarvis et al., 1998; Fig. 2, C and D). In ppi2, chloroplasts in mesophyll cells lacked thylakoid membranes and starch granules, as previously described (Bauer et al., 2000; Fig. 2E). In contrast, chloroplasts in guard cells contained significant amounts of starch granules and thylakoid membranes (Fig. 2F). These data indicated that atToc159 is essential for chloroplast biogenesis only in mesophyll cells but not in guard cells.
Figure 2.
Ultrastructure of leaf plastids in wild type and mutants. Leaves of 14-d-old seedlings were fixed and examined by transmission electron microscopy. A and B, Wild type (WT); C and D, ppi1; E and F, ppi2. A, C, and E, Plastids in mesophyll cells. B, D, and F, Plastids in guard cells. Bars = 1 μm.
Ultrastructure of Plastids in Root Cells
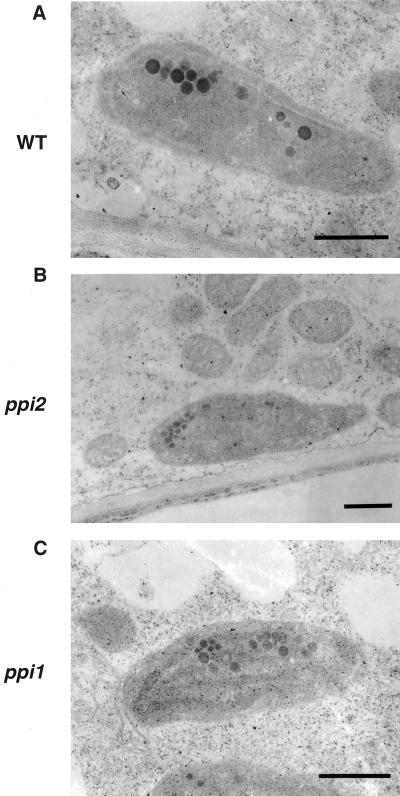
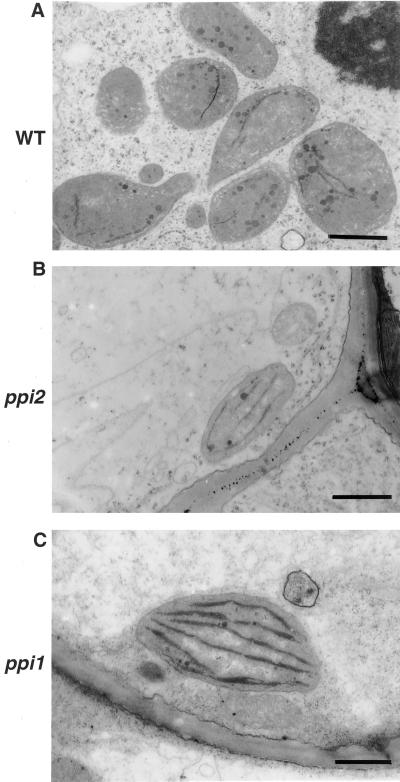
Most root tissues contain proplastids and amyloplasts rather than chloroplasts (Whatley, 1983). To know whether plastids in roots were also affected by the ppi1 and ppi2 mutations as were plastids in mesophyll cells, we examined the ultrastructure of root plastids with electron microscopy. From the samples we observed, there was no significant difference among the two mutants and the wild type (Fig. 3, A–C). Furthermore, in the two mutants and the wild type, some roots turned green after a prolonged exposure to light due to growth on the surface of agar media. Proplastids in these root tissues developed into chloroplasts (Whatley, 1983; Fig. 4, A–C). These results suggested that the ppi1 and the ppi2 mutations had little effect on protein import into root plastids. Previous suggestions that the ppi2 mutant was universally defective in importing photosynthesis-related proteins (Bauer et al., 2000) were not supported by our observations. Our results showed that root and guard cell plastids of ppi2 obviously could import enough photosynthetic proteins to allow chlorophyll accumulation and thylakoid membrane development.
Figure 3.
Root plastids in the wild type and mutants. Root tissues were harvested and examined by transmission electron microscopy. A, Wild type (WT); B, ppi2; C, ppi1. Bars = 0.5 μm.
Figure 4.
Root plastids in the wild type and mutants developed into chloroplasts after prolonged light treatments. Root tissues with a light-green color were harvested from plants grown on agar media and examined by transmission electron microscopy. A, Wild type (WT); B, ppi2; C, ppi1. Bar = 0.5 μm.
Activities of Several Plastid Enzymes Were Reduced in Mutant Leaves But Not in Roots
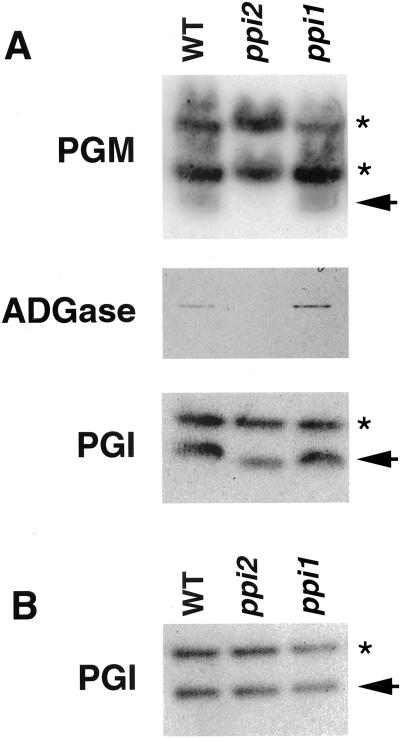
To further confirm that protein import of root plastids was functional in ppi1 and ppi2, we analyzed the activities of several plastid enzymes in the starch synthesis pathway. If plastid protein import was defective, activities of these enzymes should be reduced. In leaf tissues, the activities of chloroplast phosphoglucomutase (PGM), ADP-Glc pyrophosphorylase (ADGase), and phosphoglucoisomerase (PGI) were assayed. The ppi1 mutant showed no difference from the wild type in these enzyme activities. The ppi1 mutation may be too mild to render a clear effect on steady-state protein levels in chloroplasts. However, in the ppi2 mutant, the activities of chloroplast PGM and ADGase were almost not detectable (Fig. 5A, arrow for PGM). The activity of PGI was reduced about 30% (Fig. 5A, arrow in PGI). In contrast, plastid PGI activity in roots was similar for wild type, ppi1, and ppi2 (Fig. 5B, arrow). The activities of PGM and ADGase could not be assayed because they are not expressed in roots. The cytosolic forms of PGM and PGI were not affected either in roots or leaves (Fig. 5, A and B, asterisks). These results indicated that protein import into root plastids was relatively normal compared with leaf plastids in the absence of the atToc159 receptor.
Figure 5.
Activity assays of starch synthesis enzymes in 10-d-old seedlings. A, Leaf tissue; B, root tissue. The enzymes assayed were labeled at left. The arrows indicate the plastid forms of PGM and PGI. The asterisks indicate the cytosolic forms of the enzymes.
All Three atToc159 Homologs Are Expressed in Roots
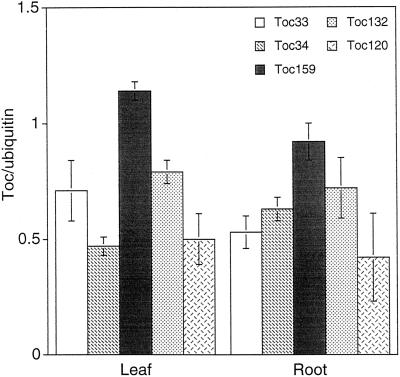
In Arabidopsis, there are two homologs of atToc159 (atToc120, and atToc132) and one homolog of atToc33 (atToc34). The simplest explanation for the tissue-specific defect of the ppi1 and ppi2 mutants is that these homologs are expressed in a tissue-specific manner. For example, atToc159 may be expressed only in leaves and atToc120 or atToc132 is expressed in roots and then the root plastids of ppi2 would be normal. Therefore, we examined the tissue expression patterns of these five genes. The expression patterns of atToc33 and atToc34 have been analyzed in detail (Gutensohn et al., 2000). We have obtained similar results, i.e. atToc33 had a higher expression level in leaves and atToc34 had a higher expression level in roots but both genes were expressed in both tissues (Fig. 6). All three atToc159 homologs similarly were expressed in leaves and roots and atToc159 was the most highly expressed gene in both tissues.
Figure 6.
Toc gene expression in leaf and root tissues. Arabidopsis leaf or root RNA was isolated from 1-month-old plants. The amounts of Toc gene transcripts were analyzed by reverse transcription PCR using gene-specific primer pairs. Quantitaion results of each Toc gene were normalized to the ubiquitin gene (UBQ 10; Sun and Callis, 1997). Lines in bars represent se, n = 3.
DISCUSSION
In the complete absence of atToc159, as in the ppi2 mutant, although chloroplasts in mesophyll cells failed to develop, chloroplasts in guard cells were relatively normal. Plastids in roots were also relatively normal in terms of starch accumulation, PGI enzyme activity, and the potential to differentiate into chloroplasts. The effect of the ppi1 mutation was much milder. However, the only place we observed an effect was also in the mesophyll cell chloroplasts. These results suggest that there may be other Toc complexes functioning in other cell types like guard cells and root cells. Different complexes may preferentially function in different cell types.
The two atToc159 homologs, atToc120 and atToc132, are the most likely candidates to function in place of atToc159 in cells like guard cells and root cells. However, our data indicate that the expression of these three genes are not tissue specific, i.e. all three genes are expressed in all tissues and atToc159 is the most highly expressed one in all tissues. This result suggests that if the three atToc159 homologs form three different Toc complexes, all three complexes are present in all tissues. They may even be present on the same plastid at the same time. Therefore, the reason that plastids in different cell types are affected to different degrees by the absence of atToc159 may be that different complexes have different affinities for different group of precursor proteins. For example, atToc159 may have a higher affinity for photosynthetic proteins and therefore mesophyll cells are most severely affected by the absence of atToc159. However, if “different affinity” is the only reason, then the atToc120- and atToc132-containing Toc complexes in mesophyll cells should allow plastids in mesophyll cells to develop to the extent we observed for guard cell chloroplasts.
Therefore, we hypothesize that there may be a “cell type-specific activator or assisting factor” for the translocon complexes. All three Toc complexes can import most, if not all, plastid proteins when associated with this “assisting factor.” This factor may preferentially associate with atToc159 in mesophyll cells because atToc159-containing Toc complex will be the “designated complex” in mesophyll cells due to the higher affinity of atToc159 for photosynthetic proteins. Without this factor, the atToc120 and atToc132 complexes have some, but very low, activities. In guard cells and root cells, the assisting factor is preferentially associated with atToc120 or atToc132, so they can actively import whatever plastid proteins are expressed in that cell type, including photosynthetic proteins. This cell type-specific assisting factor may be one protein, or more likely, a family of proteins with different members preferentially expressed in different cell types. Several cytosolic factors that can increase the efficiency of chloroplast protein import have been identified (Waegemann et al., 1990; May and Soll, 2000). They could be candidates of the “assisting factors” we are hypothesizing. However, whether they can interact with the Toc complex directly and in a tissue-specific manner remain to be tested. In the future, it will be important to analyze the tissue and cell type expression of all translocon components. It also will be interesting to isolate individual mutants in members of a gene family, e.g. mutants in atToc120 or atToc132, and generate double mutants between these mutants to study the interplay between these homologs.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis ppi2 mutant (CS11072) was obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). Arabidopsis seeds were surface sterilized with commercial bleach and grown on 1× Murashige and Skoog-agar with 2% (w/v) Suc at 25°C under 16 h light/8 h dark in a growth chamber.
Transmission Electron Microscopy
Arabidopsis leaf and root tissues were fixed in 2.5% (v/v) glutaraldehyde and 0.1 m sodium phosphate (pH 7.2) and a secondary fixation of 1% (v/v) OsO4. The fixed specimens were dehydrated and embedded in Spurr resin. Samples were section and stained with uranyl acetate and lead acetate, and viewed in a transmission electron microscope.
Enzyme Assays
Leaf and root tissues were harvested from the same batch of 10-d-old seedlings. Total enzymes were extracted with an enzyme extraction buffer (100 mm Tris [pH 7.0], 100 mm KCl, 10 mm MgCl2, 40 mm β-mercaptoethanol, and 15% [v/v] glycerol). The extracts were separated by SDS-PAGE on 12% (w/v) Tris-Gly gels and the gels were incubated at 37°C in solution containing 100 mm Tris (pH 8.0), 100 mm MgCl2, 0.15% (w/v) Fru-6-phosphate, 0.2 mm NADP, 0.02% (w/v) 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide, 0.004% (w/v) PMS, and 0.6 unit mL−1 Glc-6-phosphate dehydrogenase for PGI; 100 mm Tris (pH 7.0), 100 mm MgCl2, 0.6% (w/v) Glc-1-phosphate, 0.2 mm NADP, 0.016% (w/v) nitroblue tetrazolium, 0.0008% (w/v) PMS, and 0.8 unit mL−1 Glc-6-phosphate dehydrogenase for PGM; and 100 mm Tris (pH 8.0), 5 mm CaCl2, 5 mm Glc-1-phosphate, 5 mm β-mercaptoethanol, 5 mm ATP, and 10 mm 3-phosphoglyceric acid for ADGase enzyme assay (Caspar et al., 1991).
Reverse Transcription-PCR Analysis
First-strand cDNA was synthesized using the Superscript Pre-amplification System (Gibco BRL, Rockville, MD) with total RNA isolated from root or leaf tissues. Primer specific for each Toc genes were amplified with 25 cycles of PCR reactions. The PCR products were analyzed by 1% (w/v) agarose gel, stained with SYBR Green (Molecular Probes, Eugene, OR), and quantified by Luminescent Image Analyzer LAS1000 plus (Fujifilm, Tokyo). Specific primer pairs for each Toc genes were as follows: ubiquitin forward CTTCG TCAAG ACTTT GACCG and reverse CTTCT TAAGC ATAAC AGAGA CGAG, atToc33 forward TCTTA TCGGC GAACA AGTCG TCCGT and reverse GTTTG TTGCT ACATC AGTTA TCGCC, atToc34 forward CTACC TTGGT CTCTC GCACA AGATC and reverse TGTCA ACATG AATCG CCTTG TTGCC, atToc159 forward CACAG TCTTG CTCTA GCTAG CCGGT TC and reverse GCTGT ACTTG TCGTT CGTCG CTTC, atToc132 forward GATTC GGTTT CTGCG GGGTT G and reverse TCATT GTCCA TATTG CGTTT GCGG, and atToc120 forward AATGC TGGGA AGGAA TTAGC GTACA CTA and reverse TCAGT GTCCA TATTG CATTT GCTCA GG.
ACKNOWLEDGMENTS
We thank Soo-ping Lee for assistance with electron microscopy. We thank Jenny Dorl and Dr. Kathy Archer for critical reading of the manuscript.
Footnotes
This work was supported by the National Science Council (grant no. NSC 89–2321–B–001–005 to H.-m.L.) and by Academia Sinica of Taiwan (grant to H.-m.L.).
LITERATURE CITED
- Bauer J, Chen K, Hiltbunner A, Wehrli E, Eugster M, Schnell D, Kessler F. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature. 2000;403:203–207. doi: 10.1038/35003214. [DOI] [PubMed] [Google Scholar]
- Boyle SA, Hemmingsen SM, Dennis DT. Uptake and processing of the precursor to the small subunit of ribulose 1,5-bisphosphate carboxylase by leucoplasts from the endosperm of developing castor oil seeds. Plant Physiol. 1986;81:817–822. doi: 10.1104/pp.81.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Lin T-P, Kakefuda G, Benbow L, Preiss J, Somerville CR. Mutation of Arabidopsis with altered regulation of starch degradation. Plant Physiol. 1991;95:1181–1188. doi: 10.1104/pp.95.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Chen X, Schnell D. Mechanism of protein import across the chloroplast envelope. Biochem Soc Trans. 2000;28:485–491. [PubMed] [Google Scholar]
- Gutensohn M, Schulz B, Nicolay P, Flügge U-I. Functional analysis of the two Arabidopsis homologues of Toc34, a component of the chloroplast protein import apparatus. Plant J. 2000;23:771–783. doi: 10.1046/j.1365-313x.2000.00849.x. [DOI] [PubMed] [Google Scholar]
- Hinnah SC, Hill K, Wagner R, Schlicher T, Soll J. Reconstitution of a chloroplast protein import channel. EMBO J. 1997;16:7351–7360. doi: 10.1093/emboj/16.24.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Muckel E, Heemeyer F, von Heijne G, Soll J. A receptor component of the chloroplast protein translocation machinery. Science. 1994;266:1989–1992. doi: 10.1126/science.7801125. [DOI] [PubMed] [Google Scholar]
- Jarvis P, Chen L-J, Li H-m, Peto CA, Fankhauser C, Chory J. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science. 1998;282:100–103. doi: 10.1126/science.282.5386.100. [DOI] [PubMed] [Google Scholar]
- Kessler F, Blobel G, Patel H, Schnell DJ. Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science. 1994;266:1035–1039. doi: 10.1126/science.7973656. [DOI] [PubMed] [Google Scholar]
- Klšsgen RB, Saedler H, Weil J-H. Subcellular location and expression level of a chimeric protein consisting of the maize waxy transit peptide and the β-glucuronidase of Escherichia coli in transgenic potato plants. Mol Gen Genet. 1989;217:155–161. doi: 10.1007/BF00269862. [DOI] [PubMed] [Google Scholar]
- Kouranov A, Schnell DJ. Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J Cell Biol. 1997;139:1677–1685. doi: 10.1083/jcb.139.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YK, Kouranov A, LaSala SE, Schnell DJ. Two components of the chloroplast protein import apparatus IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor protein at the outer envelope. J Cell Biol. 1996;134:315–327. doi: 10.1083/jcb.134.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May T, Soll J. 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell. 2000;12:53–63. doi: 10.1105/tpc.12.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SE, Keegstra K. Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell. 1994;6:93–105. doi: 10.1105/tpc.6.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S, Mache R, Reinbothe C. A second, substrate-dependent site of protein import into chloroplasts. Proc Natl Acad Sci USA. 2000;97:9795–9800. doi: 10.1073/pnas.160242597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Davila-Aponte J, Keegstra K. The evolutionary origin of the protein-translocating channel of chloroplastic envelope membranes: identification of a cyanobacterial homolog. Proc Natl Acad Sci USA. 1999;96:784–789. doi: 10.1073/pnas.96.2.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Soll J. Protein transport in intact, purified pea etioplasts. Arch Biochem Biophys. 1986;247:211–220. doi: 10.1016/0003-9861(86)90550-3. [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Blobel G, Keegstra K, Kessler F, Ko K, Soll J. A consensus nomenclature for the protein-import components of the chloroplast envelope. Trends Cell Biol. 1997;7:303–304. doi: 10.1016/S0962-8924(97)01111-2. [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Kessler F, Blobel G. Isolation of components of the chloroplast protein import machinery. Science. 1994;266:1007–1012. doi: 10.1126/science.7973649. [DOI] [PubMed] [Google Scholar]
- Seedorf M, Waegemann K, Soll J. A constituent of the chloroplast import complex represents a new type of GTP-binding protein. Plant J. 1995;7:401–411. doi: 10.1046/j.1365-313x.1995.7030401.x. [DOI] [PubMed] [Google Scholar]
- Soll J, Tien R. Protein translocation into and across the chloroplastic envelope. Plant Mol Biol. 1998;38:191–207. [PubMed] [Google Scholar]
- Strzalka K, Ngernprasirtsiri J, Watanabe A, Akazawa T. Sycamore amyloplasts can import and process precursors of nuclear encoded chloroplast proteins. Biochem Biophys Res Commun. 1987;149:799–806. doi: 10.1016/0006-291x(87)90438-4. [DOI] [PubMed] [Google Scholar]
- Sun C-W, Callis J. Independent modulation of Arabidopsis thaliana polyubiquitin mRNAs in different organs and in response to environmental changes. Plant J. 1997;11:1017–1027. doi: 10.1046/j.1365-313x.1997.11051017.x. [DOI] [PubMed] [Google Scholar]
- Sveshnikova N, Soll J, Schleiff E. Toc34 is a preprotein receptor regulated by GTP and phosphorylation. Proc Natl Acad Sci USA. 2000;97:4973–4978. doi: 10.1073/pnas.080491597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WW, Whatley JM. Development of nongreen plastids. Annu Rev Plant Physiol. 1980;31:375–394. [Google Scholar]
- Wan J, Blakeley SD, Dennis DT, Ko K. Transit peptides play a major role in the preferential import of proteins into leucoplasts and chloroplasts. J Biol Chem. 1996;271:31227–31233. doi: 10.1074/jbc.271.49.31227. [DOI] [PubMed] [Google Scholar]
- Waegemann K, Paulsen H, Soll J. Translocation of proteins into chloroplasts requires cytosolic factors to obtain import competence. FEBS Lett. 1990;261:89–92. [Google Scholar]
- Whatley JM. The ultrastructure of plastids in roots. Int Rev Cytol. 1983;85:175–220. [Google Scholar]
- Yu T-S, Lue W-L, Wang S-M, Chen J. Mutation of Arabidopsis plastid phosphoglucose isomerase affects leaf starch synthesis and floral initiation. Plant Physiol. 2000;123:319–325. doi: 10.1104/pp.123.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]