Abstract
Arabidopsis (ecotype Columbia-0) genes, AtDEF1and AtDEF2, represent eukaryotic homologs of the essential prokaryotic gene encoding peptide deformylase. Both deduced proteins contain three conserved protein motifs found in the active site of all eubacterial peptide deformylases, and N-terminal extensions identifiable as chloroplast-targeting sequences. Radiolabeled full-length AtDEF1 was imported and processed by isolated pea (Pisum sativum L. Laxton's Progress No. 9) chloroplasts and AtDEF1 and 2 were immunologically detected in Arabidopsis leaf and chloroplast stromal protein extracts. The partial cDNAs encoding the processed forms of Arabidopsis peptide deformylase 1 and 2 (pAtDEF1 and 2, respectively) were expressed in Escherichia coli and purified using C-terminal hexahistidyl tags. Both recombinant Arabidopsis peptide deformylases had peptide deformylase activity with unique kinetic parameters that differed from those reported for the E. coli enzyme. Actinonin, a specific peptide deformylase inhibitor, was effective in vitro against Arabidopsis peptide deformylase 1 and 2 activity, respectively. Exposure of several plant species including Arabidopsis to actinonin resulted in chlorosis and severe reductions in plant growth and development. The results suggest an essential role for peptide deformylase in protein processing in all plant plastids.
Initially thought to be exclusively present in prokaryotes, peptide deformylase catalyzes the removal of the formyl group from N-formyl-Met at the amino termini of nascent polypeptides. Due to extraordinary lability (t1/2 [half-life] approximately 1 min at room temperature; Rajagopalan and Pei, 1998) as a consequence of oxidation of the active site ferrous ion and an essential Cys residue, the enzyme was originally characterized only after the gene was cloned (Mazel et al., 1994) and the protein overexpressed with catalytically competent replacement metals (Becker et al., 1998; Groche et al., 1998; Rajagopalan et al., 1997a, 1997b; Ragusa et al., 1998). The significance and importance of peptide deformylase is primarily 2-fold: (a) It is the first step in cotranslational protein processing for all prokaryotically synthesized proteins, given the requirement for N-formyl-Met as the initiating residue during translation; and (b) it is restricted to prokaryotic organisms and null-def mutants are lethal in eubacteria (Mazel et al., 1994); thus, it is an ideal molecular target for the development of broad-spectrum antibiotics with little to no mammalian toxicity.
Corroborating the widely accepted endosymbiotic theory (McFadden, 1999), chloroplastic protein initiation is prokaryotic in nature (Bianchetti et al., 1971; Lucchini and Bianchetti, 1980), as evidenced by several chloroplast genome-encoded proteins with amino-terminal N-formyl-Mets (Sigrist-Nelson et al., 1978; Scheller et al., 1989; Sharma et al., 1997a). Nevertheless, the N termini of other chloroplast genome-encoded proteins (Michel et al., 1988; Sharma et al., 1997a, 1997b, 1997c), including the large subunit of Rubisco (Houtz et al., 1989), are extensively processed and do not contain an N-formyl group.
The acquisition and availability of Arabidopsis (ecotype Columbia[Col]-0) genomic sequences enabled the identification of two putative eukaryotic homologs of eubacterial peptide deformylase with chloroplast-targeting sequences (Williams et al., 2000). We report the functional analyses of two Arabidopsis peptide deformylases, which are plant eukaryotic enzymes with potentially significant utility as molecular targets for the development of a new class of broad-spectrum herbicides.
RESULTS
Clone Identification
Two deduced translation products from the Arabidopsis Genome Initiative were annotated as putative peptide deformylase enzymes. The deduced proteins each contained an N-terminal chloroplast-targeting sequence with processing sites predicted by ChloroP v1.1 (Emanuelsson et al., 1999) between residues 50 and 51 and 56 and 57 for AtDEF1 and 2, respectively (Fig. 1). The homology between the two AtDEFs and the E. coli protein was relatively low, with 17% identity and 44% similarity (determined directly from the alignment in Fig. 1).
Figure 1.
Clustal W (1.8) alignment (Thompson et al., 1994) of the deduced amino acid sequences of the two annotated peptide deformylase genes from Arabidopsis (AtDEF1, cDNA accession no. AF250959, predicted translation accession no. AAD39667.1, BAC F9L1, gene no. 34; and AtDEF2, cDNA accession no. AF269165, predicted translation accession no. CAB87633.1, BAC T15N1, gene no. 150) with the Escherichia coli peptide deformylase (accession no. CAA54826.1). For the plant peptide deformylases, the predicted transit peptides are highlighted in green. The three conserved, active-site motifs including the metal binding ligands (Meinnel, 2000) are highlighted in blue and red, respectively. Conserved amino acids in all putative peptide deformylases (Meinnel, 2000) are in bold.
Chloroplast Import and Processing
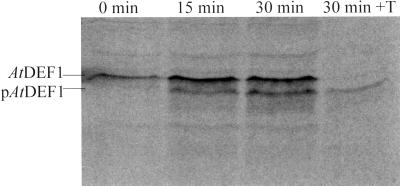
In vitro uptake and processing of full-length proteins (AtDEF1 and 2) using isolated intact chloroplasts tests whether the predicted (Emanuelsson et al., 1999) chloroplast-targeting sequences are functionally sufficient for directing these proteins to the organelle. Addition of radiolabeled AtDEF1 to intact pea (Pisum sativum L. Laxton's Progress No. 9) chloroplasts, on multiple, completely independent occasions, resulted in the uptake and subsequent proteolytic processing to the expected lower molecular mass form (Fig. 2, Δ 6 kD), consistent with an in vivo processing site as predicted. In addition, the processed form of AtDEF1 (pAtDEF1) was protected against thermolysin digestion, which is evidence that the processed form of this protein was not simply associated with, but imported into the chloroplasts. Similar experiments were attempted with AtDEF2; however, the recombinant protein, when diluted into import conditions without organelles, was determined to be insufficiently soluble to allow similar analysis as with AtDEF1.
Figure 2.
In vitro chloroplast uptake and processing of AtDEF1. Phosphorimage analysis of an electroblotted (polyvinylidene fluoride membrane) SDS-PAGE (15%, w/v) of l-[35S]Met-labeled AtDEF1 incubated with isolated and intact pea chloroplasts for 0, 15, and 30 min (as labeled) and a 30-min import followed by 20 min thermolysin (0.1 mg mg chlorophyll−1) treatment (30 min + T). Only the processed lower molecular mass form (pAtDEF1) of AtDEF1 was protected.
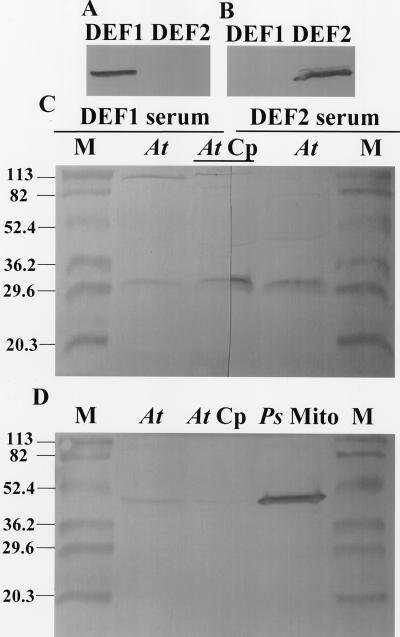
Immunoblots
Using recombinant and purified pAtDEF1 and 2 (see “Peptide Deformylase Activity” below for details) as markers, antibodies against AtDEF1 and 2 did not cross react (Fig. 3, A and B). Specific bands from Arabidopsis leaf and chloroplast stromal extracts were immunoreactive with antibodies raised against both proteins (Fig. 3C, lanes At and At Cp). Using LS and SS of Rubisco as strictly chloroplast-localized markers, the amount of enrichment expected by isolating chloroplasts from Arabidopsis leaves was estimated to be less than 20% by scanning densitometry of 50-μg protein lanes on Coomassie blue-stained SDS-PAGE (data not shown). The chloroplast preparation was examined for mitochondrial contamination using a marker antibody (mitochondrial-specific NADH-Glu dehydrogenase antibody against isoenzyme 1 of V. vinifera, provided by K.A. Roubelakis-Angelakis, University of Crete, Greece; Loulakakis and Roubelakis-Angelakis, 1990; Turano et al., 1997). With the mitochondrial marker antibody, the Arabidopsis stromal chloroplast protein extract had weak signals compared with either Arabidopsis leaf or pea mitochondrial proteins (Fig. 3D, lane At Cp compared with At and Ps Mito).
Figure 3.
Immunoreactive proteins in Arabidopsis leaf and chloroplast stromal extracts detected with antibodies raised against AtDEF1 and 2. Purified recombinant proteins (100 ng) and proteins from Arabidopsis leaves and chloroplasts (250 μg) were electrophoresed, electroblotted to a polyvinylidene fluoride membrane, and probed with primary antibody (A and C, right one-half AtDEF1 serum; and B and C, left one-half AtDEF2 serum). Pea mitochondrial and Arabidopsis chloroplast stromal proteins (250 μg) were similarly analyzed (D) with a mitochondrial-specific Vitis vinifera isoenzyme 1 NADH-Glu-dehydrogenase primary antibody (provided by K.A. Roubelakis-Angelaki). M, Prestained molecular mass markers; DEF1 and DEF2, 100 ng affinity-purified pAtDEF1 and 2; At, Arabidopsis leaf protein extract; At Cp, Arabidopsis chloroplast stromal protein extract; Ps Mito, pea mitochondrial protein extract.
Peptide Deformylase Activity
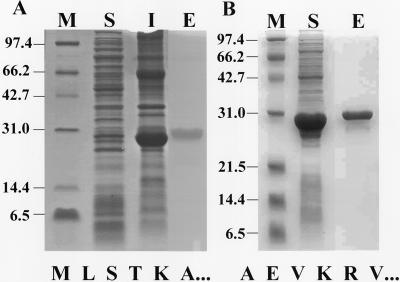
To verify the enzymatic activity of the processed forms, nucleotide sequences corresponding to the predicted chloroplast-targeting sequences were removed and the resulting clones expressed in E. coli. With expression at 37°C using 1.0 mm isopropyl β-d- thiogalactopyranoside (IPTG), both pAtDEF1 and 2 were insoluble proteins. Substantially greater amounts of soluble pAtDEF2 compared with soluble pAtDEF1 (Fig. 4) were obtained after optimizing both temperature (25°C) and IPTG concentrations (0.4 mm). A C-terminal hexahistidyl tag, which had no effects on physical and catalytic properties of the E. coli enzyme (Rajagopalan et al., 2000), was fused to both pAtDEF1 and 2 so the expressed proteins could be affinity purified with a nickel-nitrilotriacetic acid (Ni-NTA) column. Purified pAtDEF1 and 2 migrated on SDS-PAGE with an apparent molecular mass distinctly higher than predicted (30 kD on a 15% [w/v] gel for both proteins versus predicted 24.6 and 25.6 kD, respectively; Fig. 4, lane E). Edman degradative sequencing confirmed the N termini of the recombinant proteins (Fig. 4) as predicted by prokaryotic N-terminal processing (Flinta et al., 1986; Hu et al., 1999). The pAtDEF1 and 2 recovered were greater than 95% pure as determined by reverse-phase HPLC analysis (data not shown).
Figure 4.
Affinity purification of pAtDEF1 and 2. Proteins from induced bacterial cells harboring constructs designed to produce pAtDEF1 (A) and 2 (B) were electrophoresed (15% [w/v] SDS-PAGE) and the gel stained with Coomassie blue R-250. M, Molecular mass markers; S and I, soluble and insoluble proteins, respectively, from the lysate of IPTG-induced BL21(DE3) pLysS cells expressing respective construct; E, dialyzed elution from nickel-nitrilotriacetic acid agarose column. Given below their respective gels, the N-terminal sequence for each processed form (pAtDEF) after purification from a bacterial lysate confirmed N-terminal processing.
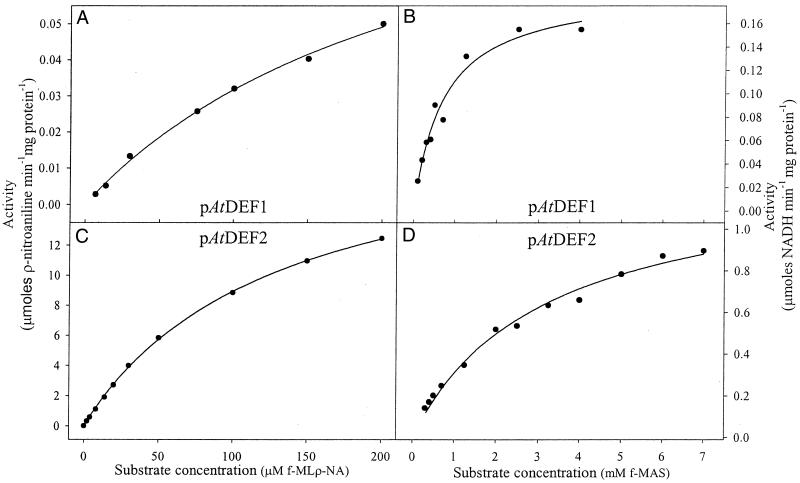
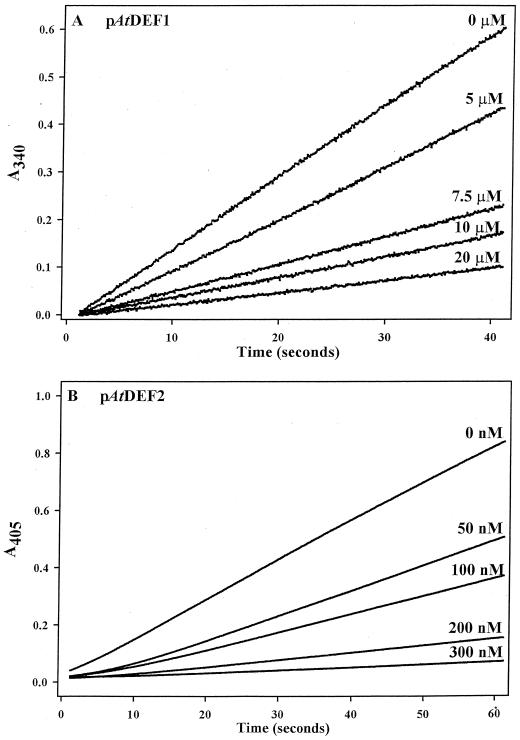
pAtDEF1 and 2 were assayed with two different continuous spectrophotometric methods for peptide deformylase (Lazennec and Meinnel, 1997; Wei and Pei, 1997). Kinetic parameters (Table I) were determined for both proteins in both assays using best fit regression lines predicted for Michaelis-Menten kinetics (V = Vmax[S]/(Km + [S]) (Fig. 5). Attempts to detect and measure activity in isolated Arabidopsis and pea chloroplasts were unsuccessful, yet unsurprising given the small amount of accumulated protein in the plant (Fig. 3), and the high lability and rapid catalytic inactivation of all peptide deformylases (Rajagopalan and Pei, 1998).
Table I.
Kinetic parameters for pAtDEF1 and 2 with two spectrophotometric assays for peptide deformylase activity
| Protein Source/Substrate Used | Vmax | Apparent Km | Kcat | Kcat/Km |
|---|---|---|---|---|
| μmols min−1 mg protein−1 ± se | μm ± se | s−1 | m−1 s−1 | |
| pAtDEF1 | ||||
| f-MLρ-NA | 0.11 ± 0.01 | 240 ± 34 | 0.044 | 1.8 × 102 |
| f-MAS | 0.20 ± 0.01 | 710 ± 120 | 0.078 | 1.1 × 102 |
| pAtDEF2 | ||||
| f-MLρ-NA | 20 ± 0.3 | 130 ± 4 | 8.7 | 6.8 × 104 |
| f-MAS | 1.3 ± 0.1 | 3,200 ± 370 | 0.54 | 1.7 × 102 |
| E. coli | ||||
| f-MLρ-NAa | – | 20.3 | 38 | 1.9 × 106 |
| f-MASb | – | 3,900 | 210 | 5.4 × 104 |
f-MLρ-NA, N-formyl-Met-Leu-ρ-nitroanilide substrate coupled with Aeromonas proteolytica aminopeptidase. f-MAS, N-formyl-Met-Ala-Ser substrate coupled with β-NAD+ and yeast formate dehydrogenase.
Figure 5.
Kinetic analyses of pAtDEF1 and 2 activity. pAtDEF1 (A and B) and pAtDEF2 (C and D) data using N-formyl-Met-Leu-ρ-nitroanilide and Aeromonas proteolytica aminopeptidase (A and C) or N-formyl-Met-Ala-Ser and formate dehydrogenase (B and D) were fitted to the Michaelis-Menten equation using SigmaPlot (Windows Version 4.0; SPSS, Inc.). See Table I for respective kinetic values derived from these fits.
Enzyme Activity Inhibition by Actinonin
The most potent inhibitors of peptide deformylase discovered thus far are actinonin, a pseudopeptide with an N-terminal Met analog (Chen et al., 2000), and a related N-formyl-hydroxylamine derivative, BB-3497 (Clements et al., 2001; not commercially available). Consistent with studies of the eubacterial enzymes, pAtDEF1 and 2 were substantially inhibited by low amounts of actinonin (Fig. 6, A and B, respectively). The amounts of enzyme required to reliably detect activity differed significantly between pAtDEF1 and 2 such that the quantities of actinonin used for inhibition represent approximately a 2-fold weaker binding of actinonin by pAtDEF1.
Figure 6.
Inhibition of pAtDEF1 and 2 by actinonin. A, pAtDEF1 (300 μg) was pre-incubated for 3 min in the absence or presence of actinonin (concentrations as indicated) prior to initiating the assay with 4 mm N-formyl-Met-Leu-Ser. B, pAtDEF2 (7.5 μg) was pre-incubated for 3 min in the absence or presence of actinonin (concentrations as indicated) prior to initiating the assay with 200 μm N-formyl-Met-Leu- ρ-nitroanilide.
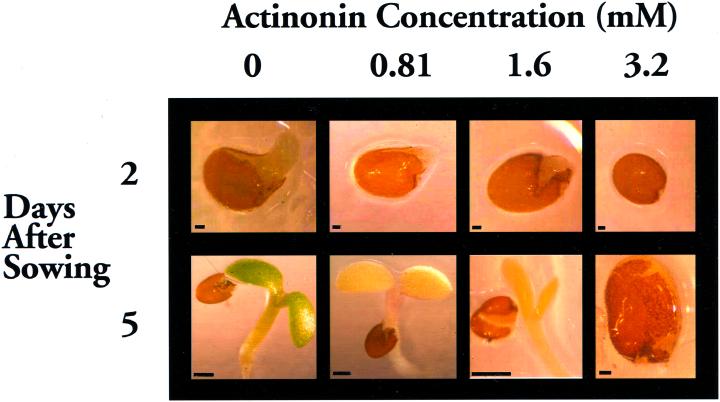
Effects on Plant Growth by Treatment with Actinonin
The potent inhibition of pAtDEFs by actinonin in vitro suggests that this compound would exhibit toxicity to plants similar to that described for eubacteria (Chen et al., 2000). When imbibing Arabidopsis seeds and growing seedlings were exposed to actinonin, there was a distinct dose response effect (Fig. 7). The lowest actinonin rate allowed cotyledon expansion, but halted further development of the morphologically normal seedlings, including any significant formation of chlorophyll. At intermediate actinonin rates, the cotyledons of the seedlings were incompletely expanded. The highest rate of actinonin completely inhibited seedling development past radicle emergence. Whereas control seedlings began to develop their first true leaves by 7 d, all actinonin-treated Arabidopsis remain unchanged beyond 5 d (data not shown). Application of actinonin to the leaves of developing Arabidopsis plants resulted in stunting and a slow bleaching of the leaves (data not shown); however, it was difficult to limit the application to only leaves. When the highest level of actinonin was applied to the leaves of pea plants, there was a significant reduction in both fresh (40%) and dry (42%) weights after 7 d and a concomitant bleaching of the leaves (data not shown).
Figure 7.
Actinonin treatment of Arabidopsis. Seeds (ecotype Col-0) were imbibed and seedlings cultured at room temperature with constant light (50 μmol m−2 s−1) in 200 μL of Murashige and Skoog basal salts with 0.2% (w/v) phytagel in the wells of a 96-well microtiter plate in the absence (0) or presence of actinonin (concentrations as indicated). Images were taken 2 and 5 d after the seeds were sown. Bars represent 50 μm in all 2-d images plus the 3.2-mm actinonin 5-d image and 5 mm in the 0-, 0.81-, and 1.6-mm actinonin 5-d images.
DISCUSSION
Peptide deformylase has been hypothesized to provide an essential enzymatic function in plant plastids (Hanson et al., 2000). The results presented here confirm that hypothesis and suggest that chloroplast-localized peptide deformylase is indispensable for plant growth and development.
The relatively low level of homology (Fig. 1; 17% identical and 44% similar) between the two pAtDEFs and the E. coli protein was not wholly unexpected because the most significant homology between all eubacterial peptide deformylases is localized to regions comprising the active site (Hao et al., 1999). However, many of these sequence differences, including several large extensions and insertions, are at sites and regions previously identified as capable of influencing the catalytic activity of E. coli peptide deformylase (Meinnel et al., 1995). The kinetic parameters determined were distinctly different than values reported for E. coli peptide deformylase (Table I). Thus, the differences in kinetic parameters and enzymatic activity between plant and bacterial peptide deformylases may be related to structural differences outside those areas comprising the catalytic site. Chloroplastic regulatory systems (e.g. thiol regulation; Ruelland and Miginiac-Maslow, 1999) may be coordinating in vivo activities of pAtDEF1 and 2 with conditions conducive for protein translation. Acylase activity of the E. coli peptide deformylase against N-acetyl or N-trifluoroacetyl-Met-Leu-ρ-nitroanilide (Wei and Pei, 1997), albeit at reduced rates, raises the possibility that one of the plant DEFs functions in such a capacity in the chloroplast. The differences between the apparent Km determined by the two assays may be explained in part by observed increases in apparent Km of the E. coli peptide deformylase when either the ρ-nitroanilide group (>40-fold; Wei and Pei, 1997) or the ρ-nitro group (>20-fold; Wei, 2000) is absent. Both pAtDEF1 and 2 also had clearly higher apparent molecular masses than that predicted by their respective amino acid composition (Fig. 4), an unexplainable observation which has also been reported for E. coli peptide deformylase (Rajagopalan et al., 1997a).
AtDEF1 was reproducibly imported and processed in vitro by isolated pea chloroplasts (Fig. 2). Non-cross-reactive AtDEF1 and 2 antibodies detected bands on immunoblots of Arabidopsis leaf and chloroplast stromal proteins at the expected molecular masses of pAtDEF1 and 2 (Fig. 3C). Thus, both proteins were determined to accumulate in vivo in the chloroplast as predicted. During the completion of these studies, AtDEF1 (referred to as PDF1A) was reported to be localized only to the mitochondria based on transient expression in onion (Allium cepa) epidermal cells of constructs representing the N-terminal 83 amino acids of AtDEF1 fused with green fluorescent protein (Giglione et al., 2000). This discrepancy may involve peptide structures in green fluorescent protein (Kunze et al., 1999) that can act in concert with the N-terminal transit sequence and the import machinery of the organelles to potentially misdirect proteins to the mitochondria instead of plastids. In addition, sequences within certain mature chloroplast proteins (e.g. chlorophyll a/b-binding protein, Kavanagh et al., 1988; Silva-Filho et al., 1996; e.g. triose phosphate 3-phosphoglycerate phosphate translocator, Silva-Filho et al., 1997) are required for proper targeting of fusion proteins (chloramphenicol acetyltransferase or β-glucuronidase). Although an exceptionally small amount of contamination by mitochondria was detected in the chloroplast preparation (Fig. 3D), this cannot account for the strong immunoreactive bands noted for both AtDEF1 and 2 antibodies (Fig. 3C). Significant mitochondrial contamination of the Arabidopsis chloroplast preparation is also unlikely given that mitochondrial preparations from Arabidopsis green tissues are difficult (Werhahn et al., 2001) and mitochondria and chloroplasts are collected from 23% (v/v)/40% (v/v) and 50% (v/v)/70% (v/v) Percoll interfaces, respectively (Rensink et al., 1998; Werhahn et al., 2001).
Searching for novel broad-spectrum antibiotics, several academic and commercial laboratories have synthesized active site-directed inhibitors of peptide deformylase based on the crystal structure and proposed catalytic mechanism (Meinnel et al., 1995, 1999; Hu et al., 1998; Durand et al., 1999; Apfel et al., 2000; Chen et al., 2000; Gordon Green et al., 2000; Huntington et al., 2000; Jayasekera et al., 2000; Margolis et al., 2000; Wei et al., 2000; Clements et al., 2001). These compounds have both bacteriostatic and bacteriocidal properties on a number of prokaryotic organisms, and represent a potentially new class of broad-spectrum antibiotics (Apfel et al., 2000; Chen et al., 2000; Huntington et al., 2000; Jayasekera et al., 2000; Margolis et al., 2000; Clements et al., 2001). The most potent inhibitors of peptide deformylase discovered thus far are actinonin, a natural product structurally resembling a pseudopeptide with an N-terminal Met analog (Chen et al., 2000; Margolis et al., 2000) and a related N-formyl-hydroxylamine derivative, BB-3497 (Clements et al., 2001; not commercially available). As with the E. coli deformylase, pAtDEF1 and 2 were completely inhibited by 20 μm and 300 nm actinonin, respectively (Fig. 6). Due to inherent differences in activity, the 67-fold greater amount of actinonin used to inhibit pAtDEF1 represented about 2-fold weaker binding.
Given that eubacterial peptide deformylase is indispensible and that protein targeting and import mechanisms are likely common to all plant plastids (Cavalier-Smith, 2000), inhibiting pAtDEF1 and 2 should compromise cotranslational polypeptide processing and thus, potentially, protein function in chloroplasts as well as all other plant plastids. Therefore, numerous aspects of plant growth and development in addition to photosynthesis should be adversely affected by inhibiting plant peptide deformylase activity. The dramatic effects on growth noted when Arabidopsis seeds were imbibed and seedlings grown in actinonin-containing Murashige and Skoog agar suggests that inhibitors of peptide deformylase are effective as pre-emergent (Fig. 7), as well as foliar, herbicides (data not shown).
The results reported here represent the discovery of a fundamental chloroplast-localized protein-processing enzyme that operates on the majority of chloroplast-encoded proteins. Similar to reports for eubacterial peptide deformylase, cloning and overexpression of pAtDEF1 and 2 was required to confirm enzymatic activity and allow partial characterization. The existence of chloroplast proteins both with and without N-formyl-Met residues at the N terminus is circumstantial evidence for the in vivo specificity of pAtDEF1 and 2; however, it is possible that protein translation by membrane-bound ribosomes could exclude N-terminal processing by soluble peptide deformylase. Identification of N-acetyl-O-phospho-Thr as the N terminus of D1 (Michel et al., 1988) and the cotranslational assembly of membrane-bound photosystem II complexes to replace photodamaged D1 polypeptides (Zhang et al., 1999) necessitates N-terminal processing by peptide deformylase, adding additional intricacy to the repair process. Species' specificity differences in peptide deformylase activity may also negatively impact attempts at Rubisco improvement and could account for the observed lack of cyanobacterial Rubisco large subunit protein in tobacco (Nicotiana tabacum) chloroplasts transformed with the Synechococcus PCC6301 rbcL gene (Kanevski et al., 1999). In addition, the limited replacement of tobacco Rubisco small subunits when the chloroplast is transformed with a construct encoding a tagged version suggests that specificity may exist for the protein sequences encoded within the chloroplast genome (Whitney and Andrews, 2001). These are simple speculations to account for two enzymes with similar activities targeted to the same organelle; however, authentic reasons are currently unknown and form the basis for further research of plant peptide deformylases.
The sensitivity of seedlings and whole plants to actinonin (Fig. 7), a well-characterized peptide deformylase inhibitor, suggests that the activity of plant peptide deformylase, as with its prokaryotic counterparts, is essential for survival. Analogous to the eubacterial enzyme, plant peptide deformylase may represent an ideal target for the development of specific inhibitors with little or no toxicity to other eukaryotic organisms. Finally, the toxicity of actinonin to eubacteria is titratable by peptide deformylase expression level (Chen et al., 2000). This observation provides the paradigm for engineering resistance in plants to peptide deformylase-specific inhibitors—conventional overexpression of plant and/or eubacterial peptide deformylase enzymes in transgenic plants.
MATERIALS AND METHODS
Plant Material
Arabidopsis (ecotype Columbia[Col]-0; Lehle Seeds, Round Rock, TX) and pea (Pisum sativum L. cv Laxton's Progress No. 9) seeds were sown in flats (25 × 52 cm and 46 × 62 cm, respectively) of MetroMix360 (Scotts-Sierra Horticulture Products Company, Marysville, OH) and the seedlings grown for 5 weeks and 8 to 10 d, respectively, in the greenhouse (minimum night temperature of 30°C).
Cloning
For both genes (AtDEF1, cDNA accession no. AF250959, predicted translation accession no. AAD39667.1, BAC F9L1, gene no. 34; and AtDEF2, cDNA accession no. AF269165, predicted translation accession no. CAB87633.1, BAC T15N1, gene no. 150), primers were designed to reverse transcribe the mRNA and amplify partial cDNAs encoding the entire putative protein and the ChloroP-predicted (Emanuelsson et al., 1999) mature protein, respectively (cAtDEF1, RLH171: 5′-GGAAGGCCATATGGAAACCCTTTTCAGAGTC-3′ and RLH167: 5′-GGAAGGCCATATGTTGTCGACAAAAGCCGGTTGG-3′, respectively with RLH172: 5′-GGCCGGGCTCGAGTCATTGAGGTCCGAGCTTAG-3′; cAtDEF2, RLH219: 5′-CATATGGCCGTCTGTAACTGCTTC-3′ and RLH220: 5′- CATATGGCAGAAGTAAAGCGCGTCTC-3′, respectively with RLH222: 5′-CTCGAGACGTTTGCCAAAACCAAC-3′; all primers were synthesized by the Macromolecular Structure Analysis Facility, University of Kentucky Markey Cancer Center, Lexington). These primers included 5′-NdeI and 3′-XhoI restriction enzyme sites (bold letters in above primers) for subcloning into expression vectors, pET23a or b (Novagen Inc., Madison, WI). Reverse transcriptase-PCR was conducted using Arabidopsis leaf RNA (isolated using TRIzol Reagent; Gibco BRL Life Technologies, Rockville, MD), oligomers and Moloney-Murine Leukemia Virus reverse transcriptase and Taq polymerase (Gibco BRL Life Technologies). The expected partial cDNAs and corresponding sizes (cAtDEF1, 780 and 630 bp; cAtDEF2, 831 and 663 bp, full length and processed, respectively) were observed. PCR products were purified with QIAquick PCR Purification Kit or QIAquick Gel Extraction Kit (Qiagen Inc., Valencia, CA), cloned into pCR2.1 (Invitrogen, Carlsbad, CA), and subcloned into the pET expression vectors (Novagen Inc.). Nucleotide sequences (all DNA was sequenced using an ABI PRISM DNA Sequencer [Applied Biosystems, Foster City, CA] by the Macromolecular Structure Analysis Facility, UK Markey Cancer Center) of the partial cDNAs, cAtDEF1 and 2, were verified by comparison with those annotated by the Arabidopsis Genome Initiative.
Chloroplast Import and Processing
Chloroplasts were isolated (Mills and Joy, 1980) from 8- to 10-d-old pea seedlings and full-length AtDEF1 and 2 were isolated as bacterially expressed inclusion bodies (Waegemann and Soll, 1995). A typical 100-μL import assay contained 330 mm sorbitol, 50 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]/KOH (pH 7.6), 3 mm MgSO4, 10 mm Met, 20 mm K-gluconate, 10 mm NaHCO3, 2% (w/v) bovine serum albumin, 3 mm ATP, 15 to 20 μg chlorophyll, and 1 to 2 μL l-[35S]Met-labeled peptide deformylase precursor protein (120,000–600,000 dpm μL−1, 5 to 6 μg μL−1 in 8 m urea). Incubations were performed at 25°C with gentle agitation for 0, 15, or 30 min. Following incubation, the chloroplasts were re-isolated through 40% (v/v) percoll gradients, lysed, resuspended in SDS sample buffer, and analyzed by SDS-PAGE (15%, w/v). To confirm chloroplast localization, intact chloroplasts were treated with thermolysin (0.1 mg mg chlorophyll−1) for 20 min on ice following the initial 30-min incubation.
Western Blot
Proteins were extracted from Arabidopsis leaves (1.5 g) by pulverizing in liquid nitrogen, suspending (1 g mL−1), and subsequent grinding in 50 mm Tris (pH 8.2), 5 mm MgCl2, 1 mm EDTA, and 100 μm NiSO4. Arabidopsis chloroplasts were isolated according to Rensink et al. (1998) and lysed in 50 mm Tris (pH 8.2), 5 mm MgCl2, and 1 mm EDTA for 10 min. Pea mitochondria were isolated according to Mulligan et al. (1991) and lysed by freeze/thawing in 0.3 m mannitol, 1 mm EDTA, 0.05% (w/v) Cys, and 10 mm MOPS [3-(N-morpholino)-propanesulfonic acid] (pH 7.2). The supernatants after centrifuging for 10 min at 14,000 rpm at 4°C were assayed for protein using Coomassie Plus Protein Assay Reagent (Pierce, Rockford, IL). Arabidopsis leaf and chloroplast stromal proteins and pea mitochondrial proteins (approximately 250 μg) with prestained low-range Mr markers (Bio-Rad Laboratories, Hercules, CA) were separated by SDS-PAGE (12.5%, w/v) and electroblotted to polyvinylidene difluoride membranes as described (Wang et al., 1995). Immunoblot analyses were conducted with 1:2,500 and 1:5,000 dilutions of primary antibodies for AtDEF1 and 2 and a mitochondrial-specific isoenzyme 1 NADH-Glu-dehydrogenase of Vitis vinifera (Loulakakis and Roubelakis-Angelakis, 1990; Turano et al., 1997; antibody provided by K.A. Roubelakis-Angelakis), respectively, for 90 min. Antibodies for AtDEF1 and 2 were raised (Strategic Biosolutions, Ramona, CA) against full-length AtDEF1 and 2 isolated as inclusion bodies. Secondary antibody (goat anti-rabbit IgG alkaline phosphatase conjugate; Bio-Rad Laboratories) was used at a 1:3,000 dilution for 90 min. Thorough distilled water and 0.1 m Tris, 0.5 m NaCl, and 0.05% (v/v) Tween 20 rinses were conducted after both the primary and secondary antibody incubations. Blots were developed using nitroblue tetrazolium (300 μg mL−1; Bio-Rad Laboratories) and 5-bromo-4-chloro-3-indolyl phosphate (150 μg mL−1; Bio-Rad Laboratories) in 0.1 m NaHCO2 and 1 mm MgCl2 at pH 9.5 until products were detected and stopped by rinsing with water as described (Zheng et al., 1998).
Expression
The partial cDNAs (cAtDEF1 and 2) encoding the mature forms of the protein (pAtDEF1 and 2) were transformed into BL21(DE3) pLysS (Novagen Inc.) and grown in Luria-Bertani broth medium with ampicillin (100 μg mL−1) and chloramphenicol (34 μg mL−1) for expression. Cells were induced with 0.4 or 1.0 mm IPTG after attaining an A600 of 0.4, and cultured for an additional 12 h at 25°C, 30°C, or 37°C. pAtDEF2 was soluble in much greater quantities compared with pAtDEF1 when using 0.4 mm IPTG at 25°C for 12 h. The cells were harvested and lysed in binding buffer plus 100 μm NiSO4 to retain an active and stable form of the enzyme (Rajagopalan and Pei, 1998). Average yield of soluble pAtDEF1 and 2 was 7 and 45 mg L culture−1, respectively. The N termini of the recombinant processed forms were sequenced utilizing conventional Edman degradative sequencing (Macromolecular Structure Analysis Facility, UK Markey Cancer Center).
Purification
Soluble pAtDEF1 and 2 were affinity purified using either: (a) His-Bind resin (Novagen Inc.; cells lysed in binding buffer, 5 mm imidazole, 1 m NaCl, 20 mm Tris-HCl [pH 7.9], and 100 μm NiSO4 and processed batchwise), or (b) HiTrap affinity columns (Amersham Pharmacia Biotech Inc., Piscataway, NJ; cells lysed in binding buffer, 10 mm each mono- and di-basic phosphate [pH 7.4], 0.5 m NaCl, 10 mm imidazole, and 100 μm NiSO4), according to the manufacturer's instructions with the inclusion of nickel to ensure preservation of enzyme activity. Prior to activity measurements, the imidazole used to elute the proteins was removed by exhaustive dialysis.
Peptide Deformylase Assays
The pH optimum of pAtDEF2 was broad and extended from pH 7.5 to 9.5 (data not shown) and presumed to be similar for pAtDEF1.
N-Formyl-Met-Leu-ρ-NA
Spectrophotometric assays of peptide deformylase activity (Wei and Pei, 1997) were conducted at 25°C in polystyrene cuvettes containing 50 mm MES (2-[N-morpholino] ethanesulfonic acid), 50 mm bis-tris propane {1,3-bis[tris (hydroxymethyl)methylamino]propane}, pH 8, 200 μm NiSO4, and 0 to 200 μm peptide substrate (N-formyl-Met-Leu-ρ-nitroanilide substrate, BACHEM Bioscience Inc., King of Prussia, PA) and 1.0 unit Aeromonas proteolytica aminopeptidase (Sigma, St. Louis). The reactions were initiated by the addition of pAtDEF1 (300 μg) or pAtDEF2 (7.5 μg) enzyme. The release of ρ-nitroaniline was measured by monitoring the increase in A405 using a UV-201 PC scanning spectrophotometer (Shimadzu Scientific Instruments, Inc., Columbia, MD) and initial velocities calculated from the early part of the reaction progression curves (<60s).
N-Formyl-Met-Ala-Ser
Spectrophotometric assays of peptide deformylase activity (Lazennec and Meinnel, 1997) were conducted at 25°C in quartz cuvettes containing 50 mm MES, 50 mm bis-tris propane (pH 8), 0 to 9,000 μm peptide substrate (N-formyl-Met-Ala-Ser substrate; BACHEM Bioscience Inc.), 12.0 mm β-NAD+, and 1.2 unit formate dehydrogenase (F8649, Sigma). The reactions were initiated by the addition of pAtDEF1 (900 μg) or pAtDEF2 (50 μg) enzyme. The formation of β-NADH was measured by monitoring the increase in A340 using UV-201 PC scanning spectrophotometer (Shimadzu Scientific Instruments, Inc.), and initial velocities calculated from the linear part of the reaction progression curves.
Peptide Deformylase Assay Inhibition
Affinity-purified pAtDEF1 (300 μg) was pre-incubated for 3 min in the absence or presence of 0 to 20 μm actinonin (50 mm MES, and 50 mm bis-tris propane, pH 8) prior to initiating the assay with 4 mm N-formyl-Met-Ala-Ser. Affinity-purified pAtDEF2 (7.5 μg) was pre-incubated for 3 min in the absence or presence of 0 to 300 nm actinonin (50 mm Tris-HCl, pH 8.2, 5 mm MgCl2, and 1 mm EDTA) prior to initiating the assay with 200 μm N-formyl-Met-Leu-ρ-nitroanilide; similar results were observed when the assay was initiated with 40, 65, and 125 μm N-formyl-Met-Leu-ρ-nitroanilide (data not shown).
Plant Treatment with Actinonin
Arabidopsis seeds (Ecotype Col-0, Lehle Seeds) were imbibed and seedlings cultured at room temperature with constant light (50 μmol m−2 s−1) in 200 μL Murashige and Skoog basal salts (Sigma) with 0.2% (w/v) phytagel in the wells of a 96-well microtiter plate in the absence or presence of actinonin (0.81, 1.6, and 3.2 mm). Images were taken with a dissecting microscope (Stemi 2000-C; Carl Zeiss, Inc., Thornwood, NY) 2 and 5 d after the seeds were sown. Seedlings were monitored for a further 2 weeks with no perceivable change in the effects of actinonin (data not shown).
ACKNOWLEDGMENTS
We are thankful to Dehua Pei (Department of Chemistry, Ohio State University, Columbus) for his selfless expertise lent to these studies. Many thanks to Carol Beach and Mike Russ at the Macromolecular Structure Analysis Facility, UK Markey Cancer Center for the protein sequence and DNA sequence data and primer synthesis, respectively. Mitochondrial-specific isoenzyme 1 NADH-Glu-dehydrogenase of V. vinifera antibody was kindly provided by K.A. Roubelakis-Angelakis. Our thanks also go to Brent W. Meier for his assistance with HPLC analysis of purified pAtDEF1 and 2.
Footnotes
This work was supported by the U.S. Department of Energy (grant no. DEFG02–92ER20075 to R.L.H.). This is Kentucky Agricultural Experiment Station article no. 00–11–202.
LITERATURE CITED
- Apfel C, Banner DW, Bur D, Dietz M, Hirata T, Hubschwerlen C, Locher H, Page MG, Pirson W, Rosse G. Hydroxamic acid derivatives as potent peptide deformylase inhibitors and antibacterial agents. J Med Chem. 2000;43:2324–2331. doi: 10.1021/jm000018k. [DOI] [PubMed] [Google Scholar]
- Becker A, Schlichting I, Kabsch W, Schultz S, Wagner AFV. Structure of peptide deformylase and identification of the substrate binding site. J Biol Chem. 1998;273:11413–11416. doi: 10.1074/jbc.273.19.11413. [DOI] [PubMed] [Google Scholar]
- Bianchetti R, Lucchini G, Sartirana ML. Endogenous synthesis of formyl-methionine peptides in isolated mitochondria and chloroplasts. Biochem Biophys Res Commun. 1971;42:97–102. doi: 10.1016/0006-291x(71)90367-6. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 2000;5:174–182. doi: 10.1016/s1360-1385(00)01598-3. [DOI] [PubMed] [Google Scholar]
- Chen DZ, Patel DV, Hackbarth CJ, Wang W, Dreyer G, Young DC, Margolis PS, Wu C, Ni ZJ, Trias J. Actinonin, a naturally occurring antibacterial agent, is a potent deformylase inhibitor. Biochemistry. 2000;39:1256–1262. doi: 10.1021/bi992245y. [DOI] [PubMed] [Google Scholar]
- Clements JM, Beckett RP, Brown A, Catlin G, Lobell M, Palan S, Thomas W, Whittaker M, Wood S, Salama S. Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor. Antimicrobrial Agents Chemother. 2001;45:563–570. doi: 10.1128/AAC.45.2.563-570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand DJ, Gordon Green B, O'Connell JF, Grant SK. Peptide aldehyde inhibitors of bacterial peptide deformylases. Arch Biochem Biophys. 1999;367:297–302. doi: 10.1006/abbi.1999.1274. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinta C, Persson B, Jornvall H, von Heijne G. Sequence determinants of cytosolic N-terminal protein processing. Eur J Biochem. 1986;154:193–196. doi: 10.1111/j.1432-1033.1986.tb09378.x. [DOI] [PubMed] [Google Scholar]
- Giglione C, Serero A, Pierre M, Boisson B, Meinnel T. Identification of eukaryotic peptide deformylases reveals universality of N-terminal protein processing mechanisms. EMBO J. 2000;19:5916–5929. doi: 10.1093/emboj/19.21.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon Green B, Toney JH, Kozarich JW, Grant SK. Inhibition of bacterial peptide deformylase by biaryl acid analogs. Arch Biochem Biophys. 2000;375:355–358. doi: 10.1006/abbi.1999.1673. [DOI] [PubMed] [Google Scholar]
- Groche D, Becker A, Schlichting I, Kabsch W, Schultz S, Wagner AFV. Isolation and crystallization of functionally competent Escherichia colipeptide deformylase forms containing either iron or nickel in the active site. Biochem Biophys Res Commun. 1998;246:342–346. doi: 10.1006/bbrc.1998.8616. [DOI] [PubMed] [Google Scholar]
- Hanson AD, Gage DA, Shachar-Hill Y. Plant one-carbon metabolism and its engineering. Trends Plant Sci. 2000;5:206–213. doi: 10.1016/s1360-1385(00)01599-5. [DOI] [PubMed] [Google Scholar]
- Hao B, Gong W, Rajagopalan PT, Zhou Y, Pei D, Chan MK. Structural basis for the design of antibiotics targeting peptide deformylase. Biochemistry. 1999;38:4712–4719. doi: 10.1021/bi982594c. [DOI] [PubMed] [Google Scholar]
- Houtz RL, Stults JT, Mulligan RM, Tolbert NE. Post-translational modifications in the large subunit of ribulose bisphosphate carboxylase/oxygenase. Proc Natl Acad Sci USA. 1989;86:1855–1859. doi: 10.1073/pnas.86.6.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YJ, Rajagopalan PT, Pei D. H-phosphonate derivatives as novel peptide deformylase inhibitors. Bioorg Med Chem Lett. 1998;8:2479–2482. doi: 10.1016/s0960-894x(98)00443-0. [DOI] [PubMed] [Google Scholar]
- Hu YJ, Wei Y, Zhou Y, Rajagopalan PT, Pei D. Determination of substrate specificity for peptide deformylase through the screening of a combinatorial peptide library. Biochemistry. 1999;38:643–650. doi: 10.1021/bi9820412. [DOI] [PubMed] [Google Scholar]
- Huntington KM, Yi T, Wei Y, Pei D. Synthesis and antibacterial activity of peptide deformylase inhibitors. Biochemistry. 2000;39:4543–4551. doi: 10.1021/bi992452y. [DOI] [PubMed] [Google Scholar]
- Jayasekera MM, Kendall A, Shammas R, Dermyer M, Tomala M, Shapiro MA, Holler TP. Novel nonpeptidic inhibitors of peptide deformylase. Arch Biochem Biophys. 2000;381:313–316. doi: 10.1006/abbi.2000.1987. [DOI] [PubMed] [Google Scholar]
- Kanevski I, Maliga P, Rhoades DF, Gutteridge S. Plastome engineering of ribulose-1,5-bisphosphate carboxylase/oxygenase in tobacco to form a sunflower large subunit and tobacco small subunit hybrid. Plant Physiol. 1999;119:133–141. doi: 10.1104/pp.119.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh TA, Jefferson RA, Bevan MW. Targeting a foreign protein to chloroplasts using fusions to the transit peptide of a chlorophyll a/bprotein. Mol Gen Genet. 1988;215:38–45. doi: 10.1007/BF00331300. [DOI] [PubMed] [Google Scholar]
- Kunze I, Hensel G, Adler K, Bernard J, Neubohn B, Nilsson C, Stoltenburg R, Kohlwein SD, Kunze G. The green fluorescent protein targets secretory proteins to the yeast vacuole. Biochim Biophys Acta. 1999;1410:287–298. doi: 10.1016/s0005-2728(99)00006-7. [DOI] [PubMed] [Google Scholar]
- Lazennec C, Meinnel T. Formate dehydrogenase-coupled spectrophotometric assay of peptide deformylase. Anal Biochem. 1997;244:180–182. doi: 10.1006/abio.1996.9910. [DOI] [PubMed] [Google Scholar]
- Loulakakis CA, Roubelakis-Angelakis KA. Immunocharacterization of NADH-glutamate dehydrogenase from Vitis viniferaL. Plant Physiol. 1990;94:109–113. doi: 10.1104/pp.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini G, Bianchetti R. Initiation of protein synthesis in isolated mitochondria and chloroplasts. Biochim Biophys Acta. 1980;608:54–61. doi: 10.1016/0005-2787(80)90133-1. [DOI] [PubMed] [Google Scholar]
- Margolis PS, Hackbarth CJ, Young DC, Wang W, Chen D, Yuan Z, White R, Trias J. Peptide deformylase in Staphylococcus aureus: resistance to inhibition is mediated by mutations in the formyltransferase gene. Antimicrobrial Agents Chemother. 2000;44:1825–1831. doi: 10.1128/aac.44.7.1825-1831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel D, Pochet S, Marliere P. Genetic characterization of polypeptide deformylase, a distinctive enzyme of eubacterial translation. EMBO J. 1994;13:914–923. doi: 10.1002/j.1460-2075.1994.tb06335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI. Endosymbiosis and evolution of the plant cell. Curr Opin Plant Biol. 1999;2:513–519. doi: 10.1016/s1369-5266(99)00025-4. [DOI] [PubMed] [Google Scholar]
- Meinnel T. Peptide deformylase of eukaryotic protists: a target for new antiparasitic agents? Parasitol Today. 2000;16:165–168. doi: 10.1016/s0169-4758(99)01627-0. [DOI] [PubMed] [Google Scholar]
- Meinnel T, Lazennec C, Blanquet S. Mapping of the active site zinc ligands of peptide deformylase. J Mol Biol. 1995;254:175–183. doi: 10.1006/jmbi.1995.0609. [DOI] [PubMed] [Google Scholar]
- Meinnel T, Patiny L, Ragusa S, Blanquet S. Design and synthesis of substrate analogue inhibitors of peptide deformylase. Biochemistry. 1999;38:4287–4295. doi: 10.1021/bi982622r. [DOI] [PubMed] [Google Scholar]
- Michel H, Hunt DF, Shabanowitz J, Bennett J. Tandem mass spectrometry reveals that three photosystem II proteins of spinach chloroplasts contain N-acetyl-O-phosphothreonine at their NH2termini. J Biol Chem. 1988;263:1123–1130. [PubMed] [Google Scholar]
- Mills WR, Joy KW. A rapid method for isolation of purified, physiologically active chloroplasts, used to study the intracellular distribution of amino acids in pea leaves. Planta. 1980;148:75–83. doi: 10.1007/BF00385445. [DOI] [PubMed] [Google Scholar]
- Mulligan RM, Leon P, Walbot V. Transcriptional and posttranscriptional regulation of maize mitochondrial gene expression. Mol Cell Biol. 1991;11:533–543. doi: 10.1128/mcb.11.1.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa S, Blanquet S, Meinnel T. Control of peptide deformylase activity by metal cations. J Mol Biol. 1998;280:515–523. doi: 10.1006/jmbi.1998.1883. [DOI] [PubMed] [Google Scholar]
- Rajagopalan PT, Datta A, Pei D. Purification, characterization, and inhibition of peptide deformylase from Escherichia coli. Biochemistry. 1997a;36:13910–13918. doi: 10.1021/bi971155v. [DOI] [PubMed] [Google Scholar]
- Rajagopalan PT, Grimme S, Pei D. Characterization of cobalt(II)-substituted peptide deformylase: function of the metal ion and the catalytic residue Glu-133. Biochemistry. 2000;39:779–790. doi: 10.1021/bi9919899. [DOI] [PubMed] [Google Scholar]
- Rajagopalan PT, Pei D. Oxygen-mediated inactivation of peptide deformylase. J Biol Chem. 1998;273:22305–22310. doi: 10.1074/jbc.273.35.22305. [DOI] [PubMed] [Google Scholar]
- Rajagopalan PT, Yu XC, Pei D. Peptide deformylase: a new type of mononuclear iron protein. J Am Chem Soc. 1997b;119:12418–12419. [Google Scholar]
- Rensink WA, Pilon M, Weisbeek P. Domains of a transit sequence required for in vivo import in Arabidopsis chloroplasts. Plant Physiol. 1998;118:691–699. doi: 10.1104/pp.118.2.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruelland E, Miginiac-Maslow M. Regulation of chloroplast enzyme activities by thioredoxins: activation or relief from inhibition? Trends Plant Sci. 1999;4:136–141. doi: 10.1016/s1360-1385(99)01391-6. [DOI] [PubMed] [Google Scholar]
- Scheller HV, Okkels JS, Hoj PB, Svendsen I, Roepstorff P, Moller BL. The primary structure of a 4.0-kDa photosystem I polypeptide encoded by the chloroplast psaIgene. J Biol Chem. 1989;264:18402–18406. [PubMed] [Google Scholar]
- Sharma J, Panico M, Barber J, Morris HR. Characterization of the low molecular weight photosystem II reaction center subunits and their light-induced modifications by mass spectrometry. J Biol Chem. 1997a;272:3935–3943. doi: 10.1074/jbc.272.7.3935. [DOI] [PubMed] [Google Scholar]
- Sharma J, Panico M, Barber J, Morris HR. Purification and determination of intact molecular mass by electrospray ionization mass spectrometry of the photosystem II reaction center subunits. J Biol Chem. 1997b;272:33153–33157. doi: 10.1074/jbc.272.52.33153. [DOI] [PubMed] [Google Scholar]
- Sharma J, Panico M, Shipton CA, Nilsson F, Morris HR, Barber J. Primary structure characterization of the photosystem II D1 and D2 subunits. J Biol Chem. 1997c;272:33158–33166. doi: 10.1074/jbc.272.52.33158. [DOI] [PubMed] [Google Scholar]
- Sigrist-Nelson K, Sigrist H, Azzi A. Characterization of the dicyclohexylcarbodiimide-binding protein isolated from chloroplast membranes. Eur J Biochem. 1978;92:9–14. doi: 10.1111/j.1432-1033.1978.tb12717.x. [DOI] [PubMed] [Google Scholar]
- Silva-Filho Mde C, Chaumont F, Leterme S, Boutry M. Mitochondrial and chloroplast targeting sequences in tandem modify protein import specificity in plant organelles. Plant Mol Biol. 1996;30:769–780. doi: 10.1007/BF00019010. [DOI] [PubMed] [Google Scholar]
- Silva-Filho Mde C, Wieers M-C, Flugge U-I, Chaumont F, Boutry M. Different in vitro and in vivotargeting properties of the transit peptide of a chloroplast envelope inner membrane protein. J Biol Chem. 1997;272:15264–15269. doi: 10.1074/jbc.272.24.15264. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turano FJ, Thakkar SS, Fang T, Weisemann JM. Characterization and expression of NAD(H)-dependent glutamate dehydrogenase genes in Arabidopsis. Plant Physiol. 1997;113:1329–1341. doi: 10.1104/pp.113.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waegemann K, Soll J. Characterization and isolation of the chloroplast protein import machinery. Methods Cell Biol. 1995;50:255–267. doi: 10.1016/s0091-679x(08)61035-3. [DOI] [PubMed] [Google Scholar]
- Wang P, Royer M, Houtz RL. Affinity purification of ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit εN-methyltransferase. Protein Expr Purif. 1995;6:528–536. doi: 10.1006/prep.1995.1070. [DOI] [PubMed] [Google Scholar]
- Wei Y. Peptide deformylase: characterization and antibacterial drug design. PhD thesis. Columbus: The Ohio State University; 2000. [Google Scholar]
- Wei Y, Pei D. Continuous spectrophotometric assay of peptide deformylase. Anal Biochem. 1997;250:29–34. doi: 10.1006/abio.1997.2194. [DOI] [PubMed] [Google Scholar]
- Wei Y, Yi T, Huntington KM, Chaudhury C, Pei D. Identification of a potent peptide deformylase inhibitor from a rationally designed combinatorial library. J Comb Chem. 2000;2:650–657. doi: 10.1021/cc000036n. [DOI] [PubMed] [Google Scholar]
- Werhahn W, Niemeyer A, Jänsch L, Kruft V, Schmitz UK, Braun H-P. Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis: identification of multiple forms of TOM20. Plant Physiol. 2001;125:943–954. doi: 10.1104/pp.125.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Andrews TJ. The gene for the ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) small subunit relocated to the plastid genome of tobacco directs the synthesis of small subunits that assemble into Rubisco. Plant Cell. 2001;13:193–205. doi: 10.1105/tpc.13.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MA, Dirk LMA, Houtz RL. Characterization of a chloroplast-localized peptide deformylase from Arabidopsis thaliana(abstract no. 621) Plant Physiol. 2000;123:S-131. [Google Scholar]
- Zhang L, Paakkarinen V, van Wijk KJ, Aro EM. Co-translational assembly of the D1 protein into photosystem II. J Biol Chem. 1999;274:16062–16067. doi: 10.1074/jbc.274.23.16062. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Simel EJ, Klein PE, Royer MT, Houtz RL. Expression, purification, and characterization of recombinant ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit εN-methyltransferase. Protein Expr Purif. 1998;14:104–112. doi: 10.1006/prep.1998.0936. [DOI] [PubMed] [Google Scholar]