Abstract
Purpose
This research aims to identify the miRNAs that could target the genes overexpressed in prostate cancer so that miRNA-based therapeutics could be developed.
Methods
A 7mer-m8 model of microRNA targeting was utilized in order to analyse the relationship between microRNAs and overexpressed genes. The efficiency of miRNA binding was investigated using various parameters namely free energy (AMFE), GC and GC3 content, translation efficiency, cosine similarity metric, mRNA stability, free energy of RNA duplex, and base compositional difference. BLAST2GO software was used to elucidate the functional roles of the genes overexpressed in prostate cancer.
Results
The current research reveals that the coding sequences of the genes were found targeted with multiple miRNAs. For instance, the HPN gene was targeted by the microRNA miR-4279 at two distinct sites i.e. 263–278 and 746–761 in the coding sequence. In the present study, it was observed that the target region of the genes exhibited a comparatively high GC and GC3 contents in comparison to the flanking regions. A low translational rate and weak relationship between RSCU and tRNA were obtained which may be due to the absence of optimal codons.
Conclusion
In this study, we have uncovered the human miRNAs that have potential for binding to the coding sequences of 14 most overexpressed genes in prostate cancer and thereby could silence those genes.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00432-023-04910-z.
Keywords: miRNA-target prediction, Overexpressed genes, Prostate cancer, Translation rate, 7mer-seed match model, Therapeutic strategy
Introduction
Cancer is a disorder that arises from the unrestrained growth and multiplication of malfunctioning cells in the body. The most common cancer-causing factors are alcohol, tobacco, toxic medicines, chemicals, and some environmental factors (Irigaray et al. 2007). However, some inherited genetic mutations also contribute to the incidence of cancer (Peto and Houlston 2001). Around 15% of cancer is due to infections caused by certain viruses (Moore and Chang 2010). Globally cancer cases had risen by 23.6 million in 2019 and cancer deaths were equal to 10 million (Kocarnik et al. 2022). The second most frequently observed malignancy amongst men is prostate cancer having escalating mortality rate (Rawla 2019). A recent study revealed around 1,276,000 new cases in the year 2018 coupled with 359,000 deaths around the world (Culp et al. 2020). The fatality rate owing to prostate cancer in men is estimated to rise up to 740,000 in 2040 (Culp et al. 2020). Prostate cancer mostly develops in men over 50 years of age, but if it occurs in younger men, the condition may become aggressive (Zheng et al. 2020). A study showed that the family history is responsible in the case of 20% of prostate cancers, which is due to inherited genes as well as the same form of exposure to some environmental carcinogens and common lifestyle habits (Rawla 2019). Studies revealed that men having inherited genetic backgrounds are susceptible to a greater risk of prostate cancer (Choudhury et al. 2012). The genetic basis of prostate cancer is associated with both acquired and inherited genetic alterations. Mutated forms of some genes like BRCA1, BRCA2, HOXB13, and RNASEL contribute to hereditary prostate cancer. However, somatic mutations in certain genes namely TP53, PTEN, and SPOP are responsible for sporadic prostate cancer (Abumsimir and Ennaji 2019). It is vital to know the genetic basis of the disorder, such that efficacious diagnostic and treatment procedures can be developed. The diagnosis period experiences various complications accompanied by prolonged treatment which include intake of numerous drugs, chemotherapy, radiotherapy etc. (Litwin and Tan 2017). Therapies for eradicating prostate cancer are based upon the stages of disease development. Despite the treatment process being a prolonged one, early diagnosis with accurate screening and treatment with maintaining preventive measures increases the efficacy of the treatment and facilitates the curable and survival rate of the patient. Prostate cancer constitutes frequent alterations in certain gene expression (Calvo et al. 2002). Identification and targeting of these overexpressed genes with microRNAs has come out as a potential targeted therapeutic method (Massillo et al. 2017). MicroRNAs are non-coding RNAs that possess both oncogenic and tumor-suppressing activity (Markopoulos et al. 2017). They are the prime regulatory agents of target gene silencing. These small molecules are single-stranded, having a length of approximately 22–23 nucleotides, and are capable to repress any unwanted gene expression (Kadri et al. 2011). The first two miRNAs to be identified were lin-4 and let-7 in 1993 (in Caenorhabditis elegans) (Dalmay 2008). In mammals, miRNA undergoes complementary base pairing with target mRNA, and regulates genes at post-transcriptional level by either mRNA degradation or down-regulation (Panda et al. 2014). Usually, the miRNAs bind to the target mRNAs at 3ˈ UTR regions (Lee et al. 1993). However, from several studies it became evident that the coding sequences of genes also contain binding sites for miRNA (Gu et al. 2013). Several enzymes namely RNA polymerase II, Drosha, Exportin-5, and Dicer are involved in catalyzing the biosynthesis of miRNA (Vishnoi and Rani 2017). For silencing a gene, the miRNA molecule coheres with Argonaute protein and forms the RNA-induced silencing complex (RISC), which pairs with its complementary mRNA and prevents gene expression (Reddy 2015). The miRNAs are very vital for cellular proliferation, differentiation, and apoptosis (Carleton et al. 2007). The prime criteria of miRNA-target binding are the perfect seed match. Seed region of miRNA contains 7 nucleotides (2nd to 8th nt) in the 5ˈ-3ˈdirection. These 2nd to 8th nucleotides target the coding sequence of gene and thus, H-bonds are formed between miRNA and mRNA for the perfect formation of a duplex structure (Moore et al. 2015). The miRNAs are widespread throughout the body and are estimated to regulate around 30% of human genome (Wienholds and Plasterk 2005). From several studies it became evident that different molecular pathways of cancer can be regulated by miRNA targeting the oncogenes. The chief requisite for a miRNA to implement the remedial action is its entry inside the cytoplasm of the target cell.
In this study the 7mer-m8 model was used which involves the identification of miRNAs that can complementarily bind to the mRNA. Lewis et al. was the first to introduce the 7mer-m8 model in 2003 in one of their studies (Lewis et al. 2003). Research on miRNA could be a potentially effective treatment for prostate cancer; however, there have been no comprehensive studies conducted on this topic. A computational method was utilised for the purpose of predicting the binding sites of some mature miRNAs in the coding sequences of overexpressed genes in prostate cancer for the purpose of this study. In addition, the efficiency of binding was determined by using certain parameters such as free energy, GC and GC3 content, the efficiency of translation, cosine similarity metric, mRNA stability, free energy of RNA duplex formation, and base compositional difference. The BLAST2GO software was important in elucidating the roles played by the genes that had been found to be overexpressed in prostate cancer.
Materials and methodology
Data retrieval of overexpressed genes in prostate cancer
Axelsen et al. reported some genes overexpressed in prostate cancer (Axelsen et al. 2007). From these genes, the entire coding sequences of 14 most overexpressed genes were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/). A total of 2555 mature human miRNA sequences were taken from the miRBase 21 (http://www.mirbase.org). Both sequences were examined by the in-house software developed by the corresponding author (SC) to find out if the sequences were complementary.
MicroRNA target prediction: complete seed matching and variable non-seed matching
In the coding regions of the overexpressed prostate cancer genes, the microRNA targets were searched to find out their complementary binding. It was carried out by a 7mer-m8 seed match. The miRNA seed region lies in 2–8 nt positions (7-nucleotide long) in the 5’ end. It undergoes complementary binding with the target mRNA following Watson—Crick model of base-pairing (Lewis et al. 2003). The miRNA segment lying beside the seed region is called the non-seed region, which pairs up with seqflank region of mRNA. The analysis showed a perfect complementarity between seed region of miRNA and mRNA. A total of 6 mismatches were taken into consideration in binding between the non-seed region of miRNA and the seqflank of target mRNA.
For the ease of interpretation, the miRNA target site, and its upstream and downstream regions were split to obtain sequences based on codon frame (i.e. a multiple of three nucleotides), and the three regions were analysed individually. In our study, 18 nucleotides were considered (i.e. 6 codons in one frame) in the upstream and downstream regions of the target, as suggested by Kertesz et al. (Kertesz et al. 2007). It was suggested that, these regions hold the best correlation between miRNA activity and site accessibility (Kertesz et al. 2007).
Free energy of mRNA and miRNA binding
The extent to which the target sequence of mRNA is exposed for the ligation of miRNA-RISC complex is the site accessibility (Gu, Wang et al. 2012a; b). The miRNA and mRNA binding strength is estimated by the parameter free energy or minimal free energy (MFE). The free energy was measured using a 7-bp sliding window at 37 ℃ (in kcal/mol) (SantaLucia Jr 1998). The absolute value of the free energy was taken into consideration by omitting the negative sign for easy interpretation. Thus, high magnitude of free energy corresponds to a static interaction between the target mRNA and its corresponding miRNA. Moreover, the mRNA may adopt a secondary structure, and the accessibility of binding region of miRNA can be established from free energy calculations (Riffo-Campos, Riquelme et al. 2016). Longer sequences generally have a higher MFE value. Therefore, to cope with the bias, MFE value is adjusted. The adjusted minimal free energy (AMFE) of 100 nucleotides was computed using the formula (Ni et al. 2010):
The calculation of minimal free energy index was done as per (Zhang et al. 2006):
GC and GC3 content estimation of miRNA target sequences
The nucleotide composition of the miRNA target region, its downstream and upstream regions was analysed to estimate the GC and GC3 contents.
Translational efficiency (compAI) of the target sequences
The translational efficiencies of all overexpressed genes in their target, downstream and upstream regions were measured to know the difference in translational rates. The estimation is done by using the parameter compAI. An inverse relationship exists between translational rate and miRNA-based mRNA suppression, where increased translational rate is associated with lower suppression and vice versa. The value of compAI ranges from 0 to 1, where 0 represents the lowest translational rate and 1 represents the highest translational rate (Dilucca et al. 2015).
Cosine similarity metric (COSM)
The parameter COSM measures the degree of relationship between RSCU (relative synonymous codon usage) value of each codon and the tRNA species in the cytoplasm (Dilucca et al. 2015). The COSM value lies within 0–1, where 0 corresponds to least similarity and 1 corresponds to highest similarity.
Stability of mRNA (MSI)
The MSI is the measure of mRNA stability and is calculated in terms of csc (correlation stability coefficient) values (Presnyak, Alhusaini et al. 2015). MSI value ranges from − 1 to + 1. Greater positive MSI indicates high mRNA stability while greater negative MSI value indicates low mRNA stability.
Free energy of RNA duplex formation
The free energy change of the cds-miRNA duplex formation was analysed at 37 degree Celsius in order to predict how stable the RNA duplexes are (Xia, SantaLucia Jr et al. 1998). For this, we selected only the complete Watson–Crick base pairing of target with miRNA. Negative sign before the estimate was removed. Greater the free energy of RNA duplex formation, greater will be the stability.
Base compositional difference
Unpaired t test was performed to analyse the significant difference, if any, for base composition between the target and the flank regions. If the p value is less than 0.01 or 0.05, it implies that a significant difference exists between the two sequences with respect to the parameter.
Gene ontology (GO)
Gene ontology was performed using BLAST2GO software to display the biological functions, cellular components, and the molecular functions of each target gene (Conesa et al. 2005).
Results
Predicted miRNA targets in the coding sequences of overexpressed prostate cancer genes
The retrieval of the coding sequences of 14 prostate cancer overexpressed genes was done from NCBI. The 7mer-m8 model was used to make predictions regarding the locations of binding sites for human microRNAs on the mRNAs. The 2–8 nucleotides at the miRNA 5' end were examined to determine whether or not they were entirely complementary to the target region of the mRNA. On the other hand, in the pairing of the non-seed region, a maximum of 6 mismatches were allowed to be evaluated. As a result of targeting many locations in the mRNA, the microRNAs are able to hinder the translation process of overexpressed genes in cancer cells. It was also observed that a single miRNA was found to target the same gene, but at different locations. In the case of gene HPN, the miR-4279 targeted at two different positions within the gene (263–278 and 746–761). Again, for the gene USP9Y, the miR-1827, miR-4267, miR-4311, and miR-4328 were the 4 miRNAs that could bind to two different target positions; miR-1827 binding to the target positions 3244–3261 and 7501–7518, miR-4267 binding to the positions 206–221 and 5459–5474, miRNA-4311 binding to the positions 3046–3063 and 3225–3242 and miR-4328 binding to the positions 3973–3989 and 6036–6052. As a result we could conclude that a single mRNA sequence can have several regions matching the seed region of the same miRNA.
The 14 overexpressed prostate cancer genes were found to be targeted by 213 human microRNAs e.g. gene CHI3L1 was targeted by 10 miRNAs (miR-7162-5p, miR-4483, miR-4258, miR-4522, miR-3649, miR-6755-5p, miR-4318, miR-4297, miR-3125, miR-4451), gene HPN was targeted by 15 miRNAs (miR-3652, miR-4267, miR-4279, miR-4279, miR-2861, miR-151b, miR-4640-5p, miR-211-3p, miR-4478, miR-6763-5p, miR-4319, miR-6070, miR-4481, miR-4519, miR-4497), gene CDC27 was targeted by 6 miRNAs (miR-548ap-5p, miR-3606-3p, miR-3653, miR-4766-5p, miR-548ap-3p, miR-4650-5p), gene USP9Y was targeted by 45 miRNAs (miR-1827, miR-1827, miR-4456, miR-335-5p, miR-1279, miR-4641, miR-2053, miR-4646-3p, miR-1261, miR-3168, miR-4267, miR-4267, let-7a-3p, miR-4288, miR-655-3p, miR-4516, miR-4499, miR-4311, miR-4311, miR-302e, miR-4299, miR-5703, miR-4800-3p, miR-4328, miR-4328, miR-95-5p, miR-4645-5p, miR-4534, miR-6816-3p, miR-4297, let-7f-2-3p, miR-3144-3p, miR-518e-3p, miR-6774-3p, miR-3155b, miR-490-5p, miR-4795-5p, miR-1537-5p, miR-4782-3p, miR-4310, miR-4418, miR-4724-5p, miR-6077, miR-3606-5p, miR-3182), gene S100A8 was targeted by 2 miRNAs (miR-4267, miR-4296), gene SLC4A4 was targeted by 33 miRNAs (miR-3150b-3p, miR-551b-3p, miR-4419a, miR-1227-5p, miR-6870-5p, miR-4445-3p, miR-4516, miR-4499, miR-4311, miR-4300, miR-320e, miR-30b-5p, miR-30c-5p, miR-548ah-5p, miR-3606-3p, miR-4306, miR-3617-5p, miR-4296, miR-6729-3p, miR-892c-5p, miR-511-5p, miR-6512-5p, miR-3688-5p, miR-644a, miR-3145-5p, miR-4266, miR-4266, miR-4463, miR-4500, miR-588, miR-3123, miR-4519, miR-7515), gene PDLIM3 was targeted by 6 miRNAs (miR-4265, miR-4748, miR-627-3p, miR-4296, miR-326, miR-4322), gene HOXC6 by 7 miRNAs (miR-6808-3p, miR-1281, miR-668-5p, miR-718, miR-3195, miR-545-3p, miR-6134), gene ALDH3B2 by 15 miRNAs (miR-3665, miR-4267, miR-4279, miR-4748, miR-513a-5p, miR-4508, miR-4531, miR-5703, miR-484, miR-7704, miR-4514, miR-659-5p, miR-4721, miR-4798-5p, miR-920), gene ABHD2 by 13 miRNAs (miR-6809-3p, miR-4294, miR-4283, miR-4283, miR-4311, miR-6881-3p, miR-626, miR-3155b, miR-4659a-3p, miR-4766-5p, miR-6085, miR-509-5p, miR-491-3p), gene PDLIM5 by 8 miRNAs (miR-6086, miR-4279, miR-3663-5p, miR-320d, miR-4505, miR-320c, miR-4256, miR-4429), gene RFX3 by 20 miRNAs (miR-6852-3p, miR-7162-5p, miR-4419a, miR-4486, miR-597-5p, miR-620, miR-1273f, miR-548ap-5p, miR-4318, miR-3193, miR-6816-3p, miR-1825, miR-3130-5p, miR-1260a, miR-6771-3p, miR-4256, miR-432-3p, miR-1260b, miR-4275, miR-6833-3p), gene OR51E2 by 9 miRNAs (miR-1827, miR-1827, miR-1261, miR-4530, miR-4530, miR-6891-5p, miR-4668-5p, miR-4291, miR-4418), and gene PCDHGB4 by 24 miRNAs (miR-4279, miR-4286, miR-33b-5p, miR-4276, miR-548ap-5p, miR-548au-5p, miR-4508, miR-4508, miR-4492, miR-3155b, miR-3155a, miR-4419b, miR-4291, miR-4451, miR-330-5p, miR-4302, miR-4279, miR-4286, miR-33b-5p, miR-4276, miR-548ap-5p, miR-548au-5p, miR-4508, miR-4508) (Tables 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13 and 14). Since it was observed that a single mRNA may harbor different binding sites for several miRNAs, and also only one miRNA might bind to an mRNA at several distinct positions, hence all the target sequences as well as its upstream and downstream sequences were further analyzed.
Table 1.
Targeting of the prostate cancer gene CHI3L1 by 10 miRNAs through a complete seed match
| No. | miRNA name | Nucleotide sequence of miRNA (excluding 1st base at 5’ end) | Nucleotide sequence of target | Position of target sequence in coding sequence | Target and miRNA binding |
|---|---|---|---|---|---|
| 1 | miR-7162-5p | UGCUUCCUUUCUCAGCUG | SEQ-5'-GAATTTATAA|AGGAAGC-3' i.e. (SeqFlank|Seed) | 478–495 | seq-5'-GAATTTATAA|AGGAAGC-3'/miR-3'-GTCGACTCTT|TCCTTCG-5' |
| 2 | miR-4483 | GGGGUGGUCUGUUGUUG | SEQ-5'-CAGCATTTT|ACCACCC-3' i.e. (SeqFlank|Seed) | 442–458 | seq-5'-CAGCATTTT|ACCACCC-3'/miR-3'-GTTGTTGTC|TGGTGGG-5' |
| 3 | miR-4258 | CCCCGCCACCGCCUUGG | SEQ-5'-ACAGGCAGC|TGGCGGG-3' i.e. (SeqFlank|Seed) | 1028–1044 | seq-5'-ACAGGCAGC|TGGCGGG-3'/miR-3'-GGTTCCGCC|ACCGCCC-5' |
| 4 | miR-4522 | UGACUCUGCCUGUAGGCCGGU | SEQ-5'-AGCCTCCAACACC|CAGAGTC-3' i.e. (SeqFlank|Seed) | 327–347 | seq-5'-AGCCTCCAACACC|CAGAGTC-3'/miR-3'-TGGCCGGATGTCC|GTCTCAG-5' |
| 5 | miR-3649 | AGGGACCUGAGUGUCUAAG | SEQ-5'-CCTCGGCCAGC|AGGTCCC-3' i.e. (SeqFlank|Seed) | 933–951 | seq-5'-CCTCGGCCAGC|AGGTCCC-3'/miR-3'-GAATCTGTGAG|TCCAGGG-5' |
| 6 | miR-6755-5p | UAGGGUAGACACUGACAACGUU | SEQ-5'-GACCTTGCCTGGCT|CTACCCT-3' i.e. (SeqFlank|Seed) | 406–427 | seq-5'-GACCTTGCCTGGCT|CTACCCT-3'/miR-3'-TTGCAACAGTCACA|GATGGGA-5' |
| 7 | miR-4318 | CACUGUGGGUACAUGCU | SEQ-5'-TCCGCGGAG|CCACAGT-3' i.e. (SeqFlank|Seed) | 908–924 | seq-5'-TCCGCGGAG|CCACAGT-3'/miR-3'-TCGTACATG|GGTGTCA-5' |
| 8 | miR-4297 | UGCCUUCCUGUCUGUG | SEQ-5'-CAGTACCG|GGAAGGC-3' i.e. (SeqFlank|Seed) | 97–112 | seq-5'-CAGTACCG|GGAAGGC-3'/miR-3'-GTGTCTGT|CCTTCCG-5' |
| 9 | miR-3125 | UAGAGGAAGCUGUGGAGAGA | SEQ-5'-GCCCTTGACCGC|TTCCTCT-3' i.e. (SeqFlank|Seed) | 133–152 | seq-5'-GCCCTTGACCGC|TTCCTCT-3'/miR-3'-AGAGAGGTGTCG|AAGGAGA-5' |
| 10 | miR-4451 | UGGUAGAGCUGAGGACA | SEQ-5'-CTTGCCTGG|CTCTACC-3' i.e. (SeqFlank|Seed) | 409–425 | seq-5'-CTTGCCTGG|CTCTACC-3'/miR-3'-ACAGGAGTC|GAGATGG-5' |
Table 2.
Targeting of the prostate cancer gene HPN by 15 miRNAs through a complete seed match
| Sl. No. | miRNA name | Nucleotide sequence of miRNA (excluding 1st base at 5’ end) | Nucleotide sequence of target | Position of target sequence in coding sequence | Target and miRNA binding |
|---|---|---|---|---|---|
| 1 | miR-3652 | CGGCUGGAGGUGUGAGGA | SEQ-5'-ACAGAATACA|TCCAGCC-3' i.e. (SeqFlank|Seed) | 808–825 | seq-5'-ACAGAATACA|TCCAGCC-3'/miR-3'-AGGAGTGTGG|AGGTCGG-5' |
| 2 | miR-4267 | UCCAGCUCGGUGGCAC | SEQ-5'-CCCACTCC|GAGCTGG-3' i.e. (SeqFlank|Seed) | 299–314 | seq-5'-CCCACTCC|GAGCTGG-3'/miR-3'-CACGGTGG|CTCGACC-5' |
| 3 | miR-4279 | CUCUCCUCCCGGCUUC | SEQ-5'-TCAGCTGC|GAGGAGA-3' i.e. (SeqFlank|Seed) | 263–278 | seq-5'-TCAGCTGC|GAGGAGA-3'/miR-3'-CTTCGGCC|CTCCTCT-5' |
| 4 | miR-4279 | CUCUCCUCCCGGCUUC | SEQ-5'-CCAACAGC|GAGGAGA-3' i.e. (SeqFlank|Seed) | 746–761 | seq-5'-CCAACAGC|GAGGAGA-3'/miR-3'-CTTCGGCC|CTCCTCT-5' |
| 5 | miR-2861 | GGGGCCUGGCGGUGGGCGG | SEQ-5'-CAGCTGCCGGC|CAGGCCC-3' i.e. (SeqFlank|Seed) | 836–854 | seq-5'-CAGCTGCCGGC|CAGGCCC-3'/miR-3'-GGCGGGTGGCG|GTCCGGG-5' |
| 6 | miR-151b | UCGAGGAGCUCACAGUCU | SEQ-5'-GGCTGCTGTG|CTCCTCG-3' i.e. (SeqFlank|Seed) | 221–238 | seq-5'-GGCTGCTGTG|CTCCTCG-3'/miR-3'-TCTGACACTC|GAGGAGC-5' |
| 7 | miR-4640-5p | UGGGCCAGGGAGCAGCUGGUGGG | SEQ-5'-GGCACTGGCTGTGCC|CTGGCCC-3' i.e. (SeqFlank|Seed) | 1132–1154 | seq-5'-GGCACTGGCTGTGCC|CTGGCCC-3'/miR-3'-GGGTGGTCGACGAGG|GACCGGG-5' |
| 8 | miR-211-3p | CGUGGGGAAACGACAGGGACG | SEQ-5'-TGGCCCAGGCCTC|TCCCCAC-3' i.e. (SeqFlank|Seed) | 665–685 | seq-5'-TGGCCCAGGCCTC|TCCCCAC-3'/miR-3'-GCAGGGACAGCAA|AGGGGTG-5' |
| 9 | miR-4478 | GAGGCUGAGCUGAGGAG | SEQ-5'-CGTGGCAAG|TCAGCCT-3' i.e. (SeqFlank|Seed) | 524–540 | seq-5'-CGTGGCAAG|TCAGCCT-3'/miR-3'-GAGGAGTCG|AGTCGGA-5' |
| 10 | miR-6763-5p | CUGGGGAGUGGCUGGGGAG | SEQ-5'-GGCCCAGGCCT|CTCCCCA-3' i.e. (SeqFlank|Seed) | 666–684 | seq-5'-GGCCCAGGCCT|CTCCCCA-3'/miR-3'-GAGGGGTCGGT|GAGGGGT-5' |
| 11 | miR-4319 | UCCCUGAGCAAAGCCAC | SEQ-5'-ATGGGCTTC|CTCAGGG-3' i.e. (SeqFlank|Seed) | 277–293 | seq-5'-ATGGGCTTC|CTCAGGG-3'/miR-3'-CACCGAAAC|GAGTCCC-5' |
| 12 | miR-6070 | CCGGUUCCAGUCCCUGGAG | SEQ-5'-CTTCCCGGAGC|GGAACCG-3' i.e. (SeqFlank|Seed) | 612–630 | seq-5'-CTTCCCGGAGC|GGAACCG-3'/miR-3'-GAGGTCCCTGA|CCTTGGC-5' |
| 13 | miR-4481 | GGAGUGGGCUGGUGGUU | SEQ-5'-GGGCACTGA|CCCACTC-3' i.e. (SeqFlank|Seed) | 290–306 | seq-5'-GGGCACTGA|CCCACTC-3'/miR-3'-TTGGTGGTC|GGGTGAG-5' |
| 14 | miR-4519 | CAGCAGUGCGCAGGGCUG | SEQ-5'-GACAGCCGCC|CACTGCT-3' i.e. (SeqFlank|Seed) | 597–614 | seq-5'-GACAGCCGCC|CACTGCT-3'/miR-3'-GTCGGGACGC|GTGACGA-5' |
| 15 | miR-4497 | CUCCGGGACGGCUGGGC | SEQ-5'-CCCACTGCT|TCCCGGA-3' i.e. (SeqFlank|Seed) | 605–621 | seq-5'-CCCACTGCT|TCCCGGA-3'/miR-3'-CGGGTCGGC|AGGGCCT-5' |
Table 3.
Targeting of the prostate cancer gene CDC27 by 6 miRNAs through a complete seed match
| No. | miRNA name | Nucleotide sequence of miRNA (excluding 1st base at 5’ end) | Nucleotide sequence of target | Position of target sequence in coding sequence | Target and miRNA binding |
|---|---|---|---|---|---|
| 1 | miR-548ap-5p | AAAAGUAAUUGCGGUCUUU | SEQ-5'-AAAGTTCAGTT|TTACTTT-3' i.e. (SeqFlank|Seed) | 806–824 | seq-5'-AAAGTTCAGTT|TTACTTT-3'/miR-3'-TTTCTGGCGTT|AATGAAA-5' |
| 2 | miR-3606-3p | GAUUCAUCACUUUCUUUAAAA | SEQ-5'-TGCAGCTGAAAGT|GATGAAT-3' i.e. (SeqFlank|Seed) | 1257–1277 | seq-5'-TGCAGCTGAAAGT|GATGAAT-3'/miR-3'-AAAATTTCTTTCA|CTACTTA-5' |
| 3 | miR-3653 | CUAAGAAGUUGACUGAAG | SEQ-5'-CCCTCGTTTA|CTTCTTA-3' i.e. (SeqFlank|Seed) | 1016–1033 | seq-5'-CCCTCGTTTA|CTTCTTA-3'/miR-3'-GAAGTCAGTT|GAAGAAT-5' |
| 4 | miR-4766-5p | UCUGAAAGAGCAGUUGGUGUU | SEQ-5'-AAAAGATGTTGCT|CTTTCAG-3' i.e. (SeqFlank|Seed) | 447–467 | seq-5'-AAAAGATGTTGCT|CTTTCAG-3'/miR-3'-TTGTGGTTGACGA|GAAAGTC-5' |
| 5 | miR-548ap-3p | UUUUCAUUAACACCAAAAA | SEQ-5'-TTTTATTTGCA|AATGAAA-3' i.e. (SeqFlank|Seed) | 941–959 | seq-5'-TTTTATTTGCA|AATGAAA-3'/miR-3'-AAAAACCACAA|TTACTTT-5' |
| 6 | miR-4650-5p | UCAGGCCUCUUUCUACCUU | SEQ-5'-AAATTCGCCAG|AGGCCTG-3' i.e. (SeqFlank|Seed) | 498–516 | seq-5'-AAATTCGCCAG|AGGCCTG-3'/miR-3'-TTCCATCTTTC|TCCGGAC-5' |
Table 4.
Targeting of the prostate cancer gene USP9Y by 45 miRNAs through a complete seed match
| No. | miRNA Name | Nucleotide sequence of miRNA (excluding 1st base at 5’ end) | Nucleotide sequence of target | Position of target sequence in coding sequence | Target and miRNA binding |
|---|---|---|---|---|---|
| 1 | miR-1827 | UGAGGCAGUAGAUUGAAU | SEQ-5'-TTTGGTCCTT|CTGCCTC-3' i.e. (SeqFlank|Seed) | 3244–3261 | seq-5'-TTTGGTCCTT|CTGCCTC-3'/miR-3'-TAAGTTAGAT|GACGGAG-5' |
| 2 | miR-1827 | UGAGGCAGUAGAUUGAAU | SEQ-5'-CCTCATTCAC|CTGCCTC-3' i.e. (SeqFlank|Seed) | 7501–7518 | seq-5'-CCTCATTCAC|CTGCCTC-3'/miR-3'-TAAGTTAGAT|GACGGAG-5' |
| 3 | miR-4456 | CCUGGUGGCUUCCUUUU | SEQ-5'-AAACTTATG|CCACCAG-3' i.e. (SeqFlank|Seed) | 3136–3152 | seq-5'-AAACTTATG|CCACCAG-3'/miR-3'-TTTTCCTTC|GGTGGTC-5' |
| 4 | miR-335-5p | UCAAGAGCAAUAACGAAAAAUGU | SEQ-5'-AATTACTTCGCTTTT|GCTCTTG-3' i.e. (SeqFlank|Seed) | 6905–6927 | seq-5'-AATTACTTCGCTTTT|GCTCTTG-3'/miR-3'-TGTAAAAAGCAATAA|CGAGAAC-5' |
| 5 | miR-1279 | UCAUAUUGCUUCUUUCU | SEQ-5'-AATAAATTT|CAATATG-3' i.e. (SeqFlank|Seed) | 2904–2920 | seq-5'-AATAAATTT|CAATATG-3'/miR-3'-TCTTTCTTC|GTTATAC-5' |
| 6 | miR-4641 | UGCCCAUGCCAUACUUUUGCCUCA | SEQ-5'-TGACTGAAATGTATTA|CATGGGC-3' i.e. (SeqFlank|Seed) | 4574–4597 | seq-5'-TGACTGAAATGTATTA|CATGGGC-3'/miR-3'-ACTCCGTTTTCATACC|GTACCCG-5' |
| 7 | miR-2053 | GUGUUAAUUAAACCUCUAUUUAC | SEQ-5'-ATTCATAGATGTATT|ATTAACA-3' i.e. (SeqFlank|Seed) | 433–455 | seq-5'-ATTCATAGATGTATT|ATTAACA-3'/miR-3'-CATTTATCTCCAAAT|TAATTGT-5' |
| 8 | miR-4646-3p | GACCCUUCCCUCUCCCUGUUA | SEQ-5'-GCACAGCAAGAGA|GAAGGGT-3' i.e. (SeqFlank|Seed) | 4094–4114 | seq-5'-GCACAGCAAGAGA|GAAGGGT-3'/miR-3'-ATTGTCCCTCTCC|CTTCCCA-5' |
| 9 | miR-1261 | AUGGAUAAGGCUUUGGCUU | SEQ-5'-AAAATAATATC|TTATCCA-3' i.e. (SeqFlank|Seed) | 1178–1196 | seq-5'-AAAATAATATC|TTATCCA-3'/miR-3'-TTCGGTTTCGG|AATAGGT-5' |
| 10 | miR-3168 | GAGUUCUACAGUCAGAC | SEQ-5'-TTCCACTTC|TAGAACT-3' i.e. (SeqFlank|Seed) | 506–522 | seq-5'-TTCCACTTC|TAGAACT-3'/miR-3'-CAGACTGAC|ATCTTGA-5' |
| 11 | miR-4267 | UCCAGCUCGGUGGCAC | SEQ-5'-CACATACT|GAGCTGG-3' i.e. (SeqFlank|Seed) | 206–221 | seq-5'-CACATACT|GAGCTGG-3'/miR-3'-CACGGTGG|CTCGACC-5' |
| 12 | miR-4267 | UCCAGCUCGGUGGCAC | SEQ-5'-TTCCTCGA|GAGCTGG-3' i.e. (SeqFlank|Seed) | 5459–5474 | seq-5'-TTCCTCGA|GAGCTGG-3'/miR-3'-CACGGTGG|CTCGACC-5' |
| 13 | let-7a-3p | CUUUCUGUCAUCUAACAUAUC | SEQ-5'-GATTCTGAAGATG|ACAGAAA-3' i.e. (SeqFlank|Seed) | 2827–2847 | seq-5'-GATTCTGAAGATG|ACAGAAA-3'/miR-3'-CTATACAATCTAC|TGTCTTT-5' |
| 14 | miR-4288 | UUGUCUGCUGAGUUUCC | SEQ-5'-CAAACCAGG|GCAGACA-3' i.e. (SeqFlank|Seed) | 2681–2697 | seq-5'-CAAACCAGG|GCAGACA-3'/miR-3'-CCTTTGAGT|CGTCTGT-5' |
| 15 | miR-655-3p | UUUCUCCAAUUGGUACAUAAUA | SEQ-5'-CAGTATTTAAGAAG|TGGAGAA-3' i.e. (SeqFlank|Seed) | 4432–4453 | seq-5'-CAGTATTTAAGAAG|TGGAGAA-3'/miR-3'-ATAATACATGGTTA|ACCTCTT-5' |
| 16 | miR-4516 | GGGAGAAGGGUCGGGGC | SEQ-5'-GGTCTTGTC|CTTCTCC-3' i.e. (SeqFlank|Seed) | 6395–6411 | seq-5'-GGTCTTGTC|CTTCTCC-3'/miR-3'-CGGGGCTGG|GAAGAGG-5' |
| 17 | miR-4499 | AAGACUGAGAGGAGGGA | SEQ-5'-AGACTTCAT|TCAGTCT-3' i.e. (SeqFlank|Seed) | 2442–2458 | seq-5'-AGACTTCAT|TCAGTCT-3'/miR-3'-AGGGAGGAG|AGTCAGA-5' |
| 18 | miR-4311 | GAAAGAGAGCUGAGUGUG | SEQ-5'-CCCAGATACA|TCTCTTT-3' i.e. (SeqFlank|Seed) | 3046–3063 | seq-5'-CCCAGATACA|TCTCTTT-3'/miR-3'-GTGTGAGTCG|AGAGAAA-5' |
| 19 | miR-4311 | GAAAGAGAGCUGAGUGUG | SEQ-5'-ACCCCTTGAC|TCTCTTT-3' i.e. (SeqFlank|Seed) | 3225–3242 | seq-5'-ACCCCTTGAC|TCTCTTT-3'/miR-3'-GTGTGAGTCG|AGAGAAA-5' |
| 20 | miR-302e | UAAGUGCUUCCAUGCUU | SEQ-5'-TACTTGTGA|AGCACTT-3' i.e. (SeqFlank|Seed) | 4608–4624 | seq-5'-TACTTGTGA|AGCACTT-3'/miR-3'-TTCGTACCT|TCGTGAA-5' |
| 21 | miR-4299 | GCUGGUGACAUGAGAGGC | SEQ-5'-GATCATTATG|TCACCAG-3' i.e. (SeqFlank|Seed) | 5943–5960 | seq-5'-GATCATTATG|TCACCAG-3'/miR-3'-CGGAGAGTAC|AGTGGTC-5' |
| 22 | miR-5703 | AGGAGAAGUCGGGAAGGU | SEQ-5'-ACTGTGCGTT|CTTCTCC-3' i.e. (SeqFlank|Seed) | 6222–6239 | seq-5'-ACTGTGCGTT|CTTCTCC-3'/miR-3'-TGGAAGGGCT|GAAGAGG-5' |
| 23 | miR-4800-3p | CACCUGUCUGCCUGCCUAC | SEQ-5'-AAACCAGGGCA|GACAGGT-3' i.e. (SeqFlank|Seed) | 2682–2700 | seq-5'-AAACCAGGGCA|GACAGGT-3'/miR-3'-CATCCGTCCGT|CTGTCCA-5' |
| 24 | miR-4328 | CCAGUUUUCCCAGGAUU | SEQ-5'-TGTCCAAGC|AAAACTG-3' i.e. (SeqFlank|Seed) | 3973–3989 | seq-5'-TGTCCAAGC|AAAACTG-3'/miR-3'-TTAGGACCC|TTTTGAC-5' |
| 25 | miR-4328 | CCAGUUUUCCCAGGAUU | SEQ-5'-GTTTGTGAA|AAAACTG-3' i.e. (SeqFlank|Seed) | 6036–6052 | seq-5'-GTTTGTGAA|AAAACTG-3'/miR-3'-TTAGGACCC|TTTTGAC-5' |
| 26 | miR-95-5p | UCAAUAAAUGUCUGUUGAAUU | SEQ-5'-ATATCAAAGGAGA|TTTATTG-3' i.e. (SeqFlank|Seed) | 5276–5296 | seq-5'-ATATCAAAGGAGA|TTTATTG-3'/miR-3'-TTAAGTTGTCTGT|AAATAAC-5' |
| 27 | miR-4645-5p | ACCAGGCAAGAAAUAUUGU | SEQ-5'-GGAAAGTTGTT|TGCCTGG-3' i.e. (SeqFlank|Seed) | 3009–3027 | seq-5'-GGAAAGTTGTT|TGCCTGG-3'/miR-3'-TGTTATAAAGA|ACGGACC-5' |
| 28 | miR-4534 | GGAUGGAGGAGGGGUCU | SEQ-5'-ATGAGCCCT|CTCCATC-3' i.e. (SeqFlank|Seed) | 7466–7482 | seq-5'-ATGAGCCCT|CTCCATC-3'/miR-3'-TCTGGGGAG|GAGGTAG-5' |
| 29 | miR-6816-3p | GCUUCCACGUCCAGGAAG | SEQ-5'-TAAGCTGGAG|GTGGAAG-3' i.e. (SeqFlank|Seed) | 3831–3848 | seq-5'-TAAGCTGGAG|GTGGAAG-3'/miR-3'-GAAGGACCTG|CACCTTC-5' |
| 30 | miR-4297 | UGCCUUCCUGUCUGUG | SEQ-5'-CCAATACT|GGAAGGC-3' i.e. (SeqFlank|Seed) | 4279–4294 | seq-5'-CCAATACT|GGAAGGC-3'/miR-3'-GTGTCTGT|CCTTCCG-5' |
| 31 | let-7f-2-3p | CCUUUCUGUCAUCUGACAUAUC | SEQ-5'-GATTCTGAAGATGA|CAGAAAG-3' i.e. (SeqFlank|Seed) | 2827–2848 | seq-5'-GATTCTGAAGATGA|CAGAAAG-3'/miR-3'-CTATACAGTCTACT|GTCTTTC-5' |
| 32 | miR-3144-3p | AUUUCUCUGGCUUGUCCAUAUA | SEQ-5'-TATGTAGAAAAGCT|AGAGAAA-3' i.e. (SeqFlank|Seed) | 1228–1249 | seq-5'-TATGTAGAAAAGCT|AGAGAAA-3'/miR-3'-ATATACCTGTTCGG|TCTCTTT-5' |
| 33 | miR-518e-3p | GUGAGACUUCCCUUCGCGAAA | SEQ-5'-GTTCTTGATGGCC|AGTCTCA-3' i.e. (SeqFlank|Seed) | 55–75 | seq-5'-GTTCTTGATGGCC|AGTCTCA-3'/miR-3'-AAAGCGCTTCCCT|TCAGAGT-5' |
| 34 | miR-6774-3p | GACACCUGUUCUCCCUGUGCU | SEQ-5'-AACACAGGTGAAA|CAGGTGT-3' i.e. (SeqFlank|Seed) | 4252–4272 | seq-5'-AACACAGGTGAAA|CAGGTGT-3'/miR-3'-TCGTGTCCCTCTT|GTCCACA-5' |
| 35 | miR-3155b | CCAGGCUCUGCAGUGGGA | SEQ-5'-TGAAGATGAA|GAGCCTG-3' i.e. (SeqFlank|Seed) | 183–200 | seq-5'-TGAAGATGAA|GAGCCTG-3'/miR-3'-AGGGTGACGT|CTCGGAC-5' |
| 36 | miR-490-5p | CCAUGGAUCUCCAGGUGGGU | SEQ-5'-AATCAGGTGGTT|ATCCATG-3' i.e. (SeqFlank|Seed) | 2422–2441 | seq-5'-AATCAGGTGGTT|ATCCATG-3'/miR-3'-TGGGTGGACCTC|TAGGTAC-5' |
| 37 | miR-4795-5p | AGAAGUGGCUAAUAAUAUUGA | SEQ-5'-CCAAGATTGGTTT|CCACTTC-3' i.e. (SeqFlank|Seed) | 495–515 | seq-5'-CCAAGATTGGTTT|CCACTTC-3'/miR-3'-AGTTATAATAATC|GGTGAAG-5' |
| 38 | miR-1537-5p | AGCUGUAAUUAGUCAGUUUUCU | SEQ-5'-AGATAAATCTCTAA|TTACAGC-3' i.e. (SeqFlank|Seed) | 2871–2892 | seq-5'-AGATAAATCTCTAA|TTACAGC-3'/miR-3'-TCTTTTGACTGATT|AATGTCG-5' |
| 39 | miR-4782-3p | CAAGAUCUAUACUUCUGUUAGU | SEQ-5'-GCTAACAGAGCTAT|AGATCTT-3' i.e. (SeqFlank|Seed) | 2359–2380 | seq-5'-GCTAACAGAGCTAT|AGATCTT-3'/miR-3'-TGATTGTCTTCATA|TCTAGAA-5' |
| 40 | miR-4310 | GCAGCAUUCAUGUCCC | SEQ-5'-GACTCAAA|AATGCTG-3' i.e. (SeqFlank|Seed) | 4679–4694 | seq-5'-GACTCAAA|AATGCTG-3'/miR-3'-CCCTGTAC|TTACGAC-5' |
| 41 | miR-4418 | CACUGCAGGACUCAGCAG | SEQ-5'-TTATTGAAGA|CTGCAGT-3' i.e. (SeqFlank|Seed) | 6869–6886 | seq-5'-TTATTGAAGA|CTGCAGT-3'/miR-3'-GACGACTCAG|GACGTCA-5' |
| 42 | miR-4724-5p | AACUGAACCAGGAGUGAGCUUCG | SEQ-5'-CGAATGACACAATTG|GTTCAGT-3' i.e. (SeqFlank|Seed) | 2726–2748 | seq-5'-CGAATGACACAATTG|GTTCAGT-3'/miR-3'-GCTTCGAGTGAGGAC|CAAGTCA-5' |
| 43 | miR-6077 | GGGAAGAGCUGUACGGCCUUC | SEQ-5'-CAAGTCCTGATAG|CTCTTCC-3' i.e. (SeqFlank|Seed) | 2924–2944 | seq-5'-CAAGTCCTGATAG|CTCTTCC-3'/miR-3'-CTTCCGGCATGTC|GAGAAGG-5' |
| 44 | miR-3606-5p | UUAGUGAAGGCUAUUUUAAUU | SEQ-5'-ACTAACAATATCT|TTCACTA-3' i.e. (SeqFlank|Seed) | 372–392 | seq-5'-ACTAACAATATCT|TTCACTA-3'/miR-3'-TTAATTTTATCGG|AAGTGAT-5' |
| 45 | miR-3182 | GCUUCUGUAGUGUAGUC | SEQ-5'-ATATAGGCG|ACAGAAG-3' i.e. (SeqFlank|Seed) | 5820–5836 | seq-5'-ATATAGGCG|ACAGAAG-3'/miR-3'-CTGATGTGA|TGTCTTC-5' |
Table 5.
Targeting of the prostate cancer gene S100A8 by 2 miRNAs through a complete seed match
| No | miRNA name | Nucleotide sequence of miRNA(excluding 1st base at 5’ end) | Nucleotide sequence of target | Position of target sequence in coding sequence | Target and miRNA binding |
|---|---|---|---|---|---|
| 1 | miR-4267 | UCCAGCUCGGUGGCAC | SEQ-5'-TGTTGACC|GAGCTGG-3' i.e. (SeqFlank|Seed) | 2–17 | seq-5'-TGTTGACC|GAGCTGG-3'/miR-3'-CACGGTGG|CTCGACC-5' |
| 2 | miR-4296 | AUGUGGGCUCAGGCUCA | SEQ-5'-GGCGTGGCA|GCCCACA-3' i.e. (SeqFlank|Seed) | 235–251 | seq-5'-GGCGTGGCA|GCCCACA-3'/miR-3'-ACTCGGACT|CGGGTGT-5' |
Table 6.
Targeting of the prostate cancer gene SLC4A4 by 33 miRNAs through a complete seed match
| No. | miRNA name | Nucleotide sequence of miRNA(excluding 1st base at 5’ end | Nucleotide sequence of target | Position of target sequence in coding sequence | Target and miRNA binding |
|---|---|---|---|---|---|
| 1 | miR-3150b-3p | GGUUGGAGCUGCUAGAGGAGU | SEQ-5'-ACTACCCCATCAA|CTCCAAC-3' i.e. (SeqFlank|Seed) | 1829–1849 | seq-5'-ACTACCCCATCAA|CTCCAAC-3'/miR-3'-TGAGGAGATCGTC|GAGGTTG-5' |
| 2 | miR-551b-3p | GACUUUGGUUCAUACCCAGCG | SEQ-5'-ACCTGGAGAACAA|CCAAAGT-3' i.e. (SeqFlank|Seed) | 2589–2609 | seq-5'-ACCTGGAGAACAA|CCAAAGT-3'/miR-3'-GCGACCCATACTT|GGTTTCA-5' |
| 3 | miR-4419a | UGAGGGAGGAGACUGCA | SEQ-5'-GGTTATATG|CTCCCTC-3' i.e. (SeqFlank|Seed) | 2490–2506 | seq-5'-GGTTATATG|CTCCCTC-3'/miR-3'-ACGTCAGAG|GAGGGAG-5' |
| 4 | miR-1227-5p | GUGGGGCCAGGCGGUGG | SEQ-5'-CACCACACT|GGCCCCA-3' i.e. (SeqFlank|Seed) | 1926–1942 | seq-5'-CACCACACT|GGCCCCA-3'/miR-3'-GGTGGCGGA|CCGGGGT-5' |
| 5 | miR-6870-5p | UGGGGGAGAUGGGGGUUGA | SEQ-5'-TGACAGCCCAG|CTCCCCC-3' i.e. (SeqFlank|Seed) | 300–318 | seq-5'-TGACAGCCCAG|CTCCCCC-3'/miR-3'-AGTTGGGGGTA|GAGGGGG-5' |
| 6 | miR-4445-3p | ACCUAACAAAGAAAACGGCAC | SEQ-5'-CTTTCATTGCCTT|TGTTAGG-3' i.e. (SeqFlank|Seed) | 893–913 | seq-5'-CTTTCATTGCCTT|TGTTAGG-3'/miR-3'-CACGGCAAAAGAA|ACAATCC-5' |
| 7 | miR-4516 | GGGAGAAGGGUCGGGGC | SEQ-5'-GGACTACCT|CTTCTCC-3' i.e. (SeqFlank|Seed) | 2970–2986 | seq-5'-GGACTACCT|CTTCTCC-3'/miR-3'-CGGGGCTGG|GAAGAGG-5' |
| 8 | miR-4499 | AAGACUGAGAGGAGGGA | SEQ-5'-ACTGGTCTG|TCAGTCT-3' i.e. (SeqFlank|Seed) | 2659–2675 | seq-5'-ACTGGTCTG|TCAGTCT-3'/miR-3'-AGGGAGGAG|AGTCAGA-5' |
| 9 | miR-4311 | GAAAGAGAGCUGAGUGUG | SEQ-5'-ATCACTTGGA|TCTCTTT-3' i.e. (SeqFlank|Seed) | 2456–2473 | seq-5'-ATCACTTGGA|TCTCTTT-3'/miR-3'-GTGTGAGTCG|AGAGAAA-5' |
| 10 | miR-4300 | UGGGAGCUGGACUACUUC | SEQ-5'-ATGACAGCCC|AGCTCCC-3' i.e. (SeqFlank|Seed) | 299–316 | seq-5'-ATGACAGCCC|AGCTCCC-3'/miR-3'-CTTCATCAGG|TCGAGGG-5' |
| 11 | miR-320e | AAAGCUGGGUUGAGAAGG | SEQ-5'-GCCACTGATG|CCAGCTT-3' i.e. (SeqFlank|Seed) | 1708–1725 | seq-5'-GCCACTGATG|CCAGCTT-3'/miR-3'-GGAAGAGTTG|GGTCGAA-5' |
| 12 | miR-30b-5p | UGUAAACAUCCUACACUCAGCU | SEQ-5'-CAGTGCAAGTAGGA|TGTTTAC-3' i.e. (SeqFlank|Seed) | 696–717 | seq-5'-CAGTGCAAGTAGGA|TGTTTAC-3'/miR-3'-TCGACTCACATCCT|ACAAATG-5' |
| 13 | miR-30c-5p | UGUAAACAUCCUACACUCUCAGC | SEQ-5'-CCAGTGCAAGTAGGA|TGTTTAC-3' i.e. (SeqFlank|Seed) | 695–717 | seq-5'-CCAGTGCAAGTAGGA|TGTTTAC-3'/miR-3'-CGACTCTCACATCCT|ACAAATG-5' |
| 14 | miR-548ah-5p | AAAAGUGAUUGCAGUGUUUG | SEQ-5'-TAACTAATGCTA|TCACTTT-3' i.e. (SeqFlank|Seed) | 1442–1461 | seq-5'-TAACTAATGCTA|TCACTTT-3'/miR-3'-GTTTGTGACGTT|AGTGAAA-5' |
| 15 | miR-3606-3p | GAUUCAUCACUUUCUUUAAAA | SEQ-5'-TATTGAAAATGCT|GATGAAT-3' i.e. (SeqFlank|Seed) | 207–227 | seq-5'-TATTGAAAATGCT|GATGAAT-3'/miR-3'-AAAATTTCTTTCA|CTACTTA-5' |
| 16 | miR-4306 | UGGAGAGAAAGGCAGUA | SEQ-5'-AACCTCTCA|TCTCTCC-3' i.e. (SeqFlank|Seed) | 245–261 | seq-5'-AACCTCTCA|TCTCTCC-3'/miR-3'-ATGACGGAA|AGAGAGG-5' |
| 17 | miR-3617-5p | AAAGACAUAGUUGCAAGAUGGG | SEQ-5'-AGTATTTGCCAACT|ATGTCTT-3' i.e. (SeqFlank|Seed) | 1943–1964 | seq-5'-AGTATTTGCCAACT|ATGTCTT-3'/miR-3'-GGGTAGAACGTTGA|TACAGAA-5' |
| 18 | miR-4296 | AUGUGGGCUCAGGCUCA | SEQ-5'-AGTTCCTGT|GCCCACA-3' i.e. (SeqFlank|Seed) | 948–964 | seq-5'-AGTTCCTGT|GCCCACA-3'/miR-3'-ACTCGGACT|CGGGTGT-5' |
| 19 | miR-6729-3p | GACUCUCCCGCUCCCCCUACU | SEQ-5'-TGCAGGGCGTGTT|GGAGAGT-3' i.e. (SeqFlank|Seed) | 1493–1513 | seq-5'-TGCAGGGCGTGTT|GGAGAGT-3'/miR-3'-TCATCCCCCTCGC|CCTCTCA-5' |
| 20 | miR-892c-5p | UAUUCAGAAAGGUGCCAGUCA | SEQ-5'-TGACTTCCTCCAG|TCTGAAT-3' i.e. (SeqFlank|Seed) | 758–778 | seq-5'-TGACTTCCTCCAG|TCTGAAT-3'/miR-3'-ACTGACCGTGGAA|AGACTTA-5' |
| 21 | miR-511-5p | GUGUCUUUUGCUCUGCAGUCA | SEQ-5'-TGCTTATAAAGCA|AAAGACA-3' i.e. (SeqFlank|Seed) | 1068–1088 | seq-5'-TGCTTATAAAGCA|AAAGACA-3'/miR-3'-ACTGACGTCTCGT|TTTCTGT-5' |
| 22 | miR-6512-5p | UACCAUUAGAAGAGCUGGAAGA | SEQ-5'-TTTACCAACCCTGA|TAATGGT-3' i.e. (SeqFlank|Seed) | 712–733 | seq-5'-TTTACCAACCCTGA|TAATGGT-3'/miR-3'-AGAAGGTCGAGAAG|ATTACCA-5' |
| 23 | miR-3688-5p | AGUGGCAAAGUCUUUCCAUAU | SEQ-5'-ATTGGCAGAGCCA|TTGCCAC-3' i.e. (SeqFlank|Seed) | 1018–1038 | seq-5'-ATTGGCAGAGCCA|TTGCCAC-3'/miR-3'-TATACCTTTCTGA|AACGGTG-5' |
| 24 | miR-644a | AGUGUGGCUUUCUUAGAGC | SEQ-5'-ATTCCTTGAAC|GCCACAC-3' i.e. (SeqFlank|Seed) | 3116–3134 | seq-5'-ATTCCTTGAAC|GCCACAC-3'/miR-3'-CGAGATTCTTT|CGGTGTG-5' |
| 25 | miR-3145-5p | AACUCCAAACACUCAAAACUCA | SEQ-5'-ATAATTTTGACTAT|TTGGAGT-3' i.e. (SeqFlank|Seed) | 1637–1658 | seq-5'-ATAATTTTGACTAT|TTGGAGT-3'/miR-3'-ACTCAAAACTCACA|AACCTCA-5' |
| 26 | miR-4266 | CUAGGAGGCCUUGGCC | SEQ-5'-GGATAGAG|CCTCCTA-3' i.e. (SeqFlank|Seed) | 1169–1184 | seq-5'-GGATAGAG|CCTCCTA-3'/miR-3'-CCGGTTCC|GGAGGAT-5' |
| 27 | miR-4266 | CUAGGAGGCCUUGGCC | SEQ-5'-TGCCAAGT|CCTCCTA-3' i.e. (SeqFlank|Seed) | 3173–3188 | seq-5'-TGCCAAGT|CCTCCTA-3'/miR-3'-CCGGTTCC|GGAGGAT-5' |
| 28 | miR-4463 | GAGACUGGGGUGGGGCC | SEQ-5'-TGACTTCCT|CCAGTCT-3' i.e. (SeqFlank|Seed) | 758–774 | seq-5'-TGACTTCCT|CCAGTCT-3'/miR-3'-CCGGGGTGG|GGTCAGA-5' |
| 29 | miR-4500 | UGAGGUAGUAGUUUCUU | SEQ-5'-AGGCATGGA|CTACCTC-3' i.e. (SeqFlank|Seed) | 2964–2980 | seq-5'-AGGCATGGA|CTACCTC-3'/miR-3'-TTCTTTGAT|GATGGAG-5' |
| 30 | miR-588 | UUGGCCACAAUGGGUUAGAAC | SEQ-5'-GAGCAAGCCCCAT|GTGGCCA-3' i.e. (SeqFlank|Seed) | 435–455 | seq-5'-GAGCAAGCCCCAT|GTGGCCA-3'/miR-3'-CAAGATTGGGTAA|CACCGGT-5' |
| 31 | miR-3123 | CAGAGAAUUGUUUAAUC | SEQ-5'-CTTTCGGCA|ATTCTCT-3' i.e. (SeqFlank|Seed) | 1408–1424 | seq-5'-CTTTCGGCA|ATTCTCT-3'/miR-3'-CTAATTTGT|TAAGAGA-5' |
| 32 | miR-4519 | CAGCAGUGCGCAGGGCUG | SEQ-5'-GTTTCCTGGG|CACTGCT-3' i.e. (SeqFlank|Seed) | 1511–1528 | seq-5'-GTTTCCTGGG|CACTGCT-3'/miR-3'-GTCGGGACGC|GTGACGA-5' |
| 33 | miR-7515 | AGAAGGGAAGAUGGUGAC | SEQ-5'-GGCCACATTG|TCCCTTC-3' i.e. (SeqFlank|Seed) | 450–467 | seq-5'-GGCCACATTG|TCCCTTC-3'/miR-3'-CAGTGGTAGA|AGGGAAG-5' |
Table 7.
Targeting of the prostate cancer gene PDLIM3 by 6 miRNAs through a complete seed match
| No. | miRNA Name | Nucleotide sequence of miRNA (excluding 1st base at 5’ end) | Nucleotide sequence of target | Position of target sequence in coding sequence | Target and miRNA binding |
|---|---|---|---|---|---|
| 1 | miR-4265 | CUGUGGGCUCAGCUCUGGG | SEQ-5'-CGAAGCAGCGA|GCCCACA-3' i.e. (SeqFlank|Seed) | 154–172 | miRNA-3'-GGGTCTCGACT|CGGGTGT-5' i.e. (Nonseed|Seed) |
| 2 | miR-4748 | GAGGUUUGGGGAGGAUUUGCU | SEQ-5'-ATAATATTCGGCC|CAAACCT-3' i.e. (SeqFlank|Seed) | 119–139 | miRNA-3'-TCGTTTAGGAGGG|GTTTGGA-5' i.e. (Nonseed|Seed) |
| 3 | miR-627-3p | UCACUCAGAGUUUCUUUUCU | SEQ-5'-AGTACCGGCACC|CTGAGTG-3' i.e. (SeqFlank|Seed) | 431–450 | miRNA-3'-TCTTTTCTTTGA|GACTCAC-5' i.e. (Nonseed|Seed) |
| 4 | miR-4296 | AUGUGGGCUCAGGCUCA | SEQ-5'-TCGGAATGA|GCCCACA-3' i.e. (SeqFlank|Seed) | 219–235 | miRNA-3'-ACTCGGACT|CGGGTGT-5' i.e. (Nonseed|Seed) |
| 5 | miR-326 | CCUCUGGGCCCUUCCUCCAG | SEQ-5'-CCGCACAAAGCC|CCCAGAG-3' i.e. (SeqFlank|Seed) | 540–559 | miRNA-3'-GACCTCCTTCCC|GGGTCTC-5' i.e. (Nonseed|Seed) |
| 6 | miR-4322 | CUGUGGGCUCAGCGCGUGGGG | SEQ-5'-GCCGAAGCAGCGA|GCCCACA-3' i.e. (SeqFlank|Seed) | 152–172 | miRNA-3'-GGGGTGCGCGACT|CGGGTGT-5' i.e. (Nonseed|Seed) |
Table 8.
Targeting of the prostate cancer gene HOXC6 by7 miRNAs through a complete seed match
| No. | miRNA Name | Nucleotide sequence of miRNA (excluding 1st base at 5’ end) | Nucleotide sequence of target | Position of target sequence in coding sequence | Target and miRNA binding |
|---|---|---|---|---|---|
| 1 | miR-6808-3p | GACGUCCUUGCCACCAGUGUG | SEQ-5'-CTCGCCGGGGGCC|AGGACGT-3' i.e. (SeqFlank|Seed) | 40–60 | seq-5'-CTCGCCGGGGGCC|AGGACGT-3'/miR-3'-GTGTGACCACCGT|TCCTGCA-5' |
| 2 | miR-1281 | UCGCCUCCUCCUCUCCC | SEQ-5'-GAGCGGACC|GGAGGCG-3' i.e. (SeqFlank|Seed) | 413–429 | seq-5'-GAGCGGACC|GGAGGCG-3'/miR-3'-CCCTCTCCT|CCTCCGC-5' |
| 3 | miR-668-5p | UGCGCCUCGGGUGAGCAUG | SEQ-5'-GGAGCGGACCG|GAGGCGC-3' i.e. (SeqFlank|Seed) | 412–430 | seq-5'-GGAGCGGACCG|GAGGCGC-3'/miR-3'-GTACGAGTGGG|CTCCGCG-5' |
| 4 | miR-718 | CUUCCGCCCCGCCGGGCGUCG | SEQ-5'-CGCCGACAGCCTG|GGCGGAA-3' i.e. (SeqFlank|Seed) | 642–662 | seq-5'-CGCCGACAGCCTG|GGCGGAA-3'/miR-3'-GCTGCGGGCCGCC|CCGCCTT-5' |
| 5 | miR-3195 | CGCGCCGGGCCCGGGUU | SEQ-5'-AACGCGGCG|CCGGCGC-3' i.e. (SeqFlank|Seed) | 498–514 | seq-5'-AACGCGGCG|CCGGCGC-3'/miR-3'-TTGGGCCCG|GGCCGCG-5' |
| 6 | miR-545-3p | CGUGUGUUAUUUACAAACGACU | SEQ-5'-ACACCTTAGGACAT|AACACAC-3' i.e. (SeqFlank|Seed) | 269–290 | seq-5'-ACACCTTAGGACAT|AACACAC-3'/miR-3'-TCAGCAAACATTTA|TTGTGTG-5' |
| 7 | miR-6134 | UGAGGUGGUAGGAUGUAGA | SEQ-5'-TCCTTATCCTG|CCACCTC-3' i.e. (SeqFlank|Seed) | 25–43 | seq-5'-TCCTTATCCTG|CCACCTC-3'/miR-3'-AGATGTAGGAT|GGTGGAG-5' |
Table 9.
Targeting of the prostate cancer gene ALDH3B2 by 15 miRNAs through a complete seed match
| No. | miRNA Name | Nucleotide sequence of miRNA (excluding 1st base at 5’ end) | Nucleotide sequence of target | Position of target sequence in coding sequence | Target and miRNA binding |
|---|---|---|---|---|---|
| 1 | miR-3665 | AGCAGGUGCGGGGCGGCG | SEQ-5'-CCCACCACCG|CACCTGC-3' i.e. (SeqFlank|Seed) | 1025–1042 | seq-5'-CCCACCACCG|CACCTGC-3'/miR-3'-GCGGCGGGGC|GTGGACG-5' |
| 2 | miR-4267 | UCCAGCUCGGUGGCAC | SEQ-5'-TCACCCTG|GAGCTGG-3' i.e. (SeqFlank|Seed) | 377–392 | seq-5'-TCACCCTG|GAGCTGG-3'/miR-3'-CACGGTGG|CTCGACC-5' |
| 3 | miR-4279 | CUCUCCUCCCGGCUUC | SEQ-5'-TGATGCAG|GAGGAGA-3' i.e. (SeqFlank|Seed) | 746–761 | seq-5'-TGATGCAG|GAGGAGA-3'/miR-3'-CTTCGGCC|CTCCTCT-5' |
| 4 | miR-4748 | GAGGUUUGGGGAGGAUUUGCU | SEQ-5'-CCCCAGAGCTCCC|CAAACCT-3' i.e. (SeqFlank|Seed) | 583–603 | seq-5'-CCCCAGAGCTCCC|CAAACCT-3'/miR-3'-TCGTTTAGGAGGG|GTTTGGA-5' |
| 5 | miR-513a-5p | UUCACAGGGAGGUGUCAU | SEQ-5'-GCTGCACCCT|CCTGTGA-3' i.e. (SeqFlank|Seed) | 1142–1159 | seq-5'-GCTGCACCCT|CCTGTGA-3'/miR-3'-TACTGTGGAG|GGACACT-5' |
| 6 | miR-4508 | GCGGGGCUGGGCGCGCG | SEQ-5'-GTCCTGTGC|AGCCCCG-3' i.e. (SeqFlank|Seed) | 505–521 | seq-5'-GTCCTGTGC|AGCCCCG-3'/miR-3'-GCGCGCGGG|TCGGGGC-5' |
| 7 | miR-4531 | AUGGAGAAGGCUUCUGA | SEQ-5'-CTGTACGCC|TTCTCCA-3' i.e. (SeqFlank|Seed) | 847–863 | seq-5'-CTGTACGCC|TTCTCCA-3'/miR-3'-AGTCTTCGG|AAGAGGT-5' |
| 8 | miR-5703 | AGGAGAAGUCGGGAAGGU | SEQ-5'-CCCTGTACGC|CTTCTCC-3' i.e. (SeqFlank|Seed) | 845–862 | seq-5'-CCCTGTACGC|CTTCTCC-3'/miR-3'-TGGAAGGGCT|GAAGAGG-5' |
| 9 | miR-484 | UCAGGCUCAGUCCCCUCCCGAU | SEQ-5'-ACGTGCAGGAGACG|GAGCCTG-3' i.e. (SeqFlank|Seed) | 725–746 | seq-5'-ACGTGCAGGAGACG|GAGCCTG-3'/miR-3'-TAGCCCTCCCCTGA|CTCGGAC-5' |
| 10 | miR-7704 | CGGGGUCGGCGGCGACGUG | SEQ-5'-GACGACAACTG|CGACCCC-3' i.e. (SeqFlank|Seed) | 415–433 | seq-5'-GACGACAACTG|CGACCCC-3'/miR-3'-GTGCAGCGGCG|GCTGGGG-5' |
| 11 | miR-4514 | ACAGGCAGGAUUGGGGAA | SEQ-5'-ACCACCGCAC|CTGCCTG-3' i.e. (SeqFlank|Seed) | 1028–1045 | seq-5'-ACCACCGCAC|CTGCCTG-3'/miR-3'-AAGGGGTTAG|GACGGAC-5' |
| 12 | miR-659-5p | AGGACCUUCCCUGAACCAAGGA | SEQ-5'-GCCAGGGCACAGAG|AAGGTCC-3' i.e. (SeqFlank|Seed) | 185–206 | seq-5'-GCCAGGGCACAGAG|AAGGTCC-3'/miR-3'-AGGAACCAAGTCCC|TTCCAGG-5' |
| 13 | miR-4721 | UGAGGGCUCCAGGUGACGGUGG | SEQ-5'-TCTTCTTCACAGGG|AGCCCTC-3' i.e. (SeqFlank|Seed) | 308–329 | seq-5'-TCTTCTTCACAGGG|AGCCCTC-3'/miR-3'-GGTGGCAGTGGACC|TCGGGAG-5' |
| 14 | miR-4798-5p | UUCGGUAUACUUUGUGAAUUGG | SEQ-5'-CCACTACCCACCCT|ATACCGA-3' i.e. (SeqFlank|Seed) | 1080–1101 | seq-5'-CCACTACCCACCCT|ATACCGA-3'/miR-3'-GGTTAAGTGTTTCA|TATGGCT-5' |
| 15 | miR-920 | GGGGAGCUGUGGAAGCAGUA | SEQ-5'-GACGACCCCCAG|AGCTCCC-3' i.e. (SeqFlank|Seed) | 577–596 | seq-5'-GACGACCCCCAG|AGCTCCC-3'/miR-3'-ATGACGAAGGTG|TCGAGGG-5' |
Table 10.
Targeting of the prostate cancer gene ABHD2 by 13 miRNAs through a complete seed match
| No. | miRNA Name | Nucleotide sequence of miRNA (excluding 1st base at 5’ end) | Nucleotide sequence of target | Position of target sequence in coding sequence | Target and miRNA binding |
|---|---|---|---|---|---|
| 1 | miR-6809-3p | GACCCUUCCUCUCUUCUCUUC | SEQ-5'-TATGGGAAGATGG|GAAGGGT-3' i.e. (SeqFlank|Seed) | 247–267 | seq-5'-TATGGGAAGATGG|GAAGGGT-3'/miR-3'-CTTCTCTTCTCTC|CTTCCCA-5' |
| 2 | miR-4294 | GGGAGUCUACAGCAGGG | SEQ-5'-CCATGCTGG|AGACTCC-3' i.e. (SeqFlank|Seed) | 8–24 | seq-5'-CCATGCTGG|AGACTCC-3'/miR-3'-GGGACGACA|TCTGAGG-5' |
| 3 | miR-4283 | UGGGGCUCAGCGAGUUU | SEQ-5'-GAACCTGAA|GAGCCCC-3' i.e. (SeqFlank|Seed) | 99–115 | seq-5'-GAACCTGAA|GAGCCCC-3'/miR-3'-TTTGAGCGA|CTCGGGG-5' |
| 4 | miR-4283 | UGGGGCUCAGCGAGUUU | SEQ-5'-CTGTTCCCC|GAGCCCC-3' i.e. (SeqFlank|Seed) | 1153–1169 | seq-5'-CTGTTCCCC|GAGCCCC-3'/miR-3'-TTTGAGCGA|CTCGGGG-5' |
| 5 | miR-4311 | GAAAGAGAGCUGAGUGUG | SEQ-5'-CATTCCAAAA|TCTCTTT-3' i.e. (SeqFlank|Seed) | 1062–1079 | seq-5'-CATTCCAAAA|TCTCTTT-3'/miR-3'-GTGTGAGTCG|AGAGAAA-5' |
| 6 | miR-6881-3p | UCACCCUUCCUGCUUUCUCCUA | SEQ-5'-TATGGGAAGATGGG|AAGGGTG-3' i.e. (SeqFlank|Seed) | 247–268 | seq-5'-TATGGGAAGATGGG|AAGGGTG-3'/miR-3'-ATCCTCTTTCGTCC|TTCCCAC-5' |
| 7 | miR-626 | AGCUGUCUGAAAAUGUCUU | SEQ-5'-TGGACACATCC|AGACAGC-3' i.e. (SeqFlank|Seed) | 225–243 | seq-5'-TGGACACATCC|AGACAGC-3'/miR-3'-TTCTGTAAAAG|TCTGTCG-5' |
| 8 | miR-3155b | CCAGGCUCUGCAGUGGGA | SEQ-5'-AGAAACCCCA|GAGCCTG-3' i.e. (SeqFlank|Seed) | 842–859 | seq-5'-AGAAACCCCA|GAGCCTG-3'/miR-3'-AGGGTGACGT|CTCGGAC-5' |
| 9 | miR-4659a-3p | GCAACGGUACAGAUUCUUCUUU | SEQ-5'-AAAGAATACATTCC|ACCGTTG-3' i.e. (SeqFlank|Seed) | 190–211 | seq-5'-AAAGAATACATTCC|ACCGTTG-3'/miR-3'-TTTCTTCTTAGACA|TGGCAAC-5' |
| 10 | miR-4766-5p | UCUGAAAGAGCAGUUGGUGUU | SEQ-5'-CATTCCAAAATCT|CTTTCAG-3' i.e. (SeqFlank|Seed) | 1062–1082 | seq-5'-CATTCCAAAATCT|CTTTCAG-3'/miR-3'-TTGTGGTTGACGA|GAAAGTC-5' |
| 11 | miR-6085 | AAGGGGCUGGGGGAGCACA | SEQ-5'-GCTGTTCCCCG|AGCCCCT-3' i.e. (SeqFlank|Seed) | 1152–1170 | seq-5'-GCTGTTCCCCG|AGCCCCT-3'/miR-3'-ACACGAGGGGG|TCGGGGA-5' |
| 12 | miR-509-5p | UACUGCAGACAGUGGCAAUCA | SEQ-5'-GGAGTGAAGCTGG|CTGCAGT-3' i.e. (SeqFlank|Seed) | 46–66 | seq-5'-GGAGTGAAGCTGG|CTGCAGT-3'/miR-3'-ACTAACGGTGACA|GACGTCA-5' |
| 13 | miR-491-3p | CAUCUUCCCUUAGAACGUAUUC | SEQ-5'-GACAGCCTTGTATG|GGAAGAT-3' i.e. (SeqFlank|Seed) | 237–258 | seq-5'-GACAGCCTTGTATG|GGAAGAT-3'/miR-3'-CTTATGCAAGATTC|CCTTCTA-5' |
Table 11.
Targeting of the prostate cancer gene PDLIM5 by 8 miRNAs through a complete seed match
| No. | miRNA Name | Nucleotide sequence of miRNA (excluding 1st base at 5’ end) | Nucleotide sequence of target | Position of target sequence in coding sequence | Target and miRNA binding |
|---|---|---|---|---|---|
| 1 | miR-6086 | GGAGGUUGGGAAGGGCAGAG | SEQ-5'-GCCTGGACAGCC|CAACCTC-3' i.e. (SeqFlank|Seed) | 11–30 | seq-5'-GCCTGGACAGCC|CAACCTC-3'/miR-3'-GAGACGGGAAGG|GTTGGAG-5' |
| 2 | miR-4279 | CUCUCCUCCCGGCUUC | SEQ-5'-GATTTGTA|GAGGAGA-3' i.e. (SeqFlank|Seed) | 395–410 | seq-5'-GATTTGTA|GAGGAGA-3'/miR-3'-CTTCGGCC|CTCCTCT-5' |
| 3 | miR-3663-5p | GCUGGUCUGCGUGGUGCUCGG | SEQ-5'-CCCAACCTCTGGC|AGACCAG-3' i.e. (SeqFlank|Seed) | 21–41 | seq-5'-CCCAACCTCTGGC|AGACCAG-3'/miR-3'-GGCTCGTGGTGCG|TCTGGTC-5' |
| 4 | miR-320d | AAAAGCUGGGUUGAGAGGA | SEQ-5'-ACCAGCCAACT|CAGCTTT-3' i.e. (SeqFlank|Seed) | 186–204 | seq-5'-ACCAGCCAACT|CAGCTTT-3'/miR-3'-AGGAGAGTTGG|GTCGAAA-5' |
| 5 | miR-4505 | AGGCUGGGCUGGGACGGA | SEQ-5'-TTGGGACAAA|CCCAGCC-3' i.e. (SeqFlank|Seed) | 202–219 | seq-5'-TTGGGACAAA|CCCAGCC-3'/miR-3'-AGGCAGGGTC|GGGTCGG-5' |
| 6 | miR-320c | AAAAGCUGGGUUGAGAGGGU | SEQ-5'-CACCAGCCAACT|CAGCTTT-3' i.e. (SeqFlank|Seed) | 185–204 | seq-5'-CACCAGCCAACT|CAGCTTT-3'/miR-3'-TGGGAGAGTTGG|GTCGAAA-5' |
| 7 | miR-4256 | AUCUGACCUGAUGAAGGU | SEQ-5'-AAGTTTGGAA|GGTCAGA-3' i.e. (SeqFlank|Seed) | 738–755 | seq-5'-AAGTTTGGAA|GGTCAGA-3'/miR-3'-TGGAAGTAGT|CCAGTCT-5' |
| 8 | miR-4429 | AAAAGCUGGGCUGAGAGGCG | SEQ-5'-CACCAGCCAACT|CAGCTTT-3' i.e. (SeqFlank|Seed) | 185–204 | seq-5'-CACCAGCCAACT|CAGCTTT-3'/miR-3'-GCGGAGAGTCGG|GTCGAAA-5' |
Table 12.
Targeting of the prostate cancer gene RFX3 by 20 miRNAs through a complete seed match
| No. | miRNA Name | Nucleotide sequence of miRNA (excluding 1st base at 5’ end) | Nucleotide sequence of target | Position of target sequence in coding sequence | Target and miRNA binding |
|---|---|---|---|---|---|
| 1 | miR-6852-3p | GACUCCUUGUCUCCUGU | SEQ-5'-AGACAGCAG|AAGGAGT-3' i.e. (SeqFlank|Seed) | 575–591 | miRNA-3'-TGTCCTCTG|TTCCTCA-5' i.e. (Nonseed|Seed) |
| 2 | miR-7162-5p | UGCUUCCUUUCUCAGCUG | SEQ-5'-AGTATGTGGA|AGGAAGC-3' i.e. (SeqFlank|Seed) | 173–190 | miRNA-3'-GTCGACTCTT|TCCTTCG-5' i.e. (Nonseed|Seed) |
| 3 | miR-4419a | UGAGGGAGGAGACUGCA | SEQ-5'-GGCGCTATT|CTCCCTC-3' i.e. (SeqFlank|Seed) | 1151–1167 | miRNA-3'-ACGTCAGAG|GAGGGAG-5' i.e. (Nonseed|Seed) |
| 4 | miR-4486 | GCUGGGCGAGGCUGGCA | SEQ-5'-AGGGAGTTC|CGCCCAG-3' i.e. (SeqFlank|Seed) | 291–307 | miRNA-3'-ACGGTCGGA|GCGGGTC-5' i.e. (Nonseed|Seed) |
| 5 | miR-597-5p | UGUGUCACUCGAUGACCACUGU | SEQ-5'-ATTCTGGTCACTCA|GTGACAC-3' i.e. (SeqFlank|Seed) | 428–449 | miRNA-3'-TGTCACCAGTAGCT|CACTGTG-5' i.e. (Nonseed|Seed) |
| 6 | miR-620 | AUGGAGAUAGAUAUAGAAAU | SEQ-5'-TTCTCAACAGCC|ATCTCCA-3' i.e. (SeqFlank|Seed) | 536–555 | miRNA-3'-TAAAGATATAGA|TAGAGGT-5' i.e. (Nonseed|Seed) |
| 7 | miR-1273f | GGAGAUGGAGGUUGCAGUG | SEQ-5'-CTTCTCAACAG|CCATCTC-3' i.e. (SeqFlank|Seed) | 535–553 | miRNA-3'-GTGACGTTGGA|GGTAGAG-5' i.e. (Nonseed|Seed) |
| 8 | miR-548ap-5p | AAAAGUAAUUGCGGUCUUU | SEQ-5'-ACTGGAGGGAA|TTACTTT-3' i.e. (SeqFlank|Seed) | 265–283 | miRNA-3'-TTTCTGGCGTT|AATGAAA-5' i.e. (Nonseed|Seed) |
| 9 | miR-4318 | CACUGUGGGUACAUGCU | SEQ-5'-GGTCTCATC|CCACAGT-3' i.e. (SeqFlank|Seed) | 318–334 | miRNA-3'-TCGTACATG|GGTGTCA-5' i.e. (Nonseed|Seed) |
| 10 | miR-3193 | UCCUGCGUAGGAUCUGAGGAGU | SEQ-5'-AAGCAGCAGTGCCT|ACGCAGG-3' i.e. (SeqFlank|Seed) | 68–89 | miRNA-3'-TGAGGAGTCTAGGA|TGCGTCC-5' i.e. (Nonseed|Seed) |
| 11 | miR-6816-3p | GCUUCCACGUCCAGGAAG | SEQ-5'-GGTGCAGTAT|GTGGAAG-3' i.e. (SeqFlank|Seed) | 168–185 | miRNA-3'-GAAGGACCTG|CACCTTC-5' i.e. (Nonseed|Seed) |
| 12 | miR-1825 | UCCAGUGCCCUCCUCUCC | SEQ-5'-AGTATGGTGG|GCACTGG-3' i.e. (SeqFlank|Seed) | 331–348 | miRNA-3'-CCTCTCCTCC|CGTGACC-5' i.e. (Nonseed|Seed) |
| 13 | miR-3130-5p | UACCCAGUCUCCGGUGCAGCC | SEQ-5'-GCAGACATCAGAG|ACTGGGT-3' i.e. (SeqFlank|Seed) | 3–23 | miRNA-3'-CCGACGTGGCCTC|TGACCCA-5' i.e. (Nonseed|Seed) |
| 14 | miR-1260a | AUCCCACCUCUGCCACCA | SEQ-5'-GGTGGGCACT|GGTGGGA-3' i.e. (SeqFlank|Seed) | 336–353 | miRNA-3'-ACCACCGTCT|CCACCCT-5' i.e. (Nonseed|Seed) |
| 15 | miR-6771-3p | GACGCCCAUCUGUCCCCAAAC | SEQ-5'-GGTGGGATTCAGA|TGGGCGT-3' i.e. (SeqFlank|Seed) | 346–366 | miRNA-3'-CAAACCCCTGTCT|ACCCGCA-5' i.e. (Nonseed|Seed) |
| 16 | miR-4256 | AUCUGACCUGAUGAAGGU | SEQ-5'-ACCGAATCGA|GGTCAGA-3' i.e. (SeqFlank|Seed) | 1192–1209 | miRNA-3'-TGGAAGTAGT|CCAGTCT-5' i.e. (Nonseed|Seed) |
| 17 | miR-432-3p | UCUGUACCUCCUCGGUAGGUC | SEQ-5'-TGCCTACGCAGGT|GGTACAG-3' i.e. (SeqFlank|Seed) | 77–97 | miRNA-3'-CTGGATGGCTCCT|CCATGTC-5' i.e. (Nonseed|Seed) |
| 18 | miR-1260b | AUCCCACCACUGCCACCAU | SEQ-5'-TGGTGGGCACT|GGTGGGA-3' i.e. (SeqFlank|Seed) | 335–353 | miRNA-3'-TACCACCGTCA|CCACCCT-5' i.e. (Nonseed|Seed) |
| 19 | miR-4275 | CCAAUUACCACUUCUUU | SEQ-5'-GAGCAAACT|GTAATTG-3' i.e. (SeqFlank|Seed) | 928–944 | miRNA-3'-TTTCTTCAC|CATTAAC-5' i.e. (Nonseed|Seed) |
| 20 | miR-6833-3p | GACUCCUUCACCUCUCUCUUU | SEQ-5'-TATGAGACAGCAG|AAGGAGT-3' i.e. (SeqFlank|Seed) | 571–591 | miRNA-3'-TTTCTCTCTCCAC|TTCCTCA-5' i.e. (Nonseed|Seed) |
Table 13.
Targeting of the prostate cancer gene OR51E2 by 9 miRNAs through a complete seed match
| Sl. No. | miRNA Name | Nucleotide sequence of miRNA (excluding 1st base at 5’ end) | Nucleotide sequence of target | Position of target sequence in coding sequence | Target and miRNA binding |
|---|---|---|---|---|---|
| 1 | miR-1827 | UGAGGCAGUAGAUUGAAU | SEQ-5'-TTTTTTCCCA|CTGCCTC-3' i.e. (SeqFlank|Seed) | 462–479 | seq-5'-TTTTTTCCCA|CTGCCTC-3'/miR-3'-TAAGTTAGAT|GACGGAG-5' |
| 2 | miR-1827 | UGAGGCAGUAGAUUGAAU | SEQ-5'-CTACCTGCTG|CTGCCTC-3' i.e. (SeqFlank|Seed) | 834–851 | seq-5'-CTACCTGCTG|CTGCCTC-3'/miR-3'-TAAGTTAGAT|GACGGAG-5' |
| 3 | miR-1261 | AUGGAUAAGGCUUUGGCUU | SEQ-5'-TTGACCTGGCC|TTATCCA-3' i.e. (SeqFlank|Seed) | 203–221 | seq-5'-TTGACCTGGCC|TTATCCA-3'/miR-3'-TTCGGTTTCGG|AATAGGT-5' |
| 4 | miR-4530 | CCCAGCAGGACGGGAGCG | SEQ-5'-ATCCACCATC|CTGCTGG-3' i.e. (SeqFlank|Seed) | 330–347 | seq-5'-ATCCACCATC|CTGCTGG-3'/miR-3'-GCGAGGGCAG|GACGACC-5' |
| 5 | miR-4530 | CCCAGCAGGACGGGAGCG | SEQ-5'-TACTGCCATT|CTGCTGG-3' i.e. (SeqFlank|Seed) | 597–614 | seq-5'-TACTGCCATT|CTGCTGG-3'/miR-3'-GCGAGGGCAG|GACGACC-5' |
| 6 | miR-6891-5p | UAAGGAGGGGGAUGAGGGG | SEQ-5'-GTTGGCTTCCC|CCTCCTT-3' i.e. (SeqFlank|Seed) | 76–94 | seq-5'-GTTGGCTTCCC|CCTCCTT-3'/miR-3'-GGGGAGTAGGG|GGAGGAA-5' |
| 7 | miR-4668-5p | AGGGAAAAAAAAAAGGAUUUGUC | SEQ-5'-GCGGATCCCTCTTTT|TTTTCCC-3' i.e. (SeqFlank|Seed) | 449–471 | seq-5'-GCGGATCCCTCTTTT|TTTTCCC-3'/miR-3'-CTGTTTAGGAAAAAA|AAAAGGG-5' |
| 8 | miR-4291 | UUCAGCAGGAACAGCU | SEQ-5'-CACTGCCT|CTGCTGA-3' i.e. (SeqFlank|Seed) | 470–485 | seq-5'-CACTGCCT|CTGCTGA-3'/miR-3'-TCGACAAG|GACGACT-5' |
| 9 | miR-4418 | CACUGCAGGACUCAGCAG | SEQ-5'-CTGCGCCATG|CTGCAGT-3' i.e. (SeqFlank|Seed) | 385–402 | seq-5'-CTGCGCCATG|CTGCAGT-3'/miR-3'-GACGACTCAG|GACGTCA-5' |
Table 14.
Targeting of the prostate cancer gene PCDHGB4 by 24 miRNAs through a complete seed match
| No. | miRNA name | Nucleotide sequence of miRNA (excluding 1st base at 5’ end) | Nucleotide sequence of target | Position of target sequence in coding sequence | Target and miRNA binding |
|---|---|---|---|---|---|
| 1 | miR-4279 | UUUCUCUGGCCAAGUGACACU | SEQ-5'-AGACACCTTTGGA|CAGAGAA-3' i.e. (SeqFlank|Seed) | 590–610 | seq-5'-AGACACCTTTGGA|CAGAGAA-3'/miR-3'-TCACAGTGAACCG|GTCTCTT-5' |
| 2 | miR-4286 | UGAGGCGGGGGGGCGAGC | SEQ-5'-TCCTCCAGCC|CCGCCTC-3' i.e. (SeqFlank|Seed) | 2146–2163 | seq-5'-TCCTCCAGCC|CCGCCTC-3'/miR-3'-CGAGCGGGGG|GGCGGAG-5' |
| 3 | miR-33b-5p | GUGGGGCCAGGCGGUGG | SEQ-5'-GGATGGCAA|GGCCCCA-3' i.e. (SeqFlank|Seed) | 2706–2722 | seq-5'-GGATGGCAA|GGCCCCA-3'/miR-3'-GGTGGCGGA|CCGGGGT-5' |
| 4 | miR-4276 | CGGCUGGAGGUGUGAGGA | SEQ-5'-GCGACGCTCC|TCCAGCC-3' i.e. (SeqFlank|Seed) | 2139–2156 | seq-5'-GCGACGCTCC|TCCAGCC-3'/miR-3'-AGGAGTGTGG|AGGTCGG-5' |
| 5 | miR-548ap-5p | CCAGGAGGCGGAGGAGGUGGAG | SEQ-5'-CTCCTCCAGCCCCG|CCTCCTG-3' i.e. (SeqFlank|Seed) | 2145–2166 | seq-5'-CTCCTCCAGCCCCG|CCTCCTG-3'/miR-3'-GAGGTGGAGGAGGC|GGAGGAC-5' |
| 6 | miR-548au-5p | UCCAGCUCGGUGGCAC | SEQ-5'-GCGCCGGG|GAGCTGG-3' i.e. (SeqFlank|Seed) | 11–26 | seq-5'-GCGCCGGG|GAGCTGG-3'/miR-3'-CACGGTGG|CTCGACC-5' |
| 7 | miR-4508 | UCCAGCUCGGUGGCAC | SEQ-5'-AGGACGCC|GAGCTGG-3' i.e. (SeqFlank|Seed) | 1067–1082 | seq-5'-AGGACGCC|GAGCTGG-3'/miR-3'-CACGGTGG|CTCGACC-5' |
| 8 | miR-4508 | UCCAGCUCGGUGGCAC | SEQ-5'-GCGCCGGG|GAGCTGG-3' i.e. (SeqFlank|Seed) | 11–26 | seq-5'-GCGCCGGG|GAGCTGG-3'/miR-3'-CACGGTGG|CTCGACC-5' |
| 9 | miR-4492 | CUCUCCUCCCGGCUUC | SEQ-5'-TAGACAGG|GAGGAGA-3' i.e. (SeqFlank|Seed) | 269–284 | seq-5'-TAGACAGG|GAGGAGA-3'/miR-3'-CTTCGGCC|CTCCTCT-5' |
| 10 | miR-3155b | ACCCCACUCCUGGUACC | SEQ-5'-GGGCTCCGT|AGTGGGG-3' i.e. (SeqFlank|Seed) | 129–145 | seq-5'-GGGCTCCGT|AGTGGGG-3'/miR-3'-CCATGGTCC|TCACCCC-5' |
| 11 | miR-3155a | GUGCAUUGCUGUUGCAUUGC | SEQ-5'-GGAATGATTGCG|CAATGCA-3' i.e. (SeqFlank|Seed) | 970–989 | seq-5'-GGAATGATTGCG|CAATGCA-3'/miR-3'-CGTTACGTTGTC|GTTACGT-5' |
| 12 | miR-4419b | CUCAGUGACUCAUGUGC | SEQ-5'-GATCATTTC|TCACTGA-3' i.e. (SeqFlank|Seed) | 526–542 | seq-5'-GATCATTTC|TCACTGA-3'/miR-3'-CGTGTACTC|AGTGACT-5' |
| 13 | miR-4291 | AAAAGUAAUUGCGGUCUUU | SEQ-5'-CAATGCCGAGA|TTACTTT-3' i.e. (SeqFlank|Seed) | 819–837 | seq-5'-CAATGCCGAGA|TTACTTT-3'/miR-3'-TTTCTGGCGTT|AATGAAA-5' |
| 14 | miR-4451 | AAAAGUAAUUGCGGUUUUUGC | SEQ-5'-GTCAATGCCGAGA|TTACTTT-3' i.e. (SeqFlank|Seed) | 817–837 | seq-5'-GTCAATGCCGAGA|TTACTTT-3'/miR-3'-CGTTTTTGGCGTT|AATGAAA-5' |
| 15 | miR-330-5p | GCGGGGCUGGGCGCGCG | SEQ-5'-CGCTCCTCC|AGCCCCG-3' i.e. (SeqFlank|Seed) | 2143–2159 | seq-5'-CGCTCCTCC|AGCCCCG-3'/miR-3'-GCGCGCGGG|TCGGGGC-5' |
| 16 | miR-4302 | GCGGGGCUGGGCGCGCG | SEQ-5'-CCATCAGCA|AGCCCCG-3' i.e. (SeqFlank|Seed) | 2391–2407 | seq-5'-CCATCAGCA|AGCCCCG-3'/miR-3'-GCGCGCGGG|TCGGGGC-5' |
| 17 | miR-4279 | GGGGCUGGGCGCGCGCC | SEQ-5'-GACGCTCCT|CCAGCCC-3' i.e. (SeqFlank|Seed) | 2141–2157 | seq-5'-GACGCTCCT|CCAGCCC-3'/miR-3'-CCGCGCGCG|GGTCGGG-5' |
| 18 | miR-4286 | CCAGGCUCUGCAGUGGGA | SEQ-5'-GCACGCTGCA|GAGCCTG-3' i.e. (SeqFlank|Seed) | 1731–1748 | seq-5'-GCACGCTGCA|GAGCCTG-3'/miR-3'-AGGGTGACGT|CTCGGAC-5' |
| 19 | miR-33b-5p | CCAGGCUCUGCAGUGGGAACU | SEQ-5'-GCCGCACGCTGCA|GAGCCTG-3' i.e. (SeqFlank|Seed) | 1728–1748 | seq-5'-GCCGCACGCTGCA|GAGCCTG-3'/miR-3'-TCAAGGGTGACGT|CTCGGAC-5' |
| 20 | miR-4276 | GAGGCUGAAGGAAGAUGG | SEQ-5'-CCCGGGCTCT|TCAGCCT-3' i.e. (SeqFlank|Seed) | 1834–1851 | seq-5'-CCCGGGCTCT|TCAGCCT-3'/miR-3'-GGTAGAAGGA|AGTCGGA-5' |
| 21 | miR-548ap-5p | UUCAGCAGGAACAGCU | SEQ-5'-CAGTGAAG|CTGCTGA-3' i.e. (SeqFlank|Seed) | 2541–2556 | seq-5'-CAGTGAAG|CTGCTGA-3'/miR-3'-TCGACAAG|GACGACT-5' |
| 22 | miR-548au-5p | UGGUAGAGCUGAGGACA | SEQ-5'-TATTCCAGT|CTCTACC-3' i.e. (SeqFlank|Seed) | 1037–1053 | seq-5'-TATTCCAGT|CTCTACC-3'/miR-3'-ACAGGAGTC|GAGATGG-5' |
| 23 | miR-4508 | UCUCUGGGCCUGUGUCUUAGGC | SEQ-5'-GCGTTTCTCTCAGG|CCCAGAG-3' i.e. (SeqFlank|Seed) | 2421–2442 | seq-5'-GCGTTTCTCTCAGG|CCCAGAG-3'/miR-3'-CGGATTCTGTGTCC|GGGTCTC-5' |
| 24 | miR-4508 | CCAGUGUGGCUCAGCGAG | SEQ-5'-GCAGCAATGC|CACACTG-3' i.e. (SeqFlank|Seed) | 2669–2686 | seq-5'-GCAGCAATGC|CACACTG-3'/miR-3'-GAGCGACTCG|GTGTGAC-5' |
Free energy of mRNA and its accessibility to miRNAs
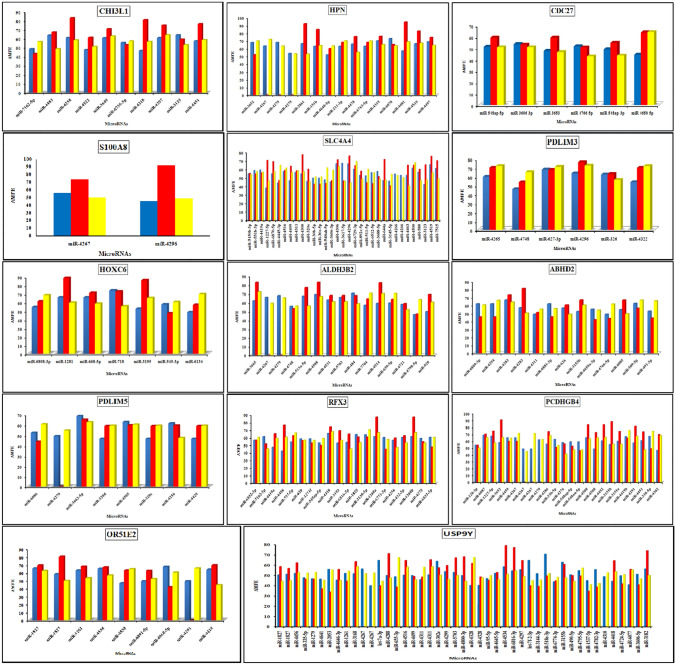
The AMFE values of the target, its upstream and downstream regions in all 14 overexpressed genes were compared so as to determine the degree of dissimilarity that exists between these regions (Fig. 1). The AMFE values for each miRNA target, as well as those for the corresponding upstream and downstream regions, are presented in Table 15. It was observed that for most of the coding sequences, the AMFE of the target region was higher than the flank regions or closely similar to that of the flank regions. But for gene HPN, USPDY, ABHD2, and PDLIM5, the miRNA targets had lower free energy values than their flank sequences. Gene S100A8 had the highest value of AMFE i.e., 83.10 in the target region, while gene USP9Y had the lowest AMFE value of 49.99 in the target region. In order to keep the miRNA–mRNA interaction in a stable state, it is necessary for the mRNA to release the maximum amount of free energy. In addition, the AMFE values also suggested that the mRNA molecules might be capable of forming intricate secondary structures. As a direct consequence of this, the microRNAs would find it difficult to access the target and flank regions of the mRNAs and binding to them.
Fig. 1.
AMFE values of 14 prostate cancer overexpressed genes for upstream, target and downstream regions. Upstream was represented by blue colour, target by red colour and downstream by yellow colour
Table 15.
AMFE (kcal/mol) of the target gene mRNAs in upstream, target and downstream regions
| Overexpressed prostate cancer genes | AMFE (Upstream) | AMFE (Target) | AMFE (Downstream) |
|---|---|---|---|
| CHI3L1 | |||
| Range | 46.5–63.9 | 43.1–83.1 | 48.4–63.9 |
| Mean | 56.51 | 66.92 | 56.53 |
| HPN | |||
| Range | 51.9–73.2 | 0.0–94.7 | 53.0–72.0 |
| Mean | 64.17 | 59.70 | 64.26 |
| CDC27 | |||
| Range | 44.3–53.6 | 50.7–63.9 | 42.8–64.3 |
| Mean | 49.57 | 56.83 | 49.82 |
| USP9Y | |||
| Range | 40.0–70.7 | 0.0–79.1 | 41–67.5 |
| Mean | 51.45 | 49.99 | 50.65 |
| S1OOA8 | |||
| Range | 45.4–56.3 | 74.1–92.1 | 49.0–50.0 |
| Mean | 50.85 | 83.10 | 49.50 |
| SLC4A4 | |||
| Range | 38.7–67.8 | 0.0–76.5 | 41.1–70.1 |
| Mean | 54.64 | 55.59 | 54.77 |
| PDLIM3 | |||
| Range | 45.7–67.8 | 53.6–69.9 | 56.3–72.4 |
| Mean | 58.82 | 66.68 | 68.08 |
| HOXC6 | |||
| Range | 48.5–74.1 | 47.3–88.7 | 55.5–69.8 |
| Mean | 59.90 | 69.26 | 62.54 |
| ALDH3B2 | |||
| Range | 46.4–70.5 | 0.0–82.9 | 52.0–72.1 |
| Mean | 61.19 | 58.99 | 62.95 |
| ABHD2 | |||
| Range | 48.9–66.8 | 41.9–81.3 | 48.8–66.6 |
| Mean | 57.03 | 55.50 | 58.39 |
| PDLIM5 | |||
| Range | 46.8–68.6 | 0–65.1 | 47.6–62.6 |
| Mean | 54.44 | 50.60 | 57.96 |
| RFX3 | |||
| Range | 42.2–65.5 | 44.7–87.3 | 45.1–67.9 |
| Mean | 57.49 | 62.25 | 58.01 |
| OR51E2 | |||
| Range | 46.6–67.5 | 0.0–80.1 | 44.3–65.3 |
| Mean | 58.59 | 57.79 | 56.40 |
| PCDHGB4 | |||
| Range | 46.8–69 | 0.0–90.9 | 41.3–75.9 |
| Mean | 59.41 | 59.78 | 60.65 |
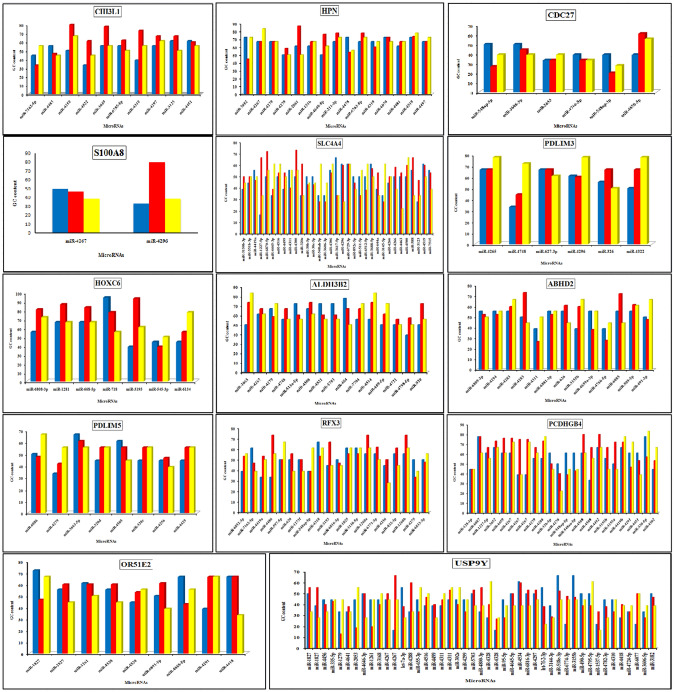
GC and GC3 contents in coding sequences of genes
The GC and GC3 contents in the target, upstream and downstream regions were evaluated for each of the 14 genes. It was found that the GC content was higher in the target region than in the flank regions. This was the case for the majority of the genes (Table 16; Fig. 2). Among all the target regions, the GC content of gene HOXC6 was the highest (73.80%). This led us to conclude that there was a strong link between the target mRNA and the miRNA. As a consequence, the mRNA would be strongly suppressed by the miRNA due to the presence of three H-bonds in each GC base-pair. The fact that the miRNA sequences were complementary to those of the targets indicated that the seed regions of the miRNAs had a high proportion of GC and a lower proportion of AT (Carmel, Shomron et al. 2012). The GC3% was calculated in order to determine if the codons (in frame) in the target region, as well as those in the upstream and downstream regions, ended in GC or AT. The GC3 content in the target regions was greater than 50% for the majority of the genes (Table 17). This indicated that the target regions of mRNAs were rich in GC-ending codons.
Table 16.
GC content (%) in prostate cancer genes
| Overexpressed prostate cancer genes | Upstream | Target | Downstream |
|---|---|---|---|
| CHI3L1 | 51.12 | 62.75 | 53.90 |
| HPN | 64.82 | 67.57 | 66.67 |
| CDC27 | 41.67 | 36.47 | 38.90 |
| USP9Y | 41.84 | 42.09 | 40.73 |
| S100A8 | 41.65 | 63.35 | 38.90 |
| SLC4A4 | 47.14 | 50.21 | 47.30 |
| PDLIM3 | 55.57 | 61.87 | 69.45 |
| HOXC6 | 58.73 | 73.80 | 64.30 |
| ALDH3B2 | 59.64 | 64.73 | 62.23 |
| ABHD2 | 50.88 | 52.76 | 54.28 |
| PDLIM5 | 48.59 | 52.44 | 53.50 |
| RFX3 | 50.28 | 54.97 | 50.83 |
| OR51E2 | 56.80 | 57.49 | 50.62 |
| PCDHGB4 | 57.18 | 63.52 | 57.40 |
Fig. 2.
GC content (%) of 14 prostate cancer overexpressed genes for upstream, target and downstream regions. Upstream was represented by blue colour, target by red colour and downstream by yellow colour
TABLE 17.
GC3 content (%) in prostate cancer genes
| Overexpressed prostate cancer genes | Upstream | Target | Downstream |
|---|---|---|---|
| CHI3L1 | 63.33 | 75.13 | 71.67 |
| HPN | 76.65 | 85.87 | 79.99 |
| CDC27 | 33.33 | 21.12 | 30.57 |
| USP9Y | 39.27 | 34.97 | 33.71 |
| S100A8 | 83.35 | 70.00 | 50.00 |
| SLC4A4 | 54.55 | 50.70 | 51.51 |
| PDLIM3 | 75.00 | 62.78 | 86.10 |
| HOXC6 | 71.43 | 83.31 | 85.71 |
| ALDH3B2 | 87.78 | 91.36 | 94.43 |
| ABHD2 | 57.69 | 64.02 | 61.55 |
| PDLIM5 | 45.81 | 49.34 | 47.90 |
| RFX3 | 56.66 | 51.16 | 45.84 |
| OR51E2 | 62.95 | 66.07 | 74.05 |
| PCDHGB4 | 68.75 | 69.89 | 71.52 |
Efficiency of target sequence translation
The variations in translational rates between the target and flank regions were analysed and computed in all the 14 genes overexpressed in prostate cancer. The average values of the compAI for the genes indicated that translation efficiency was poor (Table 18). The average compAI value in the upstream region was 0.34, followed by 0.26 for target region and 0.24 for the downstream region. It revealed that once miRNA binds to the target site, the process of translation might slow down and reduce gene expression. It was earlier shown that the number of microRNA binding regions located in an mRNA has a direct correlation to the degree of translation inhibited by the miRNAs. When there is just one target site, the potential for translational repression is only 10%; however, when there are two target sites, the potential for translational repression rises to 20% (Brümmer and Hausser 2014).
Table 18.
Translation rate (compAI) of target mRNA in the upstream, target, and downstream regions
| Overexpressed prostate cancer genes | Upstream | Target | Downstream |
|---|---|---|---|
| CHI3L1 | 0.15 | 0.16 | 0.14 |
| HPN | 0.12 | 0.14 | 0.14 |
| CDC27 | 0.20 | 0.26 | 0.23 |
| USP9Y | 0.24 | 0.22 | 0.24 |
| S100A8 | 0.34 | 0.14 | 0.14 |
| SLC4A4 | 0.22 | 0.19 | 0.21 |
| PDLIM3 | 0.12 | 0.20 | 0.13 |
| HOXC6 | 0.12 | 0.15 | 0.19 |
| ALDH3B2 | 0.15 | 0.15 | 0.20 |
| ABHD2 | 0.17 | 0.19 | 0.15 |
| PDLIM5 | 0.14 | 0.20 | 0.19 |
| RFX3 | 0.20 | 0.17 | 0.17 |
| OR51E2 | 0.17 | 0.13 | 0.18 |
| PCDHGB4 | 0.25 | 0.17 | 0.15 |
Measure of cosine similarity metric (COSM)
The COSM estimates were computed to determine the closeness of RSCU values and the tRNA molecules. From the mean values of COSM of all the three regions, a weak relationship was observed between the synonymous codons and the tRNA for all the genes, i.e., the values were close to 0 (Table 19). In the target region, gene S100A8 had the lowest COSM value of 0.09, while the highest value was 0.26 for gene PDLIM5. In the upstream region, the highest COSM value was 0.31, and in the downstream, it was 0.28. It was reported that less translational efficiency of target region could be due to lack of optimal codons and could lead to stronger repression of the overexpressed genes by miRNAs (Gu et al. 2012a; b).
Table 19.
Cosine similarity metric (COSM) of target mRNA in the upstream, target, and downstream regions
| Overexpressed prostate cancer genes | Upstream | Target | Downstream |
|---|---|---|---|
| CHI3L1 | 0.17 | 0.15 | 0.21 |
| HPN | 0.13 | 0.15 | 0.14 |
| CDC27 | 0.20 | 0.15 | 0.17 |
| USP9Y | 0.19 | 0.15 | 0.20 |
| S100A8 | 0.31 | 0.09 | 0.28 |
| SLC4A4 | 0.21 | 0.18 | 0.18 |
| PDLIM3 | 0.23 | 0.15 | 0.11 |
| HOXC6 | 0.20 | 0.15 | 0.18 |
| ALDH3B2 | 0.18 | 0.20 | 0.20 |
| ABHD2 | 0.18 | 0.19 | 0.18 |
| PDLIM5 | 0.23 | 0.26 | 0.19 |
| RFX3 | 0.22 | 0.14 | 0.20 |
| OR51E2 | 0.20 | 0.10 | 0.26 |
| PCDHGB4 | 0.20 | 0.14 | 0.20 |
Measure of mRNA stability (MSI)
The degree to which mRNA is stable in its upstream, target and downstream regions was evaluated. Unstable mRNAs possess non-optimal codons, which cause destabilized protein expression, whereas optimal codons are great contributors to mRNA stability (Presnyak et al. 2015). The rate at which mRNA degrades is a major factor in gene expression. We calculated the mRNA stability index of target, upstream and downstream regions and the results are presented in Table 20 along with the mean value for each gene. The downstream region of the gene PDLIM3 had the highest negative value of -0.188, while the target region had the highest negative value of − 0.322. For the upstream region, the gene S100A8 displayed the highest negative value (− 0.165). It was found that the majority of the genes examined in this study had acquired a negative mRNA stability index value. The findings imply that the stability of the mRNA is low; hence, we could conclude that the identified miRNAs could easily degrade the mRNAs. When the mRNA stability index is negative, this implies that the mRNA is less stable, whereas a positive value indicates high stability of the mRNA.
Table 20.
Mean mRNA stability (MSI) of target mRNA in the upstream, target, and downstream regions
| Overexpressed prostate cancer genes | Upstream | Target | Downstream |
|---|---|---|---|
| CHI3L1 | 0.016 | − 0.041 | 0.015 |
| HPN | – 0.128 | – 0.020 | – 0.103 |
| CDC27 | – 0.125 | – 0.008 | 0.063 |
| USP9Y | – 0.086 | – 0.034 | – 0.057 |
| S100A8 | – 0.165 | – 0.175 | 0.085 |
| SLC4A4 | – 0.003 | 0.071 | – 0.021 |
| PDLIM3 | – 0.025 | – 0.322 | – 0.188 |
| HOXC6 | 0.063 | – 0.191 | – 0.018 |
| ALDH3B2 | 0.002 | – 0.067 | – 0.065 |
| ABHD2 | – 0.015 | – 0.145 | – 0.097 |
| PDLIM5 | – 0.101 | 0.242 | 0.050 |
| RFX3 | 0.067 | – 0.102 | – 0.012 |
| OR51E2 | 0.047 | – 0.062 | – 0.058 |
| PCDHGB4 | – 0.014 | 0.007 | 0.007 |
Free energy of RNA duplex formation
Analysis of free energy of RNA duplex was performed for each of the overexpressed genes with their respective miRNAs (Fig. 3). Greater the free energy of RNA duplex formation, greater will be its stability. The greater stability also indicates that the miRNA-mRNA duplex formed is highly stable and can highly suppress the translation of the overexpressed gene (Hibio et al. 2012). For the gene CHI3L1, it was found that 10 miRNAs targeted the gene, among which the miR-6755-5p had the highest free energy of duplex formation (i.e. 24.36 kcal/mol). Hence the duplex formed by this miRNA and the target gene mRNA was highly stable. For the genes HPN, CDC27, USP9Y, S100A8, SLC4A4, PDLIM3, HOXC6, ALDH3B2, ABHD2, PDLIM5, RFX3, OR51E2, and PCDHGB4, the miRNAs namely miR-4640-5p, miR-4650-5p, miR-4641, miR-4296, miR-588, miR-326, miR-6808-3p, miR-659-5p, miR-491-3p, miR-6086, miR-3193, miR-4668-5p, miR-4459 respectively had the highest free energy of duplex formation. From all the above-mentioned miRNAs, the miR-4640-5p was found to have the highest free energy of duplex formation (27.45 kcal/mol). Thus, we could infer that the miR-4640-5p along with its target gene HPN formed the highly stable RNA duplex.
Fig. 3.
Free energy (kcal/mol) of RNA duplex in 14 overexpressed genes
Unpaired t test for base compositional difference
An unpaired t test was performed to know the difference in base composition between the target and its flank regions. From our study, a remarkably significant difference was noticed between the target and upstream regions for AT content at p < 0.0001, for A content at p < 0.0042, for T content at p < 0.0016, and for G content at p < 0.0038 (Table 21). Between the target and downstream regions, a notable significant difference was seen for AT content at p < 0.0001, for A content at p < 0.0003, and for G content at p < 0.0016 (Table 22).
Table 21.
Unpaired t test between target and upstream regions
| Base composition | p value |
|---|---|
| AT content | < 0.0001 |
| A content | < 0.0042 |
| T content | < 0.0016 |
| G content | < 0.0038 |
Table 22.
Unpaired t test between target and downstream regions
| Base composition | p value |
|---|---|
| AT content | < 0.0001 |
| A content | < 0.0003 |
| G content | < 0.0016 |
Annotation of the overexpressed genes
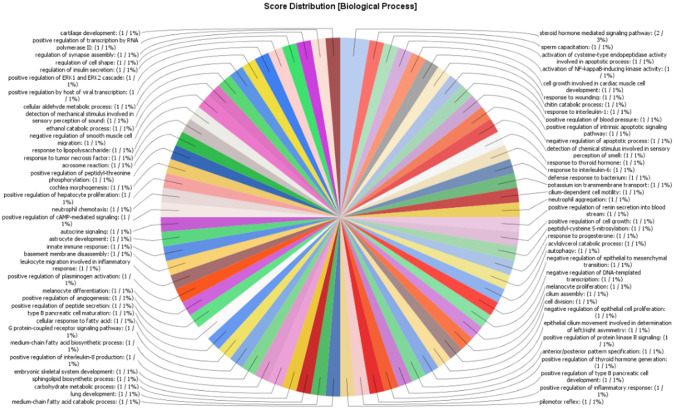
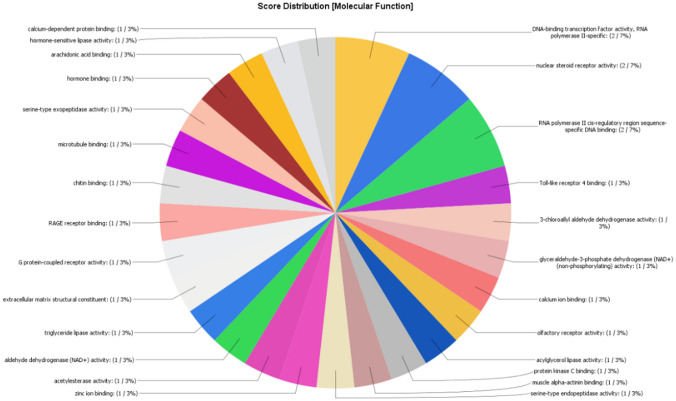
The 14 overexpressed prostate cancer genes were annotated by using the software BLAST2GO. The annotation was carried out to disclose the functions of each of the genes. The outcomes of the analysis were sorted into three categories: biological processes, cellular components, and molecular functions. It was found that 1% of the targeted overexpressed genes were associated with biological processes such as negative regulation of apoptotic process, acrosome reaction, innate immune response, melanocyte differentiation, etc. (Fig. 4). Furthermore, 4% of the targeted genes were related to cellular components such as endoplasmic reticulum membrane, sperm flagellum, transcription regulator complex, 7% were related to extracellular space and chromatin, and 11% to the cytosol (Fig. 5). Again, 3% of the targeted genes had molecular functions such as hormone binding, calcium-dependent protein binding, serine-type endopeptidase activity, while 7% were involved in nuclear steroid receptor activity (Fig. 6).
Fig. 4.
Number and percentage of target genes involved in different biological processes
Fig. 5.
Number and percentage of target genes involved in different cellular components
Fig. 6.
Number and percentage of target genes involved in different molecular functions
Discussion
MicroRNAs stand out among all other types of noncoding RNAs because of the significant roles they play in gene regulation. They are primarily associated with inhibition or repression of mRNA. Studies revealed that animal microRNAs are the prime components for weakening the mRNA stability. In this study, we evaluated the coding sequences of 14 overexpressed genes in prostate cancer and predicted the regions for possible binding of miRNAs based on nucleotide complementarity. The analysis helped us understand how miRNAs target the overexpressed genes and silence their expression. For this, the coding sequences of 14 most overexpressed genes (Axelsen et al. 2007) were downloaded from NCBI and the complete set of human mature miRNAs were retrieved from miRBase 21 database. The analysis of both the sequences was accomplished on the basis of 7mer-m8 model.
According to this model, the seed region of miRNA comprising of 7 nt (2–8 position) at the 5’ end undergoes perfect Watson–Crick base pairing with the target mRNA (Friedman et al. 2009). The base pairing in non-seed region of miRNA possesses some mismatches, and in this study, a maximum of 6 mismatches were taken into consideration in the non-seed region.
When the seed region of microRNA goes for complementary base pairing with the target region in mRNA, it results in the breakdown of mRNA or inhibition of translation (Liu and Wang 2019). Some genes overexpressed in prostate cancer were targeted by several miRNAs. Many parameters were employed to examine the affinity of miRNA towards the target gene mRNA. Once the target region of miRNA was identified in a coding sequence, the upstream, target and downstream regions were extracted and each parameter was determined in all the three regions. It was observed that a single microRNA can bind to multiple target positions on a single gene, and likewise different microRNAs can bind to a single gene on distinct sites.
The parameter free energy helps determine the degree of stability between miRNA and mRNA (Yue et al. 2009). In our study we estimated AMFE in all the three regions, and found that for most of the genes, the AMFE value was higher in the target region than the flank regions. This implies a significant stability between miRNA and mRNA. Also it states that how strongly the mRNA could be folded into a complex structure thus making it difficult for the miRNA to approach and bind to it. In a previous study on lung cancer, low free energy was reported in the downstream, target and upstream regions of each gene (Chakraborty and Nath 2022).
GC content of the three regions in the overexpressed gene reveals the intensity of coupling between miRNA and the target gene. Higher the GC content of the target gene, more stable is the bond between gene and miRNA (Grimson et al. 2007). From our analysis we perceived the abundance of GC and GC3 contents in the target gene, which indicates a strong coupling of miRNAs with the gene.
The rate of translation was estimated using the parameter compAI for the upstream, target and downstream regions of all the overexpressed genes. A direct relationship exists between translational efficiency and compAI value, i.e. higher the value of compAI, higher is the translation rate. From our analysis low values of compAI for all three regions were obtained which indicate low translational efficiency in those regions. Therefore efficient miRNA binding could take place leading to mRNA repression. Moreover, the cosine similarity metric values indicated low similarity between synonymous codons and tRNA pool. This also signifies low translational efficiency which could arise from the absence of optimal codons.
The mRNA stability estimation helps us find out how steady the mRNA molecules are. Our analysis showed the mean MSI value to be negative, which inferred low stability. Higher the negative value, lower is the stability. Hence, we could infer that miRNA could easily degrade the mRNA.
Free energy of RNA duplex formation predicts the stability of RNA duplexes. With an increase in the value of free energy of duplex formation, the stability of the duplex correspondingly increases. From our analysis we found that the duplex formed by miR-4640-5p and its target gene HPN mRNA had the highest value of free energy of duplex. Thus we could conclude that this duplex is the most stable one, thereby the miR-4640-5p can easily degrade the mRNA of HPN gene.
Unpaired t-test was performed to test the difference of base composition between the target and the flank regions. Our result showed a significant difference between the target and upstream regions for AT (p < 0.0001), A (p < 0.0042), T (p < 0.0016), and G (p < 0.0038). Between target and downstream regions, significant difference in base composition was found for AT (p < 0.0001), A (p < 0.0003), and G (p < 0.0016).
In the present study, 14 overexpressed genes were analyzed using BLAST2GO software. The results suggest acrosome reaction, negative regulation of apoptotic process etc. as certain biological functions of the genes. Hormone binding, nuclear steroid receptor activity, serine-type endopeptidase activity are some of the molecular functions, and a few cellular components linked to the genes are endoplasmic reticulum membrane, sperm flagellum, and transcription regulator complex.
Conclusion
Prostate cancer is one of most frequently occurring diseases in human males and is associated with a high mortality rate. It generally occurs due to mutations in some specific genes. This often results in the overexpression of some genes and hence overproduction of specific proteins. In this study we identified the human miRNAs which could potentially bind to the coding sequences of 14 most overexpressed genes in prostate cancer and could suppress their activity by silencing them. Several parameters were utilized to analyze the binding intensity of miRNA and its target gene mRNA. The data acquired from this work may help other researchers in designing experiments for confirmatory analysis and thus, regulating the disease by making use of the identified miRNAs as therapeutics.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contribution
AP: original draft, data curation, analysis of data, investigation, interpretation, writing, review and editing, validation. AR: investigation, analysis of data, interpretation, writing, review and editing. SKB, SB: investigation, analysis of data, interpretation, writing, review and editing. SC: conceptualization, project administration, formal analysis, software, visualization, supervision, methodology, writing, review and final editing.
Funding
None.
Data availability
Data are available in the manuscript and supplementary files.
Declarations
Conflict of interest
Authors declare no conflict of interests in the manuscript.
Ethical approval
Not applicable, the analysis is solely based on the coding sequences of 14 overexpressed genes of prostate cancer and the mature miRNA sequence data publicly available in databases.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abumsimir B, Ennaji MM (2019) Major candidate genes associated with risk of hereditary and sporadic prostate cancer. Asian Oncol Res J 2(1):1–13 [Google Scholar]
- Axelsen JB et al (2007) Genes overexpressed in different human solid cancers exhibit different tissue-specific expression profiles. Proc Natl Acad Sci 104(32):13122–13127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brümmer A, Hausser J (2014) MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. BioEssays 36(6):617–626 [DOI] [PubMed] [Google Scholar]
- Calvo A et al (2002) Alterations in gene expression profiles during prostate cancer progression: functional correlations to tumorigenicity and down-regulation of selenoprotein-P in mouse and human tumors. Can Res 62(18):5325–5335 [PubMed] [Google Scholar]
- Carleton M et al (2007) MicroRNAs and cell cycle regulation. Cell Cycle 6(17):2127–2132 [DOI] [PubMed] [Google Scholar]
- Carmel I et al (2012) Does base-pairing strength play a role in microRNA repression? RNA 18(11):1947–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Nath D (2022) A study on microRNAs targeting the genes overexpressed in lung cancer and their codon usage patterns. Mol Biotechnol 64(10):1095–1119 [DOI] [PubMed] [Google Scholar]
- Choudhury AD et al (2012) The role of genetic markers in the management of prostate cancer. Eur Urol 62(4):577–587 [DOI] [PubMed] [Google Scholar]
- Conesa A et al (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676 [DOI] [PubMed] [Google Scholar]
- Culp MB et al (2020) Recent global patterns in prostate cancer incidence and mortality rates. Eur Urol 77(1):38–52 [DOI] [PubMed] [Google Scholar]
- Dalmay T (2008) MicroRNAs and cancer. J Intern Med 263(4):366–375 [DOI] [PubMed] [Google Scholar]
- Dilucca M et al (2015) Codon bias patterns of E. coli’s interacting proteins. PLoS ONE 10(11):e0142127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC et al (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19(1):92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A et al (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27(1):91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W et al (2012a) Selection on synonymous sites for increased accessibility around miRNA binding sites in plants. Mol Biol Evol 29(10):3037–3044 [DOI] [PubMed] [Google Scholar]
- Gu W et al (2012b) Translation efficiency in upstream region of microRNA targets in Arabidopsisthaliana. Evol Bioinform 8:EBOS10362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W et al (2013) Biological basis of miRNA action when their targets are located in human protein coding region. PLoS ONE 8(5):e63403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibio N et al (2012) Stability of miRNA 5′ terminal and seed regions is correlated with experimentally observed miRNA-mediated silencing efficacy. Sci Rep 2(1):1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irigaray P et al (2007) Lifestyle-related factors and environmental agents causing cancer: an overview. Biomed Pharmacother 61(10):640–658 [DOI] [PubMed] [Google Scholar]
- Kadri S et al (2011) RNA deep sequencing reveals differential microRNA expression during development of sea urchin and sea star. PLoS ONE 6(12):e29217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M et al (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39(10):1278–1284 [DOI] [PubMed] [Google Scholar]
- Kocarnik JM et al (2022) Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019: a systematic analysis for the global burden of disease study 2019. JAMA Oncol 8(3):420–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC et al (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854 [DOI] [PubMed] [Google Scholar]
- Lewis BP et al (2003) Prediction of mammalian microRNA targets. Cell 115(7):787–798 [DOI] [PubMed] [Google Scholar]
- Litwin MS, Tan H-J (2017) The diagnosis and treatment of prostate cancer: a review. JAMA 317(24):2532–2542 [DOI] [PubMed] [Google Scholar]
- Liu W, Wang X (2019) Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol 20:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markopoulos GS et al (2017) A step-by-step microRNA guide to cancer development and metastasis. Cell Oncol 40:303–339 [DOI] [PubMed] [Google Scholar]
- Massillo C et al (2017) Implications of microRNA dysregulation in the development of prostate cancer. Reproduction 154(4):R81–R97 [DOI] [PubMed] [Google Scholar]
- Moore PS, Chang Y (2010) Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer 10(12):878–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ et al (2015) miRNA–target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat Commun 6(1):8864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M et al (2010) Correlation between sequence conservation and structural thermodynamics of microRNA precursors from human, mouse, and chicken genomes. BMC Evol Biol 10:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda D et al (2014) Computational identification and characterization of conserved miRNAs and their target genes in garlic (Alliumsativum L.) expressed sequence tags. Gene 537(2):333–342 [DOI] [PubMed] [Google Scholar]
- Peto J, Houlston R (2001) Genetics and the common cancers. Eur J Cancer 37:88–96 [DOI] [PubMed] [Google Scholar]
- Presnyak V et al (2015) Codon optimality is a major determinant of mRNA stability. Cell 160(6):1111–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawla P (2019) Epidemiology of prostate cancer. World J Oncol 10(2):63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KB (2015) MicroRNA (miRNA) in cancer. Cancer Cell Int 15(1):1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riffo-Campos ÁL et al (2016) Tools for sequence-based miRNA target prediction: what to choose? Int J Mol Sci 17(12):1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia J Jr (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci 95(4):1460–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnoi A, Rani S (2017) “MiRNA biogenesis and regulation of diseases: an overview.” MicroRNA profiling. Methods Prot 1509:1–10 [DOI] [PubMed] [Google Scholar]
- Wienholds E, Plasterk RH (2005) MicroRNA function in animal development. FEBS Lett 579(26):5911–5922 [DOI] [PubMed] [Google Scholar]
- Xia T et al (1998) Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson− Crick base pairs. Biochemistry 37(42):14719–14735 [DOI] [PubMed] [Google Scholar]
- Yue D et al (2009) Survey of computational algorithms for microRNA target prediction. Curr Genomics 10(7):478–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B et al (2006) Evidence that miRNAs are different from other RNAs. Cell Mol Life Sci CMLS 63:246–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y et al (2020) Clinicopathological characteristics of localized prostate cancer in younger men aged≤ 50 years treated with radical prostatectomy in the PSA era: a systematic review and meta-analysis. Cancer Med 9(18):6473–6484 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in the manuscript and supplementary files.