Abstract
The first committed step in the biosynthesis of indole glucosinolates is the conversion of indole-3-acetaldoxime into an indole-3-S-alkyl-thiohydroximate. The initial step in this conversion is catalyzed by CYP83B1 in Arabidopsis (S. Bak, F.E. Tax, K.A. Feldmann, D.A. Galbraith, R. Feyereisen [2001] Plant Cell 13: 101–111). The knockout mutant of the CYP83B1 gene (rnt1-1) shows a strong auxin excess phenotype and are allelic to sur-2. CYP83A1 is the closest relative to CYP83B1 and shares 63% amino acid sequence identity. Although expression of CYP83A1 under control of its endogenous promoter in the rnt1-1 background does not prevent the auxin excess and indole glucosinolate deficit phenotype caused by the lack of the CYP83B1 gene, ectopic overexpression of CYP83A1 using a 35S promoter rescues the rnt1-1 phenotype. CYP83A1 and CYP83B1 heterologously expressed in yeast (Saccharomyces cerevisiae) cells show marked differences in their substrate specificity. Both enzymes convert indole-3-acetaldoxime to a thiohydroximate adduct in the presence of NADPH and a nucleophilic thiol donor. However, indole-3-acetaldoxime has a 50-fold higher affinity toward CYP83B1 than toward CYP83A1. Both enzymes also metabolize the phenylalanine- and tyrosine-derived aldoximes. Enzyme kinetic comparisons of CYP83A1 and CYP83B1 show that indole-3-acetaldoxime is the physiological substrate for CYP83B1 but not for CYP83A1. Instead, CYP83A1 catalyzes the initial conversion of aldoximes to thiohydroximates in the synthesis of glucosinolates not derived from tryptophan. The two closely related CYP83 subfamily members therefore are not redundant. The presence of putative auxin responsive cis-acting elements in the CYP83B1 promoter but not in the CYP83A1 promoter supports the suggestion that CYP83B1 has evolved to selectively metabolize a tryptophan-derived aldoxime intermediate shared with the pathway of auxin biosynthesis in Arabidopsis.
Indole-3-acetic acid (IAA) is the primary plant auxin. The biosynthetic routes resulting in IAA production and the mechanism securing an optimal IAA concentration at the cellular level are poorly understood. Several biosynthetic pathways have been proposed. Mutant studies have provided some knowledge of IAA and indole metabolism, and have led to a current picture of a metabolic grid consisting of several redundant pathways operating at different developmental stages (Normanly and Bartel, 1999). Trp-dependent as well as -independent pathways have been proposed to occur in Arabidopsis seedlings based on the ability of the Trp auxotrophic mutants trp3-1 and trp2-1 to accumulate increased levels of IAA conjugates despite reduced Trp synthesis (Normanly et al., 1993). However, pleiotropic effects caused by these mutants renders it difficult to draw conclusions with respect to IAA synthesis under normal growth conditions. Thus, mature trp3-1 plants accumulate high levels of indole-3-glycerophosphate and increased levels of Trp-derived indole glucosinolates and indole-3-acetonitrile (IAN), whereas the level of free IAA is normal (Müller and Weiler, 2000). The latter observations question the operation of the proposed Trp-independent IAA pathway because indole-3-glycerophosphate is nonenzymatically converted to IAA under the alkaline conditions used to hydrolyze IAA conjugates (Müller and Weiler, 2000). Superroot2 (sur2) was described in 1998 as an auxin mutant that accumulated elevated levels of free IAA and less conjugated IAA (Delarue et al., 1998). Based on these observations, the sur2 gene was predicted to encode a protein involved in homeostasis of IAA by controlling auxin conjugation. It has been shown recently that sur2, which is allelic to rnt1-1, encodes a cytochrome P450, CYP83B1 (Barlier et al., 2000; Bak et al., 2001), involved in the conversion of indole-3-acetaldoxime to S-alkylthiohydroxymates in the biosynthesis of indole glucosinolates (Bak et al., 2001).
Cytochromes P450 are monooxygenases catalyzing key steps in numerous metabolic pathways (Kahn and Durst, 2000). CYP83B1/RNT1/SUR2 catalyzes the initial conversion of indole-3-acetaldoxime, a proposed intermediate in IAA biosynthesis, to the corresponding S-alkylthiohydroxymate. This is the first committed step in the biosynthesis of indole glucosinolates, e.g. glucobrassicin (Bak et al., 2001). Indole-3-acetaldoxime thus constitutes a metabolic branch point in IAA and indole glucosinolate biosynthesis and the level of IAA can be regulated by the flux of indole-3-acetaldoxime through CYP83B1. IAN generally has been assumed to be a product of indole-3-acetaldoxime metabolism in IAA biosynthesis (e.g. Normanly et al., 1995; Bartel, 1997; Normanly and Bartel, 1999; Hull et al., 2000). However, the nit1-1 mutation that renders Arabidopsis seedlings insensitive to the IAA effects of exogenously applied IAN (Normanly et al., 1997) is unable to mitigate the auxin phenotype of rnt1-1 in double mutants (Bak et al., 2001). This evidence argues against a role for IAN as a direct metabolite of indole-3-acetaldoxime (Bak et al., 2001). Instead, IAN may be regarded as a degradation product derived from turnover of indole glucosinolates that are hydrolyzed by a nitrilase belonging to the NIT1-3 group (Andersen and Muir, 1966; Ludwig-Müller et al., 1999; Bak et al., 2001; Vorwerk et al., 2001).
The postoxime-metabolizing enzymes in IAA biosynthesis in Arabidopsis still await identification. The closest homolog to CYP83B1 in the Arabidopsis genome is CYP83A1, showing 63% sequence identity and 78% sequence similarity at the amino acid level (Paquette et al., 2000). Both CYP83B1 and CYP83A1 transcripts are expressed in roots, leaves, stems, flowers, and siliques (Mizutani et al., 1998; Xu et al., 2001). However, although CYP83B1 is preferentially expressed in roots and induced by wounding or by dehydration, CYP83A1 is preferentially expressed in leaves and wounding reduces its expression (Mizutani et al., 1998; Reymond et al., 2000). CYP83B1 transcription was shown recently to be induced by IAA as well (Barlier et al., 2000), strengthening the connection between indole glucosinolate and IAA synthesis.
The present study was carried out to elucidate the function of CYP83A1 in the metabolic grid of IAA and indole glucosinolate biosynthesis. We asked whether the two genes were functional equivalents, i.e. redundant genes. We studied the functional complementation of the CYP83B1 knockout mutant of Arabidopsis by ectopic overexpression of the CYP83A1 cDNA and we compared the catalytic properties and biochemical characteristics of each protein expressed in a heterologous system. The results show that overexpression of CYP83A1 does compensate for the total lack of CYP83B1. However, the expression patterns of the two genes are different and the two enzymes operate on different substrates in vivo, thereby serving different purposes. Thus, the CYP83A1 and CYP83B1 genes are not redundant.
RESULTS
CYP83A1 Functionally Complements CYP83B1 in Rnt1-1
To determine whether CYP83A1 is a functional homolog of CYP83B1, CYP83A1 cDNA was ectopically expressed in rnt1-1 under control of the ubiquitous 35S cauliflower mosaic virus promoter (CaMV; Fig. 1). Plants heterozygous for knock out of CYP83B1 (rnt1-1/RNT1) were used for transformation because the homozygous plant is not optimal for transformation due to its severe phenotype (Bak et al., 2001, Fig. 1). Out of 26 primary transformants, 15 were viable. These 15 primary transformants were selfed, and the seeds were germinated on double selection plates to select for lines containing both the 35S:: CYP83A1 construct and the T-DNA insertion in CYP83B1. Out of these 15 original viable lines, five lines did not display the characteristic rnt1-1 seedling phenotype in an rnt1-1/rnt1-1 background (Fig. 1).
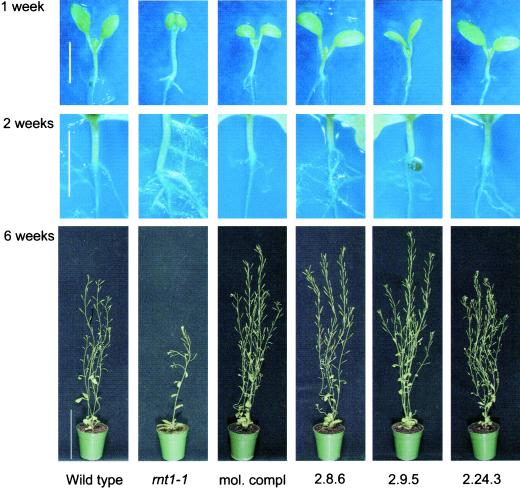
Figure 1.
Complementation of rnt1-1: comparison of wild type (wt), rnt1-1, molecularly complemented rnt1-1 (mol compl), and three independent rnt1-1 lines functionally complemented by ectopic overexpression of CYP83A1 (2.8.6, 2.9.5, and 2.24.3). Seedlings were analyzed after 1 week and after 2 weeks (bar = 3 mm); mature plants were analyzed after 6 weeks (bar = 10 cm). Hypocotyl lengths of 1-week-old seedlings: wt, 2.6 ± 0.1 mm; rnt1-1, 3.6 ± 0.2 mm; mol compl, 2.1 ± 0.1 mm; 2.8.6, 1.7 ± 0.1 mm; 2.9.5, 1.8 ± 0.1 mm; and 2.24.3, 1.7 ± 0.1 mm. Hypocotyl lengths are given with their ses of mean (n = 20).
Lines complemented by CYP83A1 under control of the 35S CaMV promoter displayed significantly shorter hypocotyls and nonepinastic cotyledons as compared with 1-week-old rnt1-1 seedlings (Fig. 1). When compared with wild-type seedlings, the hypocotyls of the CYP83A1-complemented lines were shorter. This had also been observed in rnt1-1 seedlings complemented with a genomic clone comprising the CYP83B1 gene (Bak et al., 2001, Fig. 1). The appearance of primary roots of 1-week-old rnt1-1, wild-type, or complemented seedlings did not differ. However, the characteristic extensive proliferation of root hairs and secondary roots from the primary root as well as the development of secondary roots from the vascular tissue in the hypocotyl in 2-week-old rnt1-1 seedlings (Delarue et al., 1998; Bak et al., 2001) were abolished in the complemented lines (Fig. 1).
The visual phenotypes of the complemented seedlings were very similar, whereas changes were observed in mature plants (Fig. 1). Some of the complemented lines appeared slightly bigger than wild type as shown for the lines 2.8.6 and 2.9.5, whereas other lines such as 2.24.3 were characterized by being shorter and bushier compared with e.g. the lines 2.8.6 and 2.9.5. In the latter line, up to 20 inflorescences could be observed. In addition, this line exhibited flower abnormalities and many of the siliques contained none or only a few seeds (data not shown).
Indole-3-acetaldoxime is the metabolic branch point in Trp-dependent IAA and indole glucosinolate biosynthesis. We previously have shown molecular complementation of rnt1-1 using a 5.5-kb genomic fragment comprising the CYP83B1 gene (Bak et al., 2001). In accordance with our hypothesis that indole-3-acetaldoxime is the metabolic branch point, the functionally complemented rnt1-1 lines ectopically expressing CYP83A1 cDNA complement both the high IAA phenotype and the deficiency in indole glucosinolates (Figs. 1 and 2).
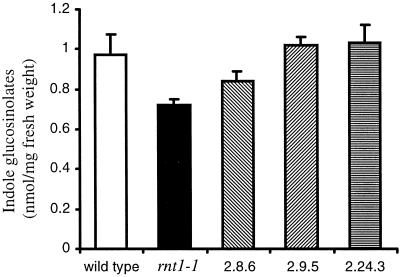
Figure 2.
Ectopic expression of CYP83A1 cDNA in rnt1-1 complements the indole glucosinolate deficiency in the CYP83B1 knockout. Indole glucosinolates were measured colorimetrically as thiocyanate (SCN−). Data are represented as mean ± se calculated per milligram fresh weight, n = 10 seedlings. The corresponding mean indole glucosinolate level per individual seedling are: wild type, 1.46 ± 0.05 nmol; rnt1-1, 0.62 ± 0.03 nmol; 2.8.6, 1.48 ± 0.15 nmol; 2.9.5, 1.60 ± 0.07 nmol; and 2.24.3, 1.15 ± 0.10 nmol.
CYP83A1 and CYP83B1 Metabolize Indole-3-Acetaldoxime with Different Affinity
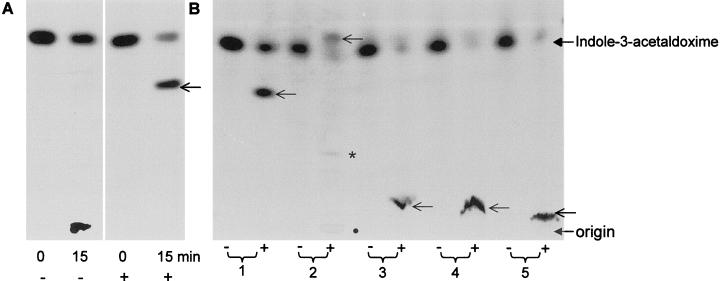
We have shown previously that CYP83B1, when co-expressed in yeast (Saccharomyces cerevisiae) with Arabidopsis NADPH cytochrome P450 reductase, metabolizes indole-3-acetaldoxime in the presence of thiol compounds to S-alkyl-thiohydroxymates (Bak et al., 2001). The nature of the initially monooxygenated product of CYP83B1 catalysis is not formally known, but it has been proposed to be an aci-nitro compound, 1-aci-nitro-2-indolyl-ethane originating from N-hydroxylation of indole-3-acetaldoxime (Ettlinger and Kjær, 1968; Bak et al., 2001). This proposed aci-nitro compound is a strong electrophile that nonenzymatically reacts preferentially with thiol compounds to form S-alkylthiohydroximate adducts (Fig. 3). In the absence of β-mercaptoethanol, the enzymatic reaction is inhibited: less indole-3-acetaldoxime is metabolized (Fig. 3A). Because the conjugate formed in the absence of a nucleophile does not migrate on thin-layer chromatography (Fig. 3A), it most likely represents the conjugate formed by the electrophilic product of the enzymatic reaction with the nucleophilic sites of the enzyme, thereby leading to the inactivation of the enzyme (Fig. 3).
Figure 3.
Products of CYP83B1 metabolism of [5-3H]indole-3-acetaldoxime in the presence and absence of nucleophiles. Reaction mixtures were analyzed by thin-layer chromatography. The components applied at the origin were focused (2 cm) in 100% methanol before development in chloroform:methanol:water (85:14:1, v/v). A, In the absence (−) of a nucleophile CYP83B1 catalysis is inhibited, and the radioactivity accumulates as an aggregate at the origin of application. In the presence (+) of β-mercaptoethanol, an adduct is formed (←). Samples were analyzed after 0 and 15 min incubation in MOPS [3-(N-morpholino)-propanesulfonic acid] buffer. B, Various structurally different nuceophiles form adducts with similar turnover. 1, β-Mercaptoethanol; 2, ethanthiol; 3, 1-thio-β-d-Glc; 4, l-Cys; 5, reduced glutathione. Samples were incubated for 15 min in the absence (−) or presence (+) of NADPH in Tris buffer. ←, The position of the adduct. Due to the volatility and immiscibility of ethanthiol in aqueous solutions adducts were identified at both the origin (●) as well as with the buffer Tris (*).
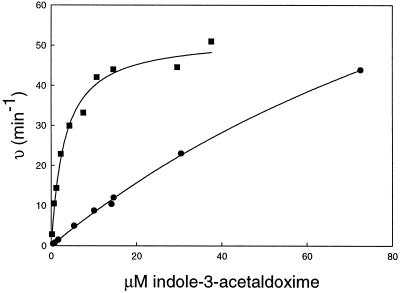
To determine if CYP83A1 metabolizes indole-3-acetaldoxime in a similar manner to CYP83B1, CYP83A1 was produced in yeast cells. Reconstitution experiments using yeast microsomes in the presence of thiol compounds showed that yeast microsomes containing CYP83A1 also metabolizes indole-3-acetaldoxime leading to thiohydroximate adducts (data not shown). Kinetics with indole-3-acetaldoxime as substrate and using Cys as thiol donor were compared for both enzymes (Fig. 4). CYP83B1 had a Km of 3.1 ± 0.4 μm and a Vmax of 52 ± 2 min−1 (Bak et al., 2001), whereas the corresponding values for CYP83A1 were 150 ± 15 μm and 140 ± 10 min−1, respectively. Based on these apparent enzyme parameters, CYP83B1 exhibits a 50-fold lower Km and a 20-fold better catalytic efficiency (Vmax/Km) compared with CYP83A1.
Figure 4.
CYP83A1 and CYP83B1 metabolize indole-3-acetaldoxime with different affinity. Kinetics with indole-3-acetaldoxime as substrate and using Cys as thiol donor were compared for both CYP83A1(●) and CYP83B1 (▪). Computed regression curves as well as the experimental data points are shown. The correlation coefficients (r2) for CYP83B1 and CYP83A1 regression analyses are 0.985 and 0.999, respectively.
Interaction with Ligands
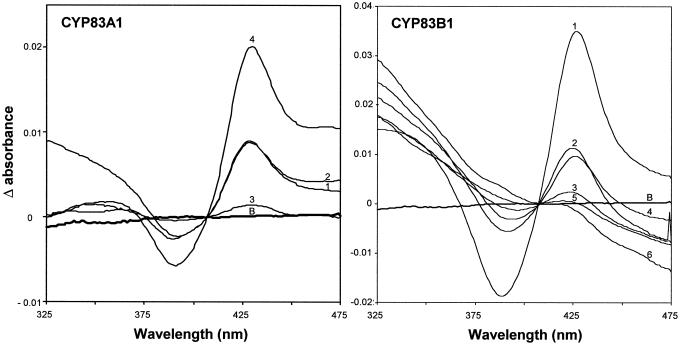
To characterize the topology of the active sites of CYP83A1 and CYP83B1, we have taken advantage of the ability of nitrogen-containing ligands like primary amines to produce type II spectra with cytochrome P450 enzymes by binding to the active site bringing the electron lone pairs of the amine group in close vicinity to the heme iron (Jefcoate, 1978). This gives rise to a characteristic spectrum with a trough around 390 nm and a peak around 425 nm. We have previously reported that tryptamine is a ligand that binds to the active site and inhibits metabolism of indole-3-acetaldoxime by CYP83B1 (Bak et al., 2001). Similar results were obtained with CYP83A1 (S. Bak, unpublished data). Likewise, type II spectra were observed for CYP83B1 and CYP83A1 with n-octylamine and the amines corresponding to Phe (β-phenylethylamine), and Tyr (tyramine; Fig. 5). IAN did not produce a type II spectrum, showing that the nitrogen atom of the indole ring system does not contribute (data not shown). Introduction of a hydroxyl group at the 5 position of tryptamine (5-OH-tryptamine/serotonin) abolished binding. Tyramine similarly produced a weak type II spectrum, whereas 3-OH-tyramine (i.e. dopamine) and histamine (data not shown) did not. This indicates that introduction of hydroxyl groups or an electronegative group in the aromatic ring causes significant reduction of ligand binding to the active site. Based on the sizes of the amplitudes of the type II spectra recorded using 200 μm ligand, the relative affinity for ligand binding to CYP83B1 is tryptamine ≫ β-phenylethylamine > n-octylamine > tyramine. CYP83A1 shows a different affinity for the same amines: n-octylamine ≫ β-phenylethylamine = tryptamine > tyramine. The observed difference in affinity for the amines tested argues that although the same ligands bind to CYP83B1 and CYP83A1, the topology of their active site differ.
Figure 5.
Spectral characterization of CYP83A1 and CYP83B1. Type II spectra were recorded with 0.15 μm of CYP83A1 or 0.44 μm of CYP83B1 using 200 μm of ligands. 1, Tryptamine; 2, β-phenylethylamine; 3, tyramine; 4, n-octylamine; 5, 5-OH-tryptamine; 6, 3-OH-tyramine; B, baseline.
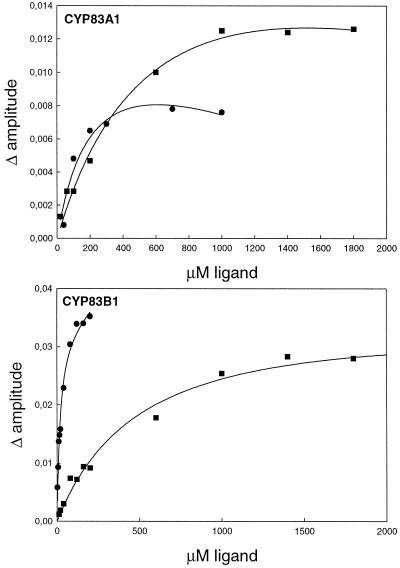
By titrating the amplitude of the type II difference spectra with increasing concentrations of ligand, Ks values were determined for tryptamine and β-phenylethylamine (Fig. 6). Ks values of 18 ± 5 μm and 240 ± 180 μm were calculated for tryptamine for CYP83B1 and CYP83A1, respectively. Ks values of 540 ± 180 μm and 390 ± 70 μm were estimated for β-phenylethylamine binding to CYP83B1 and CYP83A1, respectively. In accordance, CYP83B1 binds tryptamine 13-fold stronger compared with CYP83A1. Compared with β-phenylethylamine, tryptamine is a 30-fold stronger ligand for CYP83B1. In contrast, CYP83A1 displays similar high binding constants for tryptamine and β-phenylethylamine. Due to high absorbance and low amplitude of the type II spectra, Ks values could not be determined for tyramine.
Figure 6.
CYP83A1 and CYP83B1 have different affinity for tryptamine and β-phenyletylamine. CYP83A1 (0.15 μm) or CYP83B1 (0.44 μm) were incubated with increasing amounts of either tryptamine (●) or β-phenylethylamine (▪) and the difference in amplitude of the type II difference spectra were plotted as a function of concentration of ligand. To compensate for ligand absorbance, the experimental data were fitted to a hyperbolic curve using the equation A = Amax × X/(Ks + X) + C × X, where A is the amplitude of the spectra, X the concentration of ligand, and C the contribution from ligand absorbance. The computed regression curve is shown as well as the experimental data points. Correlation coefficients (r2) for CYP83B1 interaction with tryptamine and β-phenylethylamine are 0.983 and 0.987, respectively, and for CYP83A1 interaction with tryptamine and β-phenylethylamine 0.933 and 0.989, respectively.
Interaction with Oximes
Indole-3-acetaldoxime is a substrate for CYP83B1 and CYP83A1 as shown by heterologous expression studies and by the ability of CYP83A1 to functionally complement CYP83B1 in rnt1-1. Substrates for cytochromes P450 often give rise to the formation of a type I or reverse type I spectrum upon binding, depending on the spin state of the heme iron (Jefcoate, 1978). Besides CYP83A1 and CYP83B1, the only other plant cytochrome P450 known to metabolize an aldoxime is CYP71E1 from sorghum (Sorghum bicolor). CYP71E1 is involved in the biosynthesis of the Tyr-derived cyanogenic glucoside dhurrin and catalyzes the conversion of p-hydroxyphenylacetaldoxime to p-hydroxymandelonitrile (Kahn et al., 1997, 1999; Bak et al., 1998a). The substrate binding spectra obtained using p-hydroxyphenylacetaldoxime as a substrate for sorghum CYP71E1 were not trivial and prone to peculiar artifacts (Kahn et al., 1997, 1999). Spectral analysis of a cytochrome P450 in rat liver microsomes similarly displayed peculiar binding spectra with aryl and alkyl aldoximes (Boucher et al., 1994). Only a weak reverse type I spectrum was recorded upon indole-3-acetaldoxime binding to CYP83B1 (Bak et al., 2001). In accordance, a Ks value of 0.2 μm for indole-3-acetaldoxime binding to CYP83B1 was determined by exploiting the ability of indole-3-acetaldoxime to displace the ligand tryptamine from the active site of CYP83B1 (Bak et al., 2001). In that approach, CYP83B1 was first saturated with 100 μm tryptamine. Tryptamine was subsequently displaced from the active site by titration with increasing amounts of indole-3-acetaldoxime, causing a gradual appearance of a reverse type II spectrum. It is unfortunate that a similar approach could not be used for CYP83A1 because: (a) much higher levels of tryptamine (1,000 μm) are required to saturate CYP83A1, giving rise to interfering levels of ligand absorbance (Fig. 5); (b) the amplitude of the type II spectra produced by tryptamine binding to CYP83A1 is much weaker than for CYP83B1 (Fig. 5); and (c) indole-3-acetaldoxime absorbance interferes significantly at concentrations higher than 1 μm.
In addition to the ability to use indole-3-acetaldoxime as a substrate, we conducted reconstitution experiments to compare the ability of CYP83A1 and CYP83B1 to metabolize other oximes. The putative substrates tested were p-hydroxyphenylacetaldoxime derived from Tyr and phenylacetaldoxime derived from Phe. In all studies, β-mercaptoethanol was the thiol donor. After incubation in the presence or absence of NADPH, reaction mixtures were extracted with ethyl acetate, and the ethyl acetate phase containing both substrate and product was lyophilized, silylated, and analyzed by gas chromatography-mass spectrometry (GC-MS) as previously described (Bak et al., 2001; data not shown). As expected, the turnover of indole-3-acetaldoxime was lower using CYP83A1 compared with CYP83B1 under the experimental conditions applied (Table I). The turnover of p-hydroxyphenylacetaldoxime conversely was higher using CYP83A1 compared with CYP83B1. Phenylacetaldoxime was identified as a substrate for CYP83A1 as well as for CYP83B1 but the turnover numbers were low (Table I).
Table I.
CYP83A1 and CYP83B1 have overlapping substrate and ligand affinity in vitro
| Km Indole-3-acetaldoxime | Turnover, Indole-3-acetaldoxime | Turnover, p-Hydroxyphenyl-acetaldoxime | Turnover, Phenylethyl-acetaldoxime | Ks for Tryptamine | Ks for β-Phenylethylamine | |
|---|---|---|---|---|---|---|
| μM | min−1 | μM | ||||
| CYP83B1 | 3.1 | 26 | 9.8 | 15 | 18 | 540 |
| CYP83A1 | 150 | 10 | 25 | 7.2 | 240 | 390 |
DISCUSSION
CYP83A1 and CYP83B1 Are Not Redundant Enzymes
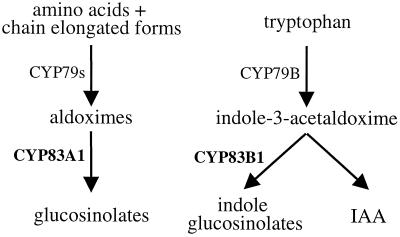
In Arabidopsis, indole-3-acetaldoxime is a metabolic branch point in IAA and indole glucosinolate biosynthesis and the level of IAA can be regulated by the flux of indole-3-acetaldoxime through CYP83B1 (Fig. 7; Bak et al., 2001). In the present study, we demonstrate that ectopic expression of CYP83A1 cDNA can functionally complement CYP83B1 by suppressing the high IAA phenotype and deficiency in indole glucosinolate of rnt1-1.
Figure 7.
CYP83A1 and CYP83A1 are not redundant enzymes. CYP83B1 is primarily involved in biosynthesis of indole glucosinolates, whereas CYP83A1 is involved in glucosinolates not derived from indole-3-acetaldoxime. The use of a separate CYP83 for indole glucosinolate biosynthesis ensures a tight control of the flux of the shared Trp-derived intermediate, indole-3-acetaldoxime, for IAA and indole glucosinolate biosynthesis.
Knockout of CYP83B1 results in plants characterized by increased apical dominance and elongated hypocotyls (Fig. 1; Barlier et al., 2000; Bak et al., 2001) due to an increase of free IAA (Delarue et al., 1998; Barlier et al., 2000). Ectopic overexpression of CYP83B1 cDNA using the 35S promoter in wild-type Arabidopsis also showed a bushier phenotype in three out of 13 transformants (S. Bak, unpublished data). Bushy phenotypes similarly were seen in two out of 18 rnt1-1 lines molecularly complemented with a genomic fragment comprising the CYP83B1 gene (Bak et al., 2001; S. Bak, unpublished data). Multiple insertions as well as position effects may result in lines that phenotypically resemble overexpression lines. The phenotype of plants like 2.24.3 is similar to the phenotype of strong alleles of axr1, characterized by decreased apical dominance and reduced hypocotyl length and fertility as a result of reduced sensing of auxin (Estelle and Sommerville, 1987; Lincoln et al., 1990; Leyser et al., 1993; Collett et al., 2000). Arabidopsis seedlings overexpressing the bacterial enzyme Trp monooxygenase (iaaM) have up to 4-fold higher IAA levels than wild type and are characterized by having elongated hypocotyls (Romano et al., 1995). Plants that overexpress iaaL conversely have reduced levels of free IAA and shorter hypocotyls due to increased conjugation of IAA to Lys (Romano et al., 1991; Jensen et al., 1998).
Although functional complementation of CYP83B1 in rnt1-1 by overexpression of CYP83A1 under the control of the 35S promoter was demonstrated, the CYP83A1 gene is not redundant compared with CYP83B1 because CYP83A1 cannot prevent the rnt1-1 phenotype when expressed under the control of its native promoter in the rnt1-1 background.
Indole-3-Acetaldoxime Metabolism by CYP83A1 and CYP83B1
In accordance with the in planta complementation results, indole-3-acetaldoxime was identified as a substrate for recombinant CYP83A1. Oximes are generally unstable and considered toxic compounds that do not accumulate in the cell. To optimize and control catalytic activities, most biosynthetic enzymes have Kms in the range of the concentration of their substrate. The in vivo concentration of indole-3-acetaldoxime in Arabidopsis is not known. However, in the related cruciferous plant Chinese cabbage (Brassica campestris), the indole-3-acetaldoxime concentration has been reported to be less than 50 pmol g−1 fresh weight (Helminger et al., 1985). Indole-3-acetaldoxime constitutes a metabolic branch point between IAA and indole glucosinolate biosynthesis, and we have concluded previously that enzymes in indole glucosinolate and IAA biosynthesis utilize the same indole-3-acetaldoxime pool (Bak et al., 2001). This implies that an enzyme working in such a branch point must have a Km in the same range as CYP83B1 to efficiently compete for the substrate. The 50-fold higher Km of CYP83A1 relative to CYP83B1 thus argues that indole-3-acetaldoxime is not a substrate for CYP83A1 under normal conditions.
Overexpression of CYP83A1 cDNA in the rnt1-1 background did not result in elevated indole glucosinolate levels as compared with wild-type seedlings (Fig. 2). This is in contrast to overexpression of CYP83B1 cDNA (Bak et al., 2001), which resulted in increased levels of indole glucosinolates. These data imply that CYP83A1 cannot to the same extent as CYP83B1 compete with an indole-3-acetaldoxime-metabolizing enzyme in IAA biosynthesis. Low levels of indole glucosinolates are present in rnt1-1 seedlings (Bak et al., 2001; Fig. 2). This may imply that in rnt1-1, indole-3-acetaldoxime accumulates to levels that become available to CYP83A1 and permits a low level of indole glucosinolate production. A more likely explanation is that at least some of the indole glucosinolates present in the seedlings may not originate from de novo synthesis, but by translocation of indole glucosinolates from the seed.
There are two reasonable explanations for the ability of the CYP83A1 cDNA to functionally complement rnt1-1: (a) In rnt1-1, indole-3-acetaldoxime accumulates to levels that makes it available to CYP83A1; or (b) ectopic expression of CYP83A1 restores the channeling of indole glucosinolate biosynthesis by restoring a supra molecular enzymatic complex with e.g. CYP79B. The latter explanation satisfies the observation that increased levels of indole glucosinolates was not seen in the functionally complemented lines (Fig. 2).
The catalytic mechanism by which CYP83B1 and CYP83A1 convert aldoximes is not known. We speculate that the oxygen atom of the oxime function lodges between the heme iron and the P450 I-helix, thereby replacing a water molecule as sixth ligand to the heme iron. This replacement of water by the oxime may explain the absence of a strong type I binding spectrum. Subsequent introduction of an additional hydroxyl group at the nitrogen atom of the oxime function generates a highly reactive aci-nitro compound. The α-carbon atom of the aci-nitro compound is a target for a nucleophilic attack from a sulfhydryl group, resulting in the formation of indole-3-S-alkylthiohydroxymate with a dehydration reaction taking place either before or after adduct formation. An aci-nitro compound previously has been proposed as an intermediate in glucosinolate biosynthesis (Ettlinger and Kjær, 1968). Liver microsomes have been suggested in a similar manner to catalyze the conversion of n-butyraldoxime to nitrobutane via an aci-nitro compound (DeMaster et al., 1992). The observed ability to form S-alkylthiohy-droximate adducts with a wide range of structurally very different thiol compounds in vitro suggests that formation of the adduct proceeds nonenzymatically outside the active site (Fig. 3). In accordance with this proposed mechanism, indole-3-acetaldoxime metabolism in the absence of a nucleophile eventually inactivates the enzyme (Fig. 3A).
CYP83B1 has recently been shown to be induced by IAA (Delarue et al., 1998). In accordance, we analyzed in silico 2.5 kb upstream of the start codon of CYP83B1 for cis-acting elements (Higo et al., 1999; http://www.dna.affrc.go.jp/htdocs/PLACE), and identified four putative auxin-responsive cis-acting elements (AuxREs; Guilfoyle et al., 1998; Ulmasov et al., 1999). We have previously shown that a 5.5-kb genomic fragment comprising this putative CYP83B1 promoter is sufficient to achieve molecular complementation of rnt1-1 (Bak et al., 2001). In a converse manner, no AuxREs could be identified 2.5 kb region upstream of CYP83A1. In accordance, cDNA micro array data show that in rnt1-1 seedlings CYP83A1 transcripts are not induced but down-regulated 3.5-fold (W. Xu and D.W. Galbraith, personal communication). This suggests that CYP83B1, but not CYP83A1, is under the regulation of auxin.
CYP83A1 and CYP83B1 Have Overlapping Substrate Specificity
Indole-3-acetaldoxime, phenylacetaldoxime, and p-hydroxyphenylacetaldoxime are all substrates for CYP83A1 and CYP83B1 in vitro. Based on the turnover numbers using high substrate concentrations (1 mm), p-hydroxyphenylacetaldoxime is the preferred substrate for CYP83A1 as compared with CYP83B1. Arabidopsis contains at least 24 glucosinolates derived from Trp and chain-elongated homologs of Phe and Met (Hogge et al., 1988; Petersen et al., 2001). In Arabidopsis, seven functional CYP79 homologs and six CYP79 pseudogenes (http://www.biobase.dk/P450) have been identified. These CYP79s most likely catalyze the conversion of amino acids and chain-elongated amino acids to their corresponding aldoximes (Bak et al., 1998b), as has been documented for CYP79A2, CYP79B2, CYP79B3, and CYP79F1 (Hull et al., 2000; Mikkelsen et al., 2000; Wittstock and Halkier, 2000; Hansen et al., 2001; Reintanz et al., 2001). These CYP79 homologs are highly substrate specific and are thought to determine the substrate specificity of glucosinolate biosynthesis. In contrast, only two CYP83 homologs are present in the Arabidopsis genome (http://www.biobase.dk/P450). The substrate specificity of these two enzymes toward aliphatic aldoximes is an open question. Detailed biochemical analysis of the recombinant enzymes is hampered by lack of aldoxime substrates. Glucosinolate profile analysis of CYP83A1 and CYP83B1 overexpression lines as well as of molecularly and functionally complemented rnt1-1 plants are in progress and should serve to clarify the substrate specificity of CYP83A1 and CYP83B1 toward naturally occurring aldoximes and their effect on glucosinolate profiles.
We propose that CYP83B1 is primarily involved in the biosynthesis of indole glucosinolates, whereas CYP83A1 is involved in biosynthesis of those glucosinolates that are not derived from Trp (Fig. 7). Use of a separate CYP83 for indole glucosinolate biosynthesis insures tight control of the flux of the shared intermediate, indole-3-acetaldoxime, for indole glucosinolate and IAA biosynthesis as is also indicated by the presence of putative AuxREs in the CYP83B1 but not in the CYP83A1 promoter. The evidence now available demonstrates that CYP83s and other postoxime enzymes have a low substrate specificity. Thus, cell suspension cultures of Brassica juncea produce artificial and novel glucosinolates from p-nitrobenzaldoxime (Grootwassink et al., 1990). Likewise, ectopic expression of sorghum CYP79A1 catalyzing the conversion of Tyr to p-hydroxyphenylacetaldoxime resulted in Arabidopsis plants with high levels of Tyr-derived p-hydroxybenzylglucosinolate, which is not a naturally occurring glucosinolate in this species (Bak et al., 1999).
It has often been suggested that glucosinolate biosynthesis has evolved from a cyanogenic predisposition (e.g. Ettlinger and Kjær, 1968; Poulton and Møller, 1993; Halkier and Du, 1997). In contrast to glucosinolates that are primarily found in the order Capparales, cyanogenic glucosides are widespread in nature and represent an evolutionary ancient trait. Cyanogenic glucosides are derived from the precursor amino acids Val, iso-Leu, Phe, and Tyr and have an oxime as intermediate just as in glucosinolate biosynthesis. Likewise, the amino acid to aldoxime conversion is catalyzed by a CYP79 homolog (Halkier et al., 1995; Andersen et al., 2000; Nielsen and Møller, 2000). It is striking that no cyanogenic glucosides are known to be derived from Trp. An explanation may be that for a natural product biosynthetic pathway to share an intermediate in the biosynthesis of an essential hormone, a tight and controlled regulation is mandatory. CYP83B1 fulfills these requirements and this may be a clue to the predominance of glucosinolates in cruciferous plants.
MATERIALS AND METHODS
Plants
Plants were grown at a photosynthetic flux of 100 to 120 μmol photons m−2 s−1 and 70% humidity, 22°C for a 12-h photoperiod. For morphometric analyses, seedlings were grown vertically on Murashige and Skoog agar plates without addition of antibiotics and grown for a 16-h photoperiod. Morphometric analyses are shown with their se of the mean.
The molecularly complemented rnt1-1 line used in this study was line 3.25.11 (Bak et al., 2001). For functional complementation of rnt1-1, overexpression constructs comprising the CYP83A1 cDNA under control of a CaMV 35S promoter and polyadenylation site were made in pPZP221 (Hajdukiewicz et al., 1994). Primary transformants were selected on Murashige and Skoog plates supplemented with 2% (w/v) Suc, 0.9% (w/v) Bacto agar, 50 μg mL−1 kanamycin, and 200 μg mL−1 gentamycin. Lines homozygous for the T-DNA insertion in CYP83B1 and homozygous for the introduced 35S::CYP83A1 construct were identified by cosegregation analysis on selective Murashige and Skoog agar plates.
Indole glucosinolate content in 10-d-old seedlings grown as described for the morphometric analyses were quantified colorimetrically as the degradation product thiocyanate as previously described by Bak et al. (1999, 2001).
Analysis of Recombinant CYP83A1 and CYP83B1 Enzyme
Microsomes from yeast (Saccharomyces cerevisiae) WAT11 cells expressing the CYP83A1 and CYP83B1 cDNA using the pYeDP60 vector were isolated and the amount of functional enzyme quantified essentially according to Pompon et al. (1996). Indole-3-acetaldoxime and radiolabeled indole-3-acetaldoxime were prepared as described by Bak et al. (2001) and references therein. Vmax and Km were determined as previously described using 2.2 nm of CYP83A1 or CYP83B1 and 50 mm l-Cys as thiol donor (Bak et al., 2001). Type II spectra were recorded using 0.44 μm CYP83B1 or 0.15 μm CYP83A1 and in the presence of 200 μm ligand and using a Lambda19 spectrophotometer (Perkin Elmer, Shelton, CT). Vmax, Km, and Ks were calculated using SigmaPlot 5.0 (SPSS Inc., Chicago). For analysis of CYP83B1 activity in the presence or absence of thiol donors, recombinant CYP83B1 was reconstituted and analyzed as previously described (Bak et al., 2001).
Identification and Quantification of Substrates and Products by GC-MS
For structural analysis of the products of CYP83A1 and CYP83B1 catalysis, 0.5 μm recombinant enzyme was reconstituted using 1 mm of either indole-3-acetaldoxime, p-hydroxyphenylacetaldoxime, or phenylacetaldoxime and incubated for 20 min at 28°C and analyzed using GC-MS essentially as previously described (Bak et al., 2001). Turnover numbers were calculated based on the relative areas under the substrate and product peaks. Silylated substrates and products were identified by their fragmentation pattern in both electron impact mode and chemical ionization mode.
Fragments identified by chemical ionization mode were as follows: silylated indole-3-acetaldoxime (15.447 min), [M+H]+ m/z 319, major fragmentation ion m/z 202; silylated S-mercaptoyl-indole-3-acetaldoxime (21.032 min), [M+H]+ m/z 467, major fragmentation ions m/z 229, and m/z 202; silylated (E+Z)- p-hydroxyphenylacetaldoxime (11.860 and 11.942 min), [M+H]+ m/z 296 major fragmentation ion m/z 179; silylated S-mercaptoyl-p-hydroxyphenylacetaldoxime (17.433 min), [M+H]+ m/z 444, major fragmentation ions m/z 206 and m/z 179; silylated phenylethylacetaldoxime (11.066 min), [M+H]+ m/z 209; and silylated S-mercaptoyl-phenylacetaldoxime (14.666 min), [M+H]+ m/z 356, major fragmentation ions m/z 226, m/z 206, m/z 118, and m/z 91.
ACKNOWLEDGMENTS
We thank Drs. Denis Pompon and Philippe Urban for the gift of pYeD60 and WAT11, Drs. Birger Lindberg Møller and Barbara Ann Halkier for the gift of p-hydroxyphenylacetaldoxime and phenylacetaldoxime, Dr. Birger Lindberg Møller for critically reading the manuscript, Adria Decker for technical assistance, Dr. Marat Murataliev for helpful discussions on characterization of recombinant CYP83A1 and CYP83B1, Dr. Frans E. Tax for critically reading the manuscript and for helpful discussions on the transgenic lines, Dr. Arpad Somogyi for performing the GC-MS analysis, and Dr. Kirsten Jørgensen for taking the seedling pictures. Images of 6-week-old wild-type, rnt1-1, and molecularly complemented plants are copyrighted by the American Society of Plant Biologists and are reprinted with permission.
Footnotes
This work was supported by The Human Frontier Science Program (grant no. RG0280/1999M) and by the U.S. Department of Agriculture (grant no. NRICGP 97 01472). S.B. was supported by The Danish Veterinary and Agricultural Research Council (grant no. 970265) and by the Danish National Science Research Foundation.
LITERATURE CITED
- Andersen AS, Muir R. Auxin activity of glucobrassicin. Plant Physiol. 1966;19:1038–1048. [Google Scholar]
- Andersen MD, Busk PK, Svendsen I, Møller BL. Cytochromes P450 from cassava (Manihot esculenta Crantz) catalyzing the first steps in the biosynthesis of the cyanogenic glucosides linamarin and lotaustralin: cloning, functional expression in Pichia pastoris, and substrate specificity of the isolated recombinant enzymes. J Biol Chem. 2000;275:1966–1977. doi: 10.1074/jbc.275.3.1966. [DOI] [PubMed] [Google Scholar]
- Bak S, Kahn RA, Nielsen HL, Møller BL, Halkier BA. Cloning of three A-type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol Biol. 1998a;36:393–405. doi: 10.1023/a:1005915507497. [DOI] [PubMed] [Google Scholar]
- Bak S, Nielsen HL, Halkier BA. The presence of CYP79 homologoues in glucosinolate-producing plants shows evolutionary conservation of the enzymes in the conversion of amino acids to aldoxime in the biosynthesis of cyanogenic glucosides and glucosinolates. Plant Mol Biol. 1998b;38:725–734. doi: 10.1023/a:1006064202774. [DOI] [PubMed] [Google Scholar]
- Bak S, Olsen CE, Petersen BL, Møller BL, Halkier BA. Metabolic engineering of p-hydroxybenzylglucosinolate in Arabidopsis by expression of the cyanogenic CYP79A1 from Sorghum bicolor. Plant J. 1999;20:663–672. doi: 10.1046/j.1365-313x.1999.00642.x. [DOI] [PubMed] [Google Scholar]
- Bak S, Tax FE, Feldmann KA, Galbraith DA, Feyereisen R. CYP83B1, a cytochrome P450 at the metabolic branchpoint in auxin and indole glucosinolate biosynthesis in Arabidopsis thaliana. Plant Cell. 2001;13:101–111. doi: 10.1105/tpc.13.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, Sandberg G, Bellini C. SUR2 gene of Arabidopsis thaliana, encodes the cytochrome P450 CYP83B1: a modulator of auxin homeostasis. Proc Natl Acad Sci USA. 2000;97:14819–14824. doi: 10.1073/pnas.260502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B. Auxin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:51–66. doi: 10.1146/annurev.arplant.48.1.51. [DOI] [PubMed] [Google Scholar]
- Boucher J, Delaforge M, Mansuy D. Dehydration of alkyl- and arylaldoximes as a new cytochrome P450-catalyzed reaction: mechanism and stereochemical characteristics. Biochemistry. 1994;33:7811–7818. doi: 10.1021/bi00191a008. [DOI] [PubMed] [Google Scholar]
- Collett CE, Harberd NP, Leyser O. Hormonal inter-actions in the control of Arabidopsis hypocotyl elongation. Plant Physiol. 2000;124:553–561. doi: 10.1104/pp.124.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M, Prinsen E, Onckelen HV, Caboche M, Bellini C. Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostatis. Plant J. 1998;14:603–611. doi: 10.1046/j.1365-313x.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- DeMaster E, Shirota FN, Nagasawa HT. A beckmann-type dehydration of n-butyraldoxime catalyzed by cytochrome P450. J Org Chem. 1992;57:5074–5075. [Google Scholar]
- Estelle MA, Sommerville C. Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol Gen Genet. 1987;206:200–206. [Google Scholar]
- Ettlinger MG, Kjær A. Sulfur compounds in plants. Rec Adv Phytochem. 1968;1:49–144. [Google Scholar]
- Grootwassink JWD, Balsevich JJ, Kolenovsky AD. Formation of sulfatoglucosides from exogenous aldoximes in plant cell cultures and organs. Plant Sci. 1990;66:11–20. [Google Scholar]
- Guilfoyle T, Hagen G, Ulmasov T, Murfett J. How does auxin turn on genes? Plant Physiol. 1998;118:341–347. doi: 10.1104/pp.118.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Halkier BA, Du L. The biosynthesis of glucosinolates. Trends Plant Sci. 1997;11:425–430. doi: 10.1016/j.tplants.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Halkier BA, Nielsen HL, Koch B, Møller BL. Purification and characterization of recombinant cytochrome P450TYR expressed at high levels in Escherichia coli. Arch Biochem Biophys. 1995;322:369–377. doi: 10.1006/abbi.1995.1477. [DOI] [PubMed] [Google Scholar]
- Hansen CH, Wittstock U, Olsen CE, Hick AJ, Pickett JA, Halkier BA. Cytochrome P450 CYP79F1 from Arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J Biol Chem. 2001;276:11078–11085. doi: 10.1074/jbc.M010123200. [DOI] [PubMed] [Google Scholar]
- Helminger J, Rausch T, Hilgenberg W. Metabolism of 14C-indole-3-acetaldoxime by hypocotyls of chinese cabbage. Phytochemistry. 1985;24:2497–2502. [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogge RL, Reed DW, Underhill EW. HPLC separation of glucosinolates from leaves and seeds of Arabidopsis thalina and their identification using thermospray liquid chromotography/mass spectrometry. Chromatogr Sci. 1988;26:551–556. [Google Scholar]
- Hull AK, Vij R, Celenza JL. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci USA. 2000;97:2379–2384. doi: 10.1073/pnas.040569997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefcoate CR. Measurement of substrate and inhibitor binding to microsomal cytochrome P-450 by optical-difference spectroscopy. Methods Enzymol. 1978;27:258–279. doi: 10.1016/s0076-6879(78)52029-6. [DOI] [PubMed] [Google Scholar]
- Jensen PJ, Hangarter RP, Estelle M. Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol. 1998;116:455–462. doi: 10.1104/pp.116.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Bak S, Svendsen I, Halkier BA, Møller BL. Isolation and reconstitution of cytochrome P450OX and in vitro reconstitution of the entire biosynthetic pathway of the cyanogenic glucoside dhurrin from Sorghum. Plant Physiol. 1997;115:1661–1670. doi: 10.1104/pp.115.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Durst F. Function and evolution of plant cytochromes P450. In: Romeo JT, Ibrahim R, De Luca V, editors. Recent Advances in Phytochemistry: Evolution of Metabolic Pathways. Amsterdam: Elsevier Science Ltd; 2000. pp. 151–190. [Google Scholar]
- Kahn RA, Fahrendorf T, Halkier BA, Møller BL. Substrate specificity of the cytochrome P450 enzymes CYP79A1 and CYP71E1 involved in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. Arch Biochem Biophys. 1999;363:9–18. doi: 10.1006/abbi.1998.1068. [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. Arabidopsis auxin resistance gene AXR1 encodes a protein related to ubiquitin activating enzyme E1. Nature. 1993;364:161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Müller J, Pieper K, Ruppel M, Cohen JD, Epstein E, Kiddle G, Bennett R. Indole glucosinolate and auxin biosynthesis in Arabidopsis thaliana (L.) Heynh. glucosinolate mutants and the development of clubroot disease. Planta. 1999;208:409–419. doi: 10.1007/s004250050576. [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA. Cytochrome P450 CYP79B2 from arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J Biol Chem. 2000;275:33712–33717. doi: 10.1074/jbc.M001667200. [DOI] [PubMed] [Google Scholar]
- Mizutani M, Ward E, Ohta D. Cytochrome P450 superfamily in Arabidopsis thaliana: isolation of cDNAs, differential expression, and RFLP mapping of multiple cytochromes P450. Plant Mol Biol. 1998;37:39–52. doi: 10.1023/a:1005921406884. [DOI] [PubMed] [Google Scholar]
- Müller A, Weiler EW. Indolic constituents and indole-3-acetic acids biosynthesis in the wild-type and a tryptophan auxotroph mutant of Arabidopsis thaliana. Planta. 2000;211:855–863. doi: 10.1007/s004250000353. [DOI] [PubMed] [Google Scholar]
- Nielsen JS, Møller BL. Cloning and expression of cytochrome P450 enzymes catalyzing the conversion of tyrosine to p-hydroxyphenylacetaldoxime in the biosynthesis of cyanogenic glucosides in Triglochin maritima. Plant Physiol. 2000;122:307–317. doi: 10.1104/pp.122.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J, Bartel B. Redundancy as a way of life: IAA metabolism. Curr Opin Plant Biol. 1999;2:207–213. doi: 10.1016/s1369-5266(99)80037-5. [DOI] [PubMed] [Google Scholar]
- Normanly J, Cohen JD, Fink GR. Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc Natl Acad Sci USA. 1993;90:10355–10359. doi: 10.1073/pnas.90.21.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J, Grisafi P, Fink GR, Bartel B. Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell. 1997;9:1781–1790. doi: 10.1105/tpc.9.10.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J, Slovin JP, Cohen JD. Rethinking auxin biosynthesis and metabolism. Plant Physiol. 1995;107:323–329. doi: 10.1104/pp.107.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette SM, Bak S, Feyereisen R. Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol. 2000;19:307–317. doi: 10.1089/10445490050021221. [DOI] [PubMed] [Google Scholar]
- Petersen BL, Andreasson E, Bak S, Agerbirk N, Halkier BA. Characterization of transgenic Arabidopsis thaliana with metabolically engineered high levels of p-hydroxybenzylglucosinolate. Planta. 2001;212:612–618. doi: 10.1007/s004250000429. [DOI] [PubMed] [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- Poulton JE, Møller BL. Glucosinolates. Methods Plant Biochem. 1993;9:209–237. [Google Scholar]
- Reintanz B, Lehnen M, Reichelt M, Gershenzon J, Kowalczyk M, Sandberg G, Godde M, Uhl R, Palme K. bus, a Bushy arabidopsis CYP79F1 knockout mutant with abolished synthesis of short chain aliphatic glucosinolates. Plant Cell. 2001;13:351–367. doi: 10.1105/tpc.13.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. Differential gene expression in responds to mechanical wounding and insect feeding in Arabidopsis. Plant Cell. 2000;12:707–719. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano C, Hein M, Klee H. Inactivation of auxin in tobacco transformed with the indoleacetic acid-lysine synthetase gene of Pseudomonas savastonoi. Genes Dev. 1991;5:438–446. doi: 10.1101/gad.5.3.438. [DOI] [PubMed] [Google Scholar]
- Romano CP, Robson PRH, Smith H, Estelle M, Klee H. Transgene-mediated auxin overproduction in Arabidopsis: hypocotyl elongation phenotype and interaction with the hy6-1 hypocotyl elongation and axr1 auxin-resisteant mutants. Plant Mol Biol. 1995;27:1071–1083. doi: 10.1007/BF00020881. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- Vorwerk S, Biernacki S, Hillebrand, Janzik I, Müller A, Weiler EW, Piotrowski M. Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3 gene cluster. Planta. 2001;212:508–516. doi: 10.1007/s004250000420. [DOI] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA. Cytochrome P450 CYP79A2 from Arabidopsis thaliana L. catalyzes the conversion of L-phenylalanine to phenylacetaldoxime in the biosynthesis of benzylglucosinolates. J Biol Chem. 2000;275:14659–14666. doi: 10.1074/jbc.275.19.14659. [DOI] [PubMed] [Google Scholar]
- Xu W, Bak S, Decker A, Paquette SM, Feyereisen R, Galbraith DW (2001) Microarray-based analysis of gene expression in very large gene families: the cytochrome P450 gene superfamily of Arabidopsis thaliana. Gene (in press) [DOI] [PubMed]