Abstract
Anthranilate synthase (AS), the control enzyme of the tryptophan (Trp) biosynthetic pathway, is encoded by nuclear genes, but is transported into the plastids. A tobacco (Nicotiana tabacum) cDNA (ASA2) encoding a feedback-insensitive tobacco AS α-subunit was transformed into two different sites of the tobacco plastid genome through site-specific insertion to obtain transplastomic plants with normal phenotype and fertility. A high and uniform level of ASA2 mRNA was observed in the transplastomic plants but not in the wild type. Although the plants with the transgene insertion at ndhF-trnL only expressed one size of the ASA2 mRNA, the plants with the transgene incorporated into the region between accD and open reading frame (ORF) 184 exhibited two species of mRNA, apparently due to readthrough. The transplastomic plants exhibited a higher level of AS α-subunit protein and AS enzyme activity that was less sensitive to Trp-feedback inhibition, leading to greatly increased free Trp levels in leaves and total Trp levels in seeds. Resistance to an AS inhibitor, 5-methyl-Trp, was found during seed germination and in suspension cultures of the transplastomic plants. The resistance to the selection agent spectinomycin and to 5-methyl-Trp was transmitted maternally. These results demonstrate the feasibility of modifying the biosynthetic pathways of important metabolites through transformation of the plastid genome by relocating a native gene from the nucleus to the plastid genome. Very high and uniform levels of gene expression can be observed in different lines, probably due to the identical insertion sites, in contrast to nuclear transformation where random insertions occur.
Biosynthesis of almost all “essential” amino acids, including Trp, in higher plants takes place in plastids. Most of the enzymes involved in essential amino acid biosynthesis are encoded by nuclear genes, synthesized in the cytosol, and imported into plastids. The endosymbiont hypothesis suggests that unicellular, photosynthetic cyanobacteria were engulfed by an early eukaryotic host and became the progenitors of the chloroplast. Most of the genes originally residing in the early chloroplasts have since moved to the nucleus (Martin and Herrmann, 1998), which likely include those involved in Trp biosynthesis.
Trp biosynthesis branches off the shikimate pathway at chorismate, which is the last common precursor of many aromatic compounds (Haslam, 1993; Herrmann, 1995). Anthranilate synthase (AS) catalyzes the first committed reaction for Trp biosynthesis, converting chorismate to anthranilate, and is feedback inhibited by the end product Trp (Haslam, 1993; Radwanski and Last, 1995; Romero et al., 1995). As in most bacteria, plant AS holoenzymes so far characterized are tetramers consisting of two α- and two β-subunits encoded by separate nuclear genes and synthesized in the cytosol as precursor proteins with plastid-targeting transit peptides (Crawford, 1989; Radwanski and Last, 1995). After entering the plastids, the transit peptides are cleaved and the mature subunits are assembled into a holoenzyme (Poulsen et al., 1993; Bohlmann et al., 1995; Romero and Roberts, 1996). The β-subunit is an aminotransferase that cleaves Gln to produce ammonia, which is utilized by the α-subunit to convert chorismate to anthranilate. The α-subunit binds Trp as a feedback inhibitor, and can use free ammonia as an alternative substrate in vitro (Crawford, 1989; Bohlmann et al., 1995; Radwanski and Last, 1995). Biosynthesis of Trp and other related secondary products in plants is tightly controlled, not only through Trp feedback inhibition of the AS enzyme, but also by regulation of the abundance of AS mRNAs (Radwanski and Last, 1995).
A tobacco (Nicotiana tabacum) suspension culture selected for resistance to the Trp analog 5-methyl-Trp (5MT) was shown to contain Trp feedback-insensitive AS enzyme activity and high levels of free Trp (Brotherton et al., 1986). A cDNA encoding a naturally occurring AS α-subunit (ASA2) was isolated from a 5MT-resistant cell line (Song et al., 1998), and when expressed in Escherichia coli (Song et al., 1998) or in a transgenic forage legume, Astragalus sinicus (Cho et al., 2000), the AS activity was found to be less sensitive to Trp inhibition. In tobacco, ASA2 mRNA can be detected in the 5MT-resistant cultures, but not in the wild type or plants regenerated from the 5MT-resistant cultures (Song et al., 1998).
Chloroplast transformation (Boynton et al., 1988; Daniell et al., 1990; Svab et al., 1990) is becoming an increasingly important supplement to nuclear transformation (Maliga et al., 1993; Bogorad, 2000). We have been exploring the genetic modification of biosynthetic pathways such as that for Trp through chloroplast transformation for its advantages of specific gene targeting, the high expression potential of the plastids, and possible use as a selectable marker. In this study, we placed the native nuclear ASA2 gene in the plastid genome to obtain high expression by using the plastid transcription and translation machinery. By doing so, we hypothesized that variations of transgene expression that often accompany nuclear transformation and are caused by “position effects” or gene silencing (Bogorad, 2000; Heifetz, 2000), particularly when using an endogenous gene as a transgene, might be avoided; that the tight regulation of ASA2 transcription in the nucleus might be bypassed in plastids; and that the processes of transcription and translation in the nucleus and cytosol followed by transportation of the ASA2 protein to plastids would be circumvented. We also attempted to examine what effects a plastid-encoded ASA2 gene might have on the overall expression of endogenous AS genes in general and β-subunit genes in particular, and whether the biosynthesis of the end-product Trp could be altered by expressing only the α-subunit of the first enzyme AS of the pathway in plastids. Our overall goal was to explore the feasibility of targeting a native nuclear gene of pre-endosymbiotic origin, instead of a foreign one, to the plastid genome to modify a plastid biosynthesis pathway. Here we report the creation and analysis of ASA2 transplastomic plants for which the ASA2 transgene was independently targeted into two separate regions of the plastid genome. Our studies demonstrate that it is possible to engineer the plastid genome and to obtain plants that uniformly produce significantly higher levels of free Trp than does the wild type, which may be applicable for the biosynthesis of other important amino acids and compounds, as well.
RESULTS
Plastid Transformation Vectors
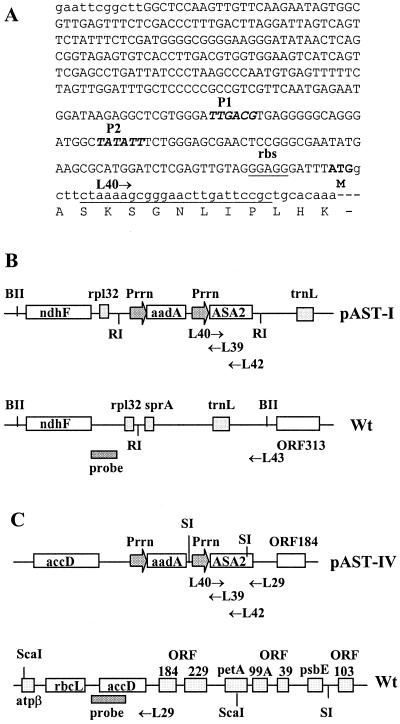
The aadA gene, which confers resistance to spectinomycin, had the Chlamydomonas rbcL 3′-untranslated region (500 bp) as the termination sequence and was driven by a chloroplast 16S rRNA promoter Prrn (Goldschmidt-Clermont, 1991; Eibl et al., 1999). The modified ASA2 gene was 1,671 bp long, encoding 556 amino acids without a putative transit peptide and containing 204 bp of 3′-non-coding region. As shown in Figure 1A, the synthetic promoter Prrn (332 bp) contains a 22-bp sequence from the tobacco rbcL 5′-non-coding region immediately upstream of the start codon, including a ribosomal-binding site GGAGG to ensure efficient mRNA transcription and translation initiation. The 3′-untranslated region of the plastid rpL32 or the intergenic region between accD and open reading frame (ORF) 184 were used as additional termination sequences for pAST-I and pAST-IV, respectively. Figure 1, B and C, shows that the Prrn-ASA2 gene was flanked by the ndhF and trnL genes for pAST-I or accD and ORF184 genes for pAST-IV, along with each of their adjacent sequences. These flanking sequences acted as anchoring regions to initiate site-specific gene targeting in the plastid genome and homologous recombination during plastid transformation.
Figure 1.
Structures of the plastid transformation vectors pAST-I and pAST-IV. A, Sequence of the modified plastid 16S rRNA operon promoter (Prrn) with a ribosomal-binding site (rbs) fused to a translation start codon ATG to initiate the coding region of the mature AS α-subunit gene (ASA2). P1 and P2 (bold italic) are transcription regulatory sequences similar to the prokaryotic −30/−10 upstream promoter elements. Only the first 16 amino acids of the predicted ASA2 protein sequence are shown. The location and direction of primer L40 is underlined. B and C, Schematic structures of pAST-I, pAST-IV, and their corresponding regions of the wild-type tobacco plastid genome. The restriction enzymes reported in this paper are mapped. RI, EcoRI; BII, BglII; and SI, SacI. Also shown are the positions and directions of primers used and the location of the DNA fragments (thick black line) as probes for Southern hybridizations. The figure size is not to scale.
It is known that the nuclear and plastid genomes have a unique base composition and codon usage (Oliver et al., 1990). It is possible that the nuclear ASA2 sequence may not be totally compatible with the plastid protein synthesis machinery and may therefore compromise the optimal protein production in plastids. However, comparison of the codon usage between ASA2 and several plastid genes does not indicate any severe bias against the ASA2 codons (data not shown).
Site-Specific Integration of the Transgene into the Plastid Genome
Four to 8 weeks after bombardment, spectinomycin-resistant green shoots or calli appeared from the bleached, enlarged white leaf pieces. Twenty independent resistant shoots/calli were recovered from 79 leaves bombarded with vector pAST-I, four of which contained the correct integration, as determined by the presence of the PCR fragments with expected sizes. Three individual resistant shoots were obtained from 60 leaves bombarded with vector pAST-IV, all of which contained the correct insertion. After three rounds of additional selection with spectinomycin, the green shoots were allowed to grow into rooted plantlets.
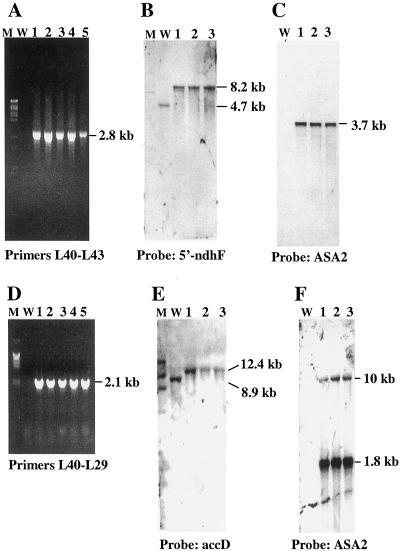
Figure 2, A and D, demonstrates confirmation of transgene integration by PCR. DNA was isolated from individual plants from four lines for pAST-I and three lines for pAST-IV. With primers L40 and L43, PCR amplified a 2.8-kb fragment only in the pAST-I-transformed tissues due to the insertion of the ASA2 gene in the region between rpL32 and ORF313 (Fig. 2A). Likewise, there could be no 2.1-kb PCR product with primers L40/L29 in the pAST-IV plants unless the ASA2 gene was inserted into the region between accD and ORF184 (Fig. 2D). No fragments were produced in the wild type since primer L40 is located in the ASA2 coding region. Therefore, the PCR strategy effectively identified the transplastomic cell lines with the expected, site-specific integration while eliminating the unwanted lines with the wrong insertion, deletion, or abnormal sequences due to recombination during the selection process. The plantlets were transferred to rooting medium to obtain whole plants.
Figure 2.
Plastid genomic analysis of site-specific integration of the ASA2 gene. A and D, PCR amplification with ASA2-specific primer L40 and plastid site-specific primers L43 or L29. Plants from every line with pAST-I (A) and pAST-IV (D) were tested (lanes 4 and 5 were from the same line in A; lanes 1 and 3 and lanes 4 and 5 were from the same line in D). B and E, DNA blots with BglII (B) or ScaI (E) hybridized to plastid site-specific probes. C and F, DNA blots with EcoRI (C) or SacI (F) hybridized to ASA2 probe. Southern-blot hybridizations show the analysis of three lines each from both transformations. W, Wild-type; lanes 1 through 5, individual transplastomic plants; M, DNA markers (Roche, Indianapolis).
The primary transplastomic tissues or plantlets were not homoplastomic based on the presence of the wild-type plastid DNA fragment by Southern-blot hybridization (data not shown). Thus, the leaf sections from the resistant tissues/plantlets were subjected to three more cycles of selection on spectinomycin until no trace of wild-type DNA was detected in the regenerated plantlets (homoplastomy). Figure 2 shows representative Southern-blot hybridizations of three lines from two independent transformation experiments. When hybridized to a 0.7-kb probe of 5′-ndhF, BglII digestion produces a 4,656-bp fragment for the wild-type chloroplast genome and an 8.2-kb band for the pAST-I-transformed chloroplast genome in T3 selfed homoplastomic plants due to the insertion of aadA and ASA2 genes that do not have a BglII site (Fig. 1B). Figure 2B shows, as expected, a 4.7-kb band in the wild-type plant and only an 8.2-kb band in the transplastomic plants, indicating that all copies of the plastid genome have the transgene incorporated in the region between ndhF and trnL. Likewise, ScaI digestion results in an 8,933-bp fragment containing accD to ORF229 in the wild-type plant, but generates a 12.4-kb band in the pAST-IV plants, with the 1.2-kb accD fragment as probe (Fig. 1C). Figure 2E shows a single 8.9- or 12.4-kb band in the wild-type and the transplastomic plants, respectively. To further prove the site of the transgene integration into the plastid genome, the cellular DNA was isolated from the pAST-I and pAST-IV leaves. PCR was carried out using gene-specific primers, one of which is located outside the transformation vectors (L43 in ORF313 for pAST-I and L36 in rbcL for pAST-IV). Sequencing of the amplified fragments confirms that the transgenes were inserted into the expected regions by homologous recombination. These results demonstrate that the regenerated plants have achieved homoplastomy and that the plastid genomes of these individual lines transformed with the same vector are identical to each other. It is expected that the individual transplastomic lines obtained by using the same vector would be identical to each other because the mechanism of plastid transformation is by site-specific insertion and homologous recombination (Maliga et al., 1993), unlike nuclear transformation where random (or semi-random) integration occurs.
Hybridizing an EcoRI-digested DNA blot with a labeled ASA2 fragment reveals a 3.7-kb band in all the transformed plants as expected, because EcoRI releases a fragment containing the aadA (1.4 kb) and ASA2 (2.3 kb) genes (Fig. 1B). Due to the relatively low amount of total cellular DNA (approximately 5 μg) used, no hybridizing band was detected in the wild-type plant under these conditions, as shown in Figure 2C, even though the tobacco nuclear genome contains a small AS α-gene family (Song et al., 1998). Figure 2F shows that when hybridized to the ASA2 probe, the SacI-digested DNA blot reveals two bands at 1.8 and 10 kb for the pAST-IV plants, a result of three SacI sites in the Prrn-ASA2 gene and downstream of ORF229 (Fig. 1C). The 1.8-kb band is much stronger because the AS probe covers most of the fragment. No signal was detected in the wild-type plant. These results demonstrate that the ASA2 gene is incorporated into the plastid genome in our transformed plants and that the insertions are identical to each other in the four pAST-I lines and in the three pAST-IV lines.
ASA2 mRNA Transcription in Transplastomic Plants
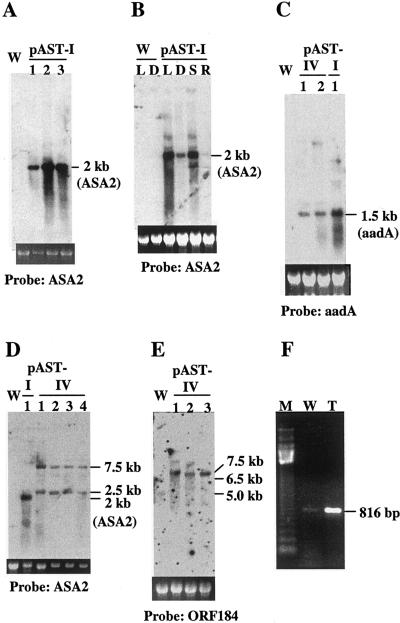
Northern-blot hybridizations were carried out to determine the expression of the Prrn-ASA2 gene in the transformed plants. No detectable ASA2 mRNA can be found in the wild-type leaves, even when the RNA was overloaded (Fig. 3, A, B, and D). However, the transplastomic plants showed a 2-kb band for the pAST-I plants (Fig. 3A) and 2.5- and 7.5-kb bands for the pAST-IV plants (Fig. 3D). This indicates a much higher level of transcription of the ASA2 transgene in the transformed plants than that of the native ASA2 gene in the wild type, which may be in part due to the high copy number of the plastid genome in leaf cells (Bendich, 1987). In contrast to the nuclear transgenic A. sinicus plants, which show a wide range of expression level of the ASA2 transgene (Cho et al., 2000), these transplastomic plants expressed a similar level of the ASA2 mRNA among individual plants and between different generations (Fig. 3 and data not shown). The small variation seen in Figure 3, A and D, can be explained by loading differences as shown by the amount of 25S rRNA in each lane.
Figure 3.
ASA2 and ORF184 transcription in wild-type and transplastomic plants. A and B, ASA2 mRNA levels in pAST-I plants. C, The same size of the aadA mRNA in pAST-I and pAST-IV plants. D exhibits two different sizes of mRNAs in pAST-IV plants. E demonstrates cotranscription of ASA2 and ORF184 in pAST-IV plants. F represents reverse transcription with primer L42 and PCR amplified with L40/L39 for wild-type (W) or pAST-IV plants (T). M, 123-bp DNA ladder (Gibco-BRL, Gaithersburg, MD). Probes used for RNA-blot hybridizations are indicated. L and D, Shoots of 2-week-old seedlings grown under normal light condition (L; see “Materials and Methods”) and in the dark for 7 d (D); S and R, shoots and roots of 2-week-old seedlings, respectively. The ethidium bromide-stained 25S rRNA is shown to indicate RNA loading.
We also found that seedlings grown under the light contained more ASA2 mRNA in the shoots than those grown in the dark, and the green shoots contained more ASA2 mRNA than did the roots (Fig. 3B). Since the promoter for the ASA2 transgene is a constitutive 16S rRNA promoter Prrn (Svab and Maliga, 1993), light or organ specificity is not expected. It has been shown that the copy number of plastid DNA per cell in tobacco roots is only 10% to 23% of that in leaves (for review, see Cannon et al., 1985). The level of plastid DNA is 2.5- to 2.8-fold higher in tobacco suspension cultures grown in the light than in those grown in the dark (Cannon et al., 1985). Therefore, the higher expression of the ASA2 gene found in green shoots grown under light is possibly due to the presence of more copies of the plastid genome in the green leaves that contain more plastids per cell and/or more DNA content per plastid than the leaves in the dark or in the roots (Zhang et al., 2001). There could also be an effect of light on the overall physiological condition of the plants since seedlings grown in the dark for 7 d had much less ASA2 mRNA (Fig. 3B).
Cotranscription of ASA2 and ORF184 in the pAST-IV Plants
It was unexpected that the ASA2 mRNA from the pAST-IV plants not only is larger in size than that from the pAST-I plants, but also occurs in two major forms of approximately 2.5 and 7.5 kb, as shown in Figure 3D. The 2.5-kb transcripts in the pAST-IV plants end in the intergenic region between accD and ORF184, whereas the 7.5-kb mRNA apparently includes sequences from other downstream genes, as shown below. Figure 3C shows that pAST-I and pAST-IV plants produced the same size aadA mRNA. However, hybridization to an ORF184 probe revealed two faint mRNA bands at approximately 5 and 6.5 kb for the wild type and a strong single 7.5-kb band with at least 10 times more intensity in the pAST-IV plants, which is the same size as the longer ASA2 mRNA (Fig. 3E). This suggests that the pAST-IV plants express a 7.5-kb mRNA in which the ASA2 gene is a part of the transcribed operon that apparently includes seven putative genes (ORF184, ORF229, petA, ORF99A, ORF39, psbE, and ORF103; Fig. 1C). This is consistent with the observation that the wild-type tobacco produced two large mRNAs (approximately 5 and 6.5 kb) that hybridized to the ORF184 probe (Fig. 3E). To our knowledge, there is no published information on the expression of the ORF184 gene, which encodes a hypothetical and yet- to-be-identified 21-kD protein. It is well known, however, that several genes are often transcribed as a long mRNA in plastids (Stern et al., 1997; Bogorad, 2000).
ASA2 mRNA Transcription in Wild-Type Tobacco Plants
ASA2 mRNA could not be detected by northern-blot analysis in the leaf, root, stem, or seed of wild-type tobacco (Song et al., 1998; Fig. 3, A, B, and D). Therefore, reverse transcription (RT)-PCR was used to determine if the ASA2 gene was expressed at all in the wild-type plant. Figure 3F shows that an 816-bp fragment was amplified by RT-PCR from the wild-type leaves much less than from leaves of the pAST-IV plant. The PCR-amplified DNA was cloned and sequenced to confirm that it was ASA2 cDNA. The genomic gene encoding the ASA2 mRNA was also cloned from the wild-type plant and was partially sequenced (data not shown). Therefore, the naturally occurring ASA2 gene appears to be expressed at a very low level or in a small population of leaf cells in the wild-type plants, reflecting tight regulation at the transcription level for this isoform of the first enzyme in the Trp biosynthetic pathway. RT-PCR experiments did not detect the expression of any other AS α-subunit gene, possibly because of wide sequence divergence.
ASA2 Protein and Enzyme Activity
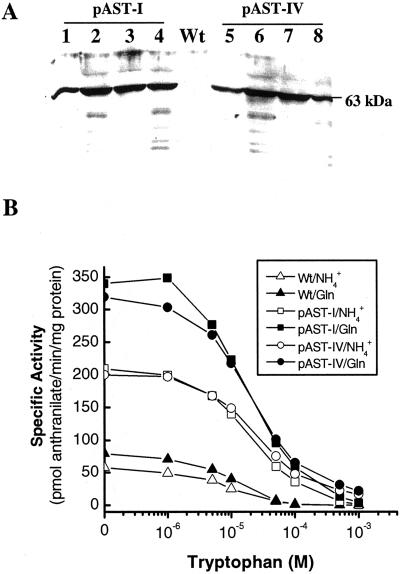
Western-blot analysis easily detected a 63-kD band, the predicted size of the mature AS α-subunit, in extracts from the transplastomic plants, but not from the wild-type plant, using polyclonal antibodies against E. coli-expressed ASA2 protein (Fig. 4A). The cellular AS enzyme activities, when measured using NH4Cl or Gln as the amino donor, were more than 4-fold higher in pAST-I and pAST-IV plants than in the wild type, as shown in Figure 4B. The ratio of NH4+-dependent activity to Gln-dependent activity was 0.60 for pAST-I and pAST-IV plants, which is similar to the wild type (0.69). These results suggest that no excess of free functional α-subunits were present in the transformed chloroplasts because the α-subunit alone can utilize NH4Cl in vitro. The AS enzyme activity from the transformed plants was less sensitive to Trp inhibition than that from the wild type. For example, at 100 μm Trp, the wild-type AS activity was almost completely inhibited, whereas the pAST-I and pAST-IV plants still retained about 20% of the total activity (Fig. 4B). The sensitivity to Trp can also be demonstrated by the apparent Ki value, i.e. the Trp concentration causing 50% inhibition of AS activity. With NH4Cl as a substrate, the estimated apparent Ki values were 8.2, 18.5, and 20.5 μm for the wild-type, pAST-I, and pAST-IV plants, respectively. When Gln was used as a substrate, the Ki values were 10.5, 19.5, and 24 μm, respectively. The greater insensitivity to Trp feedback inhibition observed in the AS from the transformed plants could lead to the increase in 5MT tolerance and Trp accumulation described below.
Figure 4.
AS protein analysis of transplastomic plants. A, Western-blot analysis of leaf soluble proteins (10 μg per lane). Wt, Wild type; lanes 1 through 4 and lanes 5 through 8, four individual pAST-I and AST-IV plants, respectively. B, Trp inhibition of AS enzyme activity in wild-type and transplastomic plants, with 100 mm NH4Cl or 10 mm Gln as substrate.
Tolerance to 5MT
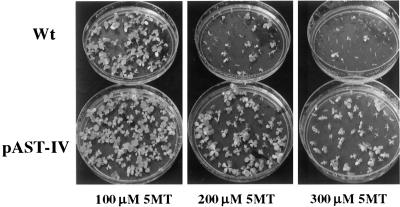
The Trp analog 5MT inhibits AS enzyme activity and plant growth. Overexpression of the ASA2 gene results in resistance to 5MT in E. coli (Song et al., 1998) and transgenic A. sinicus (Cho et al., 2000). When the wild-type and transplastomic tobacco seeds were germinated on medium with different concentrations of 5MT, pASA-I (data not shown) and pAST-IV seeds were more tolerant to high levels of 5MT (Fig. 5). At 300 μm 5MT, most of the wild-type seedlings were dead by 25 d, whereas the pAST-IV plants survived for more than 50 d. Since the plants cannot incorporate 5MT into proteins (Sasse et al., 1983), the continued accumulation of 5MT in plant cells increasingly disrupts Trp biosynthesis and eventually becomes lethal. We also tested the 5MT tolerance of suspension cultures initiated from the wild-type and transplastomic plants. The wild-type cultures did not grow in 5MT concentrations over 50 μm, whereas the transplastomic cells grew vigorously in up to 300 μm 5MT.
Figure 5.
Effect of 5MT on the germination and growth of seedlings. Seeds from wild-type or pAST-IV plants were grown on Murashige and Skoog medium containing 100, 200, or 300 μm 5MT for 30 d in 10-cm diameter petri dishes.
Maternal Inheritance
The expression of ASA2 and/or ASA2-ORF184 genes in the chloroplasts appears to have no negative effect on plant growth, fertility, or morphology. With about 150 seeds tested for each sample, all the self-pollinated seeds of transplastomic plants germinated on Murashige and Skoog-spectinomycin (500 mg L−1) medium into green, normal-looking plants, whereas the wild-type seeds emerged as small white seedlings that stopped growing (data not shown). Pollination of the wild-type plants with pollen from transplastomic plants produced progeny (approximately 100 seeds tested) sensitive to spectinomycin, whereas pollination of the transplastomic plants with the wild-type pollen produced seeds that grew into green plants on the spectinomycin medium (data not shown). Thus, spectinomycin resistance is maternally inherited, as expected for the plastids in tobacco (Svab and Maliga, 1993; Birky, 1995). The 5MT tolerance of seedlings is also transmitted maternally in the pAST-I or pAST-IV plants, showing that the ASA2 and the aadA genes are transmitted together and only from the female plant.
Trp Accumulation
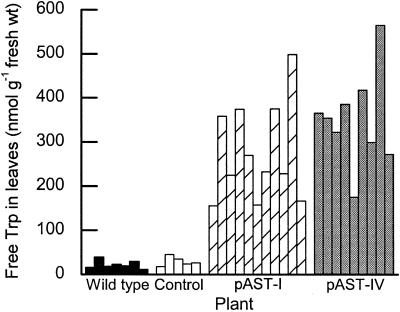
Young, expanding leaves from the same location on different plants of the same age and condition and ones grown in the same growth chamber were analyzed for free Trp content. Seven wild-type and five control plants transformed only with the aadA gene had similar Trp concentrations, averaging 22 and 30 nmol g−1 fresh weight, respectively (Fig. 6). This indicates that incorporation of the aadA gene alone in the plastid genome did not affect the leaf Trp level. The free Trp level in the wild-type tobacco leaves is comparable with that found in other plants, although the reported values have a broad range. For example, the levels vary from 15 to 18 nmol g−1 fresh weight for Arabidopsis (Li and Last, 1996), 33 for rice (Wakasa et al., 1999), 30 to 53 for maize (Anderson et al., 1997), 210 for A. sinicus (Cho et al., 2000), and 224 for Datura innoxia (Ranch et al., 1983).
Figure 6.
Free Trp content in individual third leaves from the tip of wild-type, control (aadA only), and transplastomic plants (pAST-I and pAST-IV).
Introduction and expression of the ASA2 gene in plastids dramatically increases the free Trp content in green leaves. Figure 6 shows that measurements on leaves of 11 pAST-I and nine pAST-IV plants exhibited a consistently high Trp level, averaging 296 and 350 nmol g−1 fresh weight, respectively, a 10-fold increase over the wild type. The variation in the Trp level between different transplastomic plants (Fig. 6) largely reflected the fitness of the plants, particularly the leaves, in the growth chamber and is considerably smaller than that reported previously for nuclear transgenic plants where the variation could be as high as 10-fold between different lines containing the same transgene. For example, maize plants transformed with 35S-ASA2 contained from 97 to 500 nmol Trp g−1 fresh weight in nine lines (Anderson et al., 1997), transgenic rice lines with a mutant rice AS α-subunit gene contained from 143 to 1,522 nmol Trp g−1 fresh weight (Wakasa et al., 1999), and the range of free Trp levels in the transformed A. sinicus hairy root lines was 42 to 316 nmol g−1 fresh weight (Cho et al., 2000). This demonstrates that the genetic identity of plastid transformants, due to identical insertions, results in a uniform transgene expression pattern in the progeny. In contrast, nuclear transformants almost always exhibit a wide range of variation in transgene incorporation and gene expression.
We did not find a significant difference in the free Trp level in mature, air-dried seeds between the wild-type (262 ± 9 nmol g−1 for the seeds) and the transplastomic plants (255 ± 17 nmol g−1 for pAST-I and 274 ± 9 nmol g−1 for pAST-IV). However, the total Trp concentration (free plus protein-bound) in NaOH-digested seeds was 7.75 ± 0.5 μmol g−1 for the seeds and 9.99 ± 0.7 μmol g−1 and 9.81 ± 0.3 μmol g−1 for the wild-type, pAST-I, and pAST-IV, respectively, representing a 27% to 29% increase for the transformants based on dry weight. Furthermore, the Trp content of the total seed soluble protein was 0.9%, 1.3%, and 1.4% for the wild type, pAST-I, and pAST-IV, respectively. This suggests that an increase in the free Trp pool in leaves leads to an increase in the level of protein-bound Trp in seeds, possibly resulting from enhanced synthesis of Trp-containing storage proteins in seeds.
DISCUSSION
We report here the successful integration of an endogenous, nuclear, Trp feedback-insensitive AS2A coding sequence into the tobacco chloroplast genome at two independent locations. The transplastomic plants contained a high level of ASA2 mRNA, increased accumulation of the AS α-subunit protein, and a 4-fold increase in AS enzyme activity that is less sensitive to inhibition by Trp. There was a 10-fold increase in free Trp concentration in the leaves, and a small increase in protein-bound Trp in the seeds of transformed plants without obvious phenotypic effects. Our studies demonstrate, for the first time, that it is feasible to introduce and express a native (or probably homologous as well), nuclear-encoded Trp biosynthetic control protein in chloroplasts where a functional AS enzyme can be formed.
We observed a 4-fold increase in AS activity that apparently represents an increase in AS holoenzyme activity because the ratios of NH4+-to-Gln activities in the transplastomic and wild-type plants are similar, indicating no significant excess of unassembled functional α-subunit in the plastids. A higher AS activity would be expected if the ASA2 gene product is more catalytically effective than the α-subunit encoded by other members of the native gene family expressed in wild-type cells. However, immunological probing detected a much higher level of α-subunit protein in the transplastomic plants as compared with the wild type, suggesting that the abundance of α-subunits encoded by the plastidic ASA2 may stabilize the available β-subunits, resulting in more holoenzyme. It seems less likely that the plastidic transgene is up-regulating the expression of the nuclear, native β-subunit gene, but this is the type of question that can be addressed in the future using this system when more information on the β-subunit becomes available. In a converse manner, the limiting availability of β-subunits may affect the stability of the abundant supply of α-subunits produced by the many copies of the transgene in the transplastomic plants. We speculate that introducing AS α- and β-coding genes into the plastid genome would result in a further increase in AS activity and Trp production.
The modest increase in the apparent Ki for Trp is surprising since the ASA2 gene encodes a feedback-insensitive subunit (Brotherton et al., 1986; Song et al., 1998). A similar modest increase in Ki was observed by Cho et al. (2000) when the tobacco ASA2 gene was introduced into the nuclear genome of the legume A. sinicus, where ineffective interaction between the subunits produced by the transgene and native gene might occur. However, in this case, the kinetic evidence and the ratio of NH4+-to-Gln activities also indicated that there was α- and β-subunit interaction to form a catalytically active holoenzyme. It is clear that in the case of tobacco, there would probably not be a problem with subunit interaction since the ASA2 gene is from the same plant. These results point out the potential of this system to study the mechanisms that regulate the biosynthesis, assembly, and stability of subunits of plastidial heteromeric enzymes.
In the pAST-IV plants, we unexpectedly discovered that not only is the introduced ASA2 gene highly expressed, but so are the downstream ORF184 and other putative genes. Possible reasons for the long ASA2 mRNA may be the lack of a plastid termination sequence in the Prrn-ASA2 gene and the insertion of the ASA2 gene near other putative protein coding genes, in contrast to the ASA2 gene in pAST-I, which has a long 3′-non-coding region. This may explain why a single 2-kb ASA2 mRNA was detected in the pAST-I plants, but two species (2.5 and 7.5 kb) were found in the pAST-IV plants. These results demonstrate the potential of simultaneously introducing and expressing several transgenes as an operon, or targeting a transgene to affect the transcription of neighboring genes in the genome through plastid transformation.
Although plastid transformation should be possible with many species, fertile transformed plants have only been obtained with tobacco (Bogorad, 2000). Thus far, a few foreign genes have been expressed that provide, for example, resistance to insects (McBride et al., 1995; Kota et al., 1999) or a herbicide (Daniell et al., 1998) or that allow production of a human therapeutic protein (Staub et al., 2000). Our studies demonstrate, to our knowledge, the first example of introducing a native nuclear gene of presumed pre-endosymbiotic origin into plastids to modify an endogenous biosynthetic pathway. Our results show the potential effectiveness of targeting biosynthetic pathways to the plastids by directly introducing the coding gene, instead of importing the catalyzing enzyme, as previously shown in targeting the polyhydroxybutyrate biosynthetic pathway to the plastids via nuclear transformation of Arabidopsis (Nawrath et al., 1994).
Nuclear transformation with a native or highly similar transgene may also result in variable levels of expression (“position effect,” Bogorad, 2000; Cho et al., 2000) or no expression at all (gene silencing; X.-H. Zhang, unpublished data). As an example, the Agrobacterium rhizogenes-transformed A. sinicus hairy roots with the tobacco ASA2 gene show highly variable ASA2 mRNA and free Trp levels in different lines (Cho et al., 2000). The variability associated with nuclear transformation has not been encountered in plastid transformation, at least at the transcriptional level (Heifetz, 2000; X.-H. Zhang, unpublished data). This may be in part due to the fact that in plastid transformation, the transgene is integrated into the same sites by homologous recombination, resulting in a plant with an identical plastid genome and thus eliminating variable position effects. There also appears to be no gene silencing in the plastid.
Since the expression of genes involved in many biosynthetic pathways is tightly regulated in the nucleus/cytosol and engineering a high level of the expression in the cytosol may have deleterious effects on plants (Nawrath et al., 1994), introducing the key genes into the plastid genome could avoid some of these problems as well. Plastids (chloroplasts) are the site for photosynthesis and the biosynthesis of numerous important metabolites such as amino acids, fatty acids, and phytohormones. As a consequence, hundreds of nuclear-encoded proteins are imported into the plastids, and thus are potential candidates of interest for plastid genetic engineering. This approach could also be used to convey to plastids the ability to synthesize some metabolites of interest (such as Met; Ravanel et al., 1998; Chiba et al., 1999) that are normally made in the cytosol. Furthermore, the identity and functionality of many putative plastid-encoded genes still remains to be revealed. Therefore, chloroplast transformation technology is a valuable supplement to nuclear transformation and to genomics in the investigation of gene function and nucleus-organelle interaction, as well as in the exploration of potential commercial applications where a high level of gene expression is needed.
MATERIALS AND METHODS
Construction of Plastid Transformation Vectors pAST-I and pAST-IV
The putative transit peptide coding sequence was deleted from the ASA2 cDNA and a translation start codon was introduced by PCR strategy. Oligomer A, 5′-CTCGAGTTGTAGGGAGGGATTTATGGCTTCTAAAAGCGGGAA-3′, contains an XhoI site (underlined), a ribosomal-binding site (double underlined), and codons for seven amino acids, including a start codon (bold) that initiates the putative mature AS α-subunit peptide. Oligomer B, 5′-GTAGCGACCAACACTAGAACCTCG-3′, is located in the coding sequence of the mature peptide. The PCR product (193 bp) was ligated to an XhoI-digested fragment of a modified tobacco (Nicotiana tabacum cv Petit Havana SR1) plastid 16S rRNA operon promoter, Prrn (Fig. 1A), and fused to the HindIII fragment of the ASA2 cDNA to obtain a Prrn-ASA2 gene cassette, which was confirmed by sequencing. The tobacco chloroplast expression vector, pAST-I, was constructed by ligating an HincII-cut fragment (3.7 kb) of the plasmid pFaadAII (Eibl et al., 1999) to the SmaI fragment (7.5 kb) of the Prrn-mature ASA2 clone (Fig. 1B). An aminoglycoside 3′-adenyltransferase (aadA) gene in the vector confers resistance to spectinomycin (Goldschmidt-Clermont, 1991). During chloroplast transformation, the aadA-ASA2 gene cassette is expected to be integrated into the region between the tobacco plastid ndhF gene and a gene for tRNALeu by homologous recombination (Fig. 1B).
The plasmid pFaadAII was later found to have inserted into a gene encoding a small plastid RNA in tobacco (sprA; Vera and Sugiura, 1994), the deletion of which has no effect on rRNA maturation or phenotype (Sugita et al., 1997). To avoid any gene disruption, a second vector, pAST-IV, was constructed using a presumably neutral region in the plastid genome as the targeting sequence. The aadA-Prrn-ASA2 gene cassette was inserted into a KpnI site between the accD and ORF184 genes of the tobacco plastid genome (Fig. 1C). As a control, a vector containing only the aadA gene was also used for transformation.
Transformation and Regeneration of Transplastomic Plants
Transformation of tobacco leaves was carried out as described by Svab and Maliga (1993) using 0.6-μm gold particles (Bio-Rad, Hercules, CA). Green calli and shoots resistant to spectinomycin dihydrochloride (500 mg L−1) were subjected to three additional rounds of selection on RMOP-spectinomycin medium (Svab et al., 1990). After PCR testing and Southern-blot analysis, the homoplastomic plantlets were transferred to Murashige and Skoog rooting medium containing spectinomycin (Murashige and Skoog, 1962) and then to potting soil and were grown to maturity in a growth chamber at 20°C (night) and 25°C (day) under 400 to 500 μE m−2 s−1 white incandescent light (16 h daily). Seeds were harvested and tested on Murashige and Skoog-spectinomycin (500 mg L−1) medium. All transplastomic seedlings were green and control progeny were white.
PCR, DNA, and RNA Gel-Blot Hybridization Analyses of Putative Transplastomic Plants
Total cellular DNA and RNA were extracted using Qiagen Kits (Qiagen, Valencia, CA). PCR was carried out to identify the transgene insertion, with Taq DNA polymerase (Gibco-BRL) for 30 cycles at 94°C for 45 s, 55°C for 45 s, and 72°C for 2.5 min. For pAST-I plants, primers L40 and L43 were used to amplify a 2.8-kb fragment containing the ASA2 gene that is integrated into the specific region of the plastid genome. L40, 5′-CTAAAAGCGGGAACTTGAT-TCCGC-3′, is located at the beginning of the putative mature ASA2 coding region, and L43, 5′-GGAAATCGG-GAATTGAATTCA-3′, is downstream of the trnL gene and outside the flanking region of the transformation vector (Fig. 1B). For pAST-IV plants, another primer, L29 (5′-TCATATTTCTGCGGGCATAAGAGT-3′), located upstream of the plastid gene ORF184 (Fig. 1C), was used along with primer L40, and a 2.1-kb fragment was expected. DNA and RNA blots were hybridized in ULTRAhyb Solution (Ambion, Austin, TX) to digoxigenin-11-dUTP-labeled probe, using a DIG High Prime Labeling and Detection Kit (Roche). The hybridizing signals were quantified with an Ultroscan XL Laser Densitometer (LKB Instruments, Gaithersburg, MD).
To verify the site of the transgene insertion into the plastid genome, PCR was carried out with the cellular DNA isolated from the pAST-I and pAST-IV plants as templates. Gene-specific primers were used. One of the primers was inside the transgene cassette (L40 for pAST-I and D4 for pAST-IV). Primer D4, 5′-GGAGAATCTCG-CTCTCTCCAGG-3′, is located in the aadA coding region. The other primer was outside the flanking region of the transgene vector (L43 for pAST-I and L36 for pAST-IV). Primer L36, 5′-AGTAAAAACAGTAGACATTAG-3′, is located in the rbcL 3′-non-coding region. The amplified fragments were sequenced.
cDNA Synthesis and Amplification by RT-PCR
RT-PCR was employed using DNase-treated total leaf RNA (2 μg) isolated from the wild-type or the transplastomic (pAST-IV) plants as templates. The first-strand cDNA was synthesized by using SuperScript II RNase H− Reverse Transcriptase (Gibco-BRL) and an ASA2-specific primer L42 (5′-GACTCGGCCAAGTCAATGGCTCG-3′; Fig. 1, B and C). The cDNA was then PCR amplified with primers L40 (see above) and L39 (5′-TCTGTACACTTCA-AATGGGTCAGC-3′) located in the middle of the ASA2 coding region (Fig. 1, B and C). The amplified fragment (816 bp) was cloned into a pCRII-TOPO vector (Invitrogen, Carlsbad, CA) and sequenced to confirm its identity. To isolate the corresponding genes, PCR was carried out with genomic DNA as template and L40 and L39 as primers.
AS Assays and Trp Measurement
Leaf proteins were extracted as previously described (Zhang et al., 2001). Equal amounts of protein were subjected to SDS-PAGE and western analysis. Mouse polyclonal antiserum against Eshcerichia coli-expressed tobacco ASA2 protein (Song et al., 1998) was made at the Immunological Resource Center at the University of Illinois. AS enzyme activity was measured as described by Cho et al. (2000) in crude protein extracted from leaves using the buffer of Bernasconi et al. (1994) desalted on an Econo-Pac 10DG column (Bio-Rad). Free Trp was extracted with 0.1 n HCl (Cho et al., 2000) from a 3.14-cm2 leaf circle (approximately 50–70 mg fresh weight) of the third or fourth leaves from the top of the plants grown in the growth chamber. For total Trp measurement, the air-dried seeds were ground and digested in 5 n NaOH at 110°C for 16 h. Trp analysis was carried out as described by Cho et al. (2000). Seed soluble proteins were extracted as described by Zhang et al. (2001), except for the addition of 1% (w/v) SDS. At least three independent measurements were done for each sample.
Tissue Cultures
Suspension cultures of the wild-type and the transplastomic plants were initiated by germinating seeds in solid Murashige and Skoog medium containing 1.8 μm 2,4-dichlorophenoxyacetic acid with or without 500 mg L−1 spectinomycin. The calli formed were transferred to liquid Murashige and Skoog medium with 2,4-dichlorophen-oxyacetic acid (same as above) and 5MT (100–300 μm) and cultured as described in Zhang et al. (2001).
ACKNOWLEDGMENTS
We thank Hongjian Liang (University of Illinois, Urbana) for help with the tissue culture work, Hans-Ulrich Koop (Ludwig-Maximilians Universität, Munich, Germany) for the pFaadAII plasmid, John E. Boynton (Duke University, Durham, NC) for the aadA clone, and Henry Daniell (University of Central Florida, Orlando) for advice on plastid transformation.
Footnotes
This work was supported by the Illinois Soybean Program Operating Board, by the Illinois Agricultural Experiment Station, and by the U.S. Department of Agriculture.
LITERATURE CITED
- Anderson PC, Chomet PS, Griffor MC, Kriz AL, inventors. July 24, 1997. Anthranilate synthase gene and its use thereof. World Intellectual Property Organization Patent Application No. 97/26366
- Bendich AJ. Why do chloroplasts and mitochondria contain so many copies of their genome? Bioessays. 1987;6:279–282. doi: 10.1002/bies.950060608. [DOI] [PubMed] [Google Scholar]
- Bernasconi P, Walters EW, Woodworth AR, Siehl DL, Stone TE, Subramanian MV. Functional expression of Arabidopsis thaliana anthranilate synthase subunit I in Escherichia coli. Plant Physiol. 1994;106:353–358. doi: 10.1104/pp.106.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky CW., Jr Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc Natl Acad Sci USA. 1995;92:11331–11338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogorad L. Engineering chloroplasts: an alternative site for foreign genes, proteins, reactions and products. Trends Biotechnol. 2000;18:257–263. doi: 10.1016/s0167-7799(00)01444-x. [DOI] [PubMed] [Google Scholar]
- Bohlmann J, De Luca V, Eilert U, Martin W. Purification and cDNA cloning of anthranilate synthase from Ruta graveolens: modes of expression and properties of native and recombinant enzymes. Plant J. 1995;7:491–501. doi: 10.1046/j.1365-313x.1995.7030491.x. [DOI] [PubMed] [Google Scholar]
- Boynton JE, Gillham NW, Harris EH, Hosler JP, Johnson AM, Jones AR, Randolph-Anderson BL, Robertson D, Klein TM, Shark KB. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988;240:1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- Brotherton JE, Hauptmann RM, Widholm JM. Anthranilate synthase forms in plants and cultured cells of Nicotiana tabacum L. Planta. 1986;168:214–221. doi: 10.1007/BF00402966. [DOI] [PubMed] [Google Scholar]
- Cannon G, Heinhorst S, Siedlecki J, Weissbach A. Chloroplast DNA synthesis in light and dark grown cultured Nicotiana tabacum cells as determined by molecular hybridization. Plant Cell Rep. 1985;4:41–45. doi: 10.1007/BF00269202. [DOI] [PubMed] [Google Scholar]
- Chiba Y, Ishikawa M, Kijima F, Tyson RH, Kim J, Yamamoto A, Nambara E, Leustek T, Wallsgrove RM, Naito S. Evidence for autoregulation of cystathionine γ-synthase mRNA stability in Arabidopsis. Science. 1999;286:1371–1374. doi: 10.1126/science.286.5443.1371. [DOI] [PubMed] [Google Scholar]
- Cho H-J, Brotherton JE, Song H-S, Widholm JM. Increasing tryptophan synthesis in a forage legume Astragalus sinicus by expressing the tobacco feedback-insensitive anthranilate synthase (ASA2) gene. Plant Physiol. 2000;123:1069–1076. doi: 10.1104/pp.123.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford IP. Evolution of a biosynthetic pathway: the tryptophan paradigm. Annu Rev Microbiol. 1989;43:567–600. doi: 10.1146/annurev.mi.43.100189.003031. [DOI] [PubMed] [Google Scholar]
- Daniell H, Datta R, Varma S, Gray S, Lee S-B. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat Biotechnol. 1998;16:345–348. doi: 10.1038/nbt0498-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Vivekananda J, Nielsen BL, Ye GN, Tewari KK, Sanford JC. Transient foreign gene expression in chloroplasts of cultured tobacco cells after biolistic delivery of chloroplast vectors. Proc Natl Acad Sci USA. 1990;87:88–92. doi: 10.1073/pnas.87.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibl C, Zou Z, Beck A, Kim M, Mullet J, Koop H-U. In vivo analysis of plastid psbA, rbcL and rpl32 UTR elements by chloroplast transformation: tobacco plastid gene expression is controlled by modulation of transcript levels and translation efficiency. Plant J. 1999;19:333–345. doi: 10.1046/j.1365-313x.1999.00543.x. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker for site-directed transformation of Chlamydomonas. Nucleic Acids Res. 1991;19:4083–4089. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam E. Shikimic Acid: Metabolism and Metabolites. Chichester, UK: John Wiley & Sons; 1993. [Google Scholar]
- Heifetz PB. Genetic engineering of the chloroplast. Biochimie. 2000;82:655–666. doi: 10.1016/s0300-9084(00)00608-8. [DOI] [PubMed] [Google Scholar]
- Herrmann K. The shikimate pathway as an entry to aromatic secondary metabolism. Plant Physiol. 1995;107:7–12. doi: 10.1104/pp.107.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota M, Daniell H, Varma S, Garczynski SF, Gould F, Moar WJ. Overexpression of the Bacillus thuringiensis (Bt) Cry2Aa2 protein in chloroplasts confers resistance to plants against susceptible and Bt-resistant insects. Proc Natl Acad Sci USA. 1999;96:1840–1845. doi: 10.1073/pnas.96.5.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Last RL. The Arabidopsis thaliana trp5 mutant has a feedback-resistant anthranilate synthase and elevated soluble tryptophan. Plant Physiol. 1996;110:512–559. doi: 10.1104/pp.110.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga P, Carrer H, Kanevski I, Staub J, Svab Z. Plastid engineering in land plants: a conservative genome is open to change. Philos Trans R Soc Lond B Biol Sci. 1993;342:203–208. doi: 10.1098/rstb.1993.0148. [DOI] [PubMed] [Google Scholar]
- Martin W, Herrmann RG. Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KE, Svab Z, Schaaf DJ, Hogan PS, Stalker DM, Maliga P. Amplification of a chimeric Bacillus gene in chloroplasts leads to an extraordinary level of an insecticidal protein in tobacco. Biotechnology. 1995;13:362–365. doi: 10.1038/nbt0495-362. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassay with tobacco cultures. Physiol Plant. 1962;15:473–479. [Google Scholar]
- Nawrath C, Poirier Y, Somerville C. Targeting of the polyhydroxybutyrate biosynthetic pathway to the plastids of Arabidopsis thaliana results in high levels of polymer accumulation. Proc Natl Acad Sci USA. 1994;91:12760–12764. doi: 10.1073/pnas.91.26.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver JL, Marin A, Martinez-Zapater JM. Chloroplast genes transferred to the nuclear plant genome have adjusted to nuclear base composition and codon usage. Nucleic Acids Res. 1990;18:65–69. doi: 10.1093/nar/18.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen C, Bongaerts RJM, Verpoorte R. Purification and characterization of anthranilate synthase from Catharanthus roseus. Eur J Biochem. 1993;212:431–440. doi: 10.1111/j.1432-1033.1993.tb17679.x. [DOI] [PubMed] [Google Scholar]
- Radwanski ER, Last RL. Tryptophan biosynthesis and metabolism: biochemical and molecular genetics. Plant Cell. 1995;7:921–934. doi: 10.1105/tpc.7.7.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranch JP, Rick S, Brotherton JE, Widholm JM. Expression of 5-methyltryptophan resistance in plants regenerated from resistant cell lines of Datura innoxia. Plant Physiol. 1983;71:136–140. doi: 10.1104/pp.71.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel S, Gakière B, Job D, Douce R. The specific features of methionine synthesis and metabolism in plants. Proc Natl Acad Sci USA. 1998;95:7805–7812. doi: 10.1073/pnas.95.13.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero RM, Roberts MF. Anthranilate synthase from Ailanthus altissima cell suspension cultures. Phytochemistry. 1996;41:395–402. [Google Scholar]
- Romero RM, Roberts MF, Phillipson JD. Anthranilate synthase in microorganisms and plants. Phytochemistry. 1995;39:263–276. doi: 10.1016/0031-9422(95)00010-5. [DOI] [PubMed] [Google Scholar]
- Sasse F, Buchholz M, Berlin J. Site of action of growth inhibitory tryptophan analogues in Catharanthus roseus cell suspension cultures. Z Naturforsch. 1983;38:910–915. [Google Scholar]
- Song H-S, Brotherton JE, Gonzales RA, Widholm JM. Tissue culture-specific expression of a naturally occurring tobacco feedback-insensitive anthranilate synthase. Plant Physiol. 1998;117:533–543. doi: 10.1104/pp.117.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub JM, Garcia B, Graves J, Hajdukiewicz PTJ, Hunter P, Nehra N, Paradkar V, Schlittler M, Carroll JA, Spatola L. High-yield production of a human therapeutic protein in tobacco chloroplasts. Nat Biotechnol. 2000;18:333–338. doi: 10.1038/73796. [DOI] [PubMed] [Google Scholar]
- Stern DB, Higgs DC, Yang J. Transcription and translation in chloroplasts. Trends Plant Sci. 1997;2:308–315. [Google Scholar]
- Sugita M, Svab Z, Maliga P, Sugiura M. Targeted deletion of sprA from the tobacco plastid genome indicates that the encoded small RNA is not essential for pre-16S rRNA maturation in plastids. Mol Gen Genet. 1997;257:23–27. doi: 10.1007/s004380050619. [DOI] [PubMed] [Google Scholar]
- Svab Z, Hajdukiewicz P, Maliga P. Stable transformation of plastids in higher plants. Proc Natl Acad Sci USA. 1990;87:8526–8530. doi: 10.1073/pnas.87.21.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera A, Sugiura M. A novel RNA gene in the tobacco plastid genome: its possible role in the maturation of 16S rRNA. EMBO J. 1994;13:2211–2217. doi: 10.1002/j.1460-2075.1994.tb06498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasa K, Tozawa Y, Terakawa T, Hasegawa H, inventors. March 11, 1999. Gene encoding α-subunit of rice anthranilate synthase and DNA relating thereto. World Intellectual. Property Organization Patent Application No. 99/11800
- Zhang X-H, Widholm JM, Portis AR., Jr Photosynthetic properties of two different soybean suspension cultures. J Plant Physiol. 2001;158:357–365. [Google Scholar]