Abstract
In plants, 14-3-3 proteins are key regulators of primary metabolism and membrane transport. Although the current dogma states that 14-3-3 isoforms are not very specific with regard to target proteins, recent data suggest that the specificity may be high. Therefore, identification and characterization of all 14-3-3 (GF14) isoforms in the model plant Arabidopsis are important. Using the information now available from The Arabidopsis Information Resource, we found three new GF14 genes. The potential expression of these three genes, and of two additional novel GF14 genes (Rosenquist et al., 2000), in leaves, roots, and flowers was examined using reverse transcriptase-polymerase chain reaction and cDNA library polymerase chain reaction screening. Under normal growth conditions, two of these genes were found to be transcribed. These genes were named grf11and grf12, and the corresponding new 14-3-3 isoforms were named GF14omicron and GF14iota, respectively. The gene coding for GF14omicron was expressed in leaves, roots, and flowers, whereas the gene coding for GF14iota was only expressed in flowers. Gene structures and relationships between all members of the GF14 gene family were deduced from data available through The Arabidopsis Information Resource. The data clearly support the theory that two 14-3-3 genes were present when eudicotyledons diverged from monocotyledons. In total, there are 15 14-3-3 genes (grfs 1–15) in Arabidopsis, of which 12 (grfs 1–12) now have been shown to be expressed.
14-3-3 proteins were discovered by Moore and Perez (1967) as major soluble proteins in brain tissue. Since then it has become clear that 14-3-3 proteins are present in all eukaryotic organisms (for review, see Rosenquist et al., 2000) and have a wide range of functions (for review, see Finnie et al., 1999; Fu et al., 2000). 14-3-3s typically function as dimers and bind to particular phosphorylated motifs in other proteins, thereby regulating the activity of the respective target protein. In 1992, the first four plant 14-3-3 isoforms were reported: one in Arabidopsis (Lu et al., 1992), one each in Spinacia oleracea and Oenothera hookeri (Hirsch et al., 1992), and one in Hordeum vulgare (Brandt et al., 1992). The Arabidopsis isoform was identified as part of a protein/G box complex and therefore named “G box factor 14-3-3,” in short “GF14.” This designation has been kept for the Arabidopsis isoforms, and the four additional 14-3-3 isoforms reported by Lu et al. (1994) were named GF14chi, phi, psi, and upsilon, and the original isoform received the designation GF14omega. The same year, Jarillo et al. (1994) reported two “rare cold-inducible” 14-3-3s from Arabidopsis, RCI1 and RCI2. The RCI1 was identical to GF14psi of Lu et al. (1994), whereas RCI2 was a new isoform, very closely related to GF14lambda (Wu et al., 1997) and until now regarded as identical. An additional four 14-3-3s, GF14epsilon, kappa, mu, and nu, were identified in 1997 (Wu et al., 1997). After that, the members of the 14-3-3 gene family in Arabidopsis were regarded to be 10.
Although the first Arabidopsis 14-3-3 isoform, GF14omega, was discovered as a constituent of a protein/G box complex and implicated to be involved in regulation of gene transcription (Lu et al., 1992), subsequent research also has shown plant 14-3-3s to have a broad range of functions (for review, see Finnie et al., 1999). In particular, plant 14-3-3s have been found to be important regulators of primary metabolism. For example, nitrate reductase and Suc phosphate synthase, which are key enzymes in nitrogen and carbon metabolism, respectively, are both inhibited by binding of 14-3-3 (Bachmann et al., 1996; Moorhead et al., 1996; Toroser et al., 1998), whereas binding of 14-3-3 to the plasma membrane H+ATPase activates H+ pumping (Jahn et al., 1997; Oecking et al., 1997) and hence stimulates nutrient uptake.
In a previous study, using surface plasmon resonance, we reported strong differences in binding affinity between nine GF14 isoforms and a known target, the phosphorylated C terminus of the Arabidopsis plasma membrane H+ATPase isoform 2, AHA2 (Rosenquist et al., 2000). This suggests that one reason for the large number of 14-3-3 isoforms in multicellular organisms, such as plants, is functional specificity. Thus, the regulatory role of 14-3-3s in carbon and nitrogen metabolism, in nutrient uptake, etc, may be executed by different 14-3-3 isoforms. To determine whether this is the case, it is important to first identify and then have access to all isoforms expressed. The completion of the sequencing of the Arabidopsis genome has enabled determination of gene numbers within specific gene families. Using the information now available via Internet, we recently identified two novel GF14 genes (Rosenquist et al., 2000). After further data mining of the Arabidopsis genome, we have now identified three additional GF14 genes, bringing the total number of GF14 genes to 15. Using reverse transcriptase (RT)-PCR and cDNA library PCR screening, we now demonstrate that two of the five novel genes are expressed, bringing the total number of expressed 14-3-3 isoforms in Arabidopsis to 12.
RESULTS AND DISCUSSION
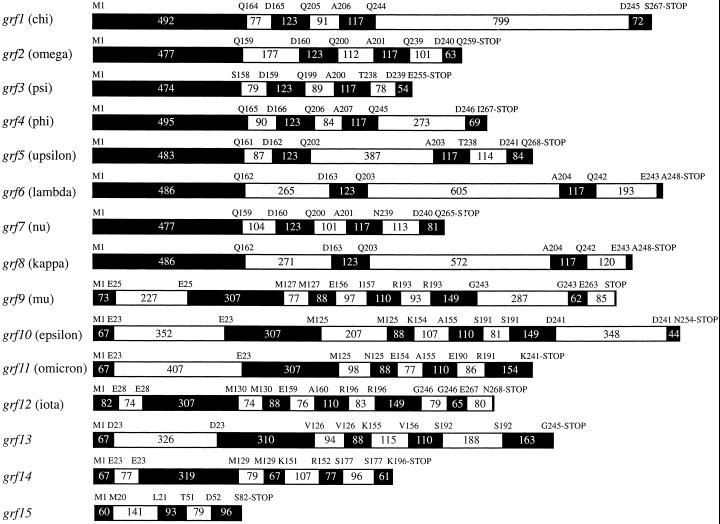
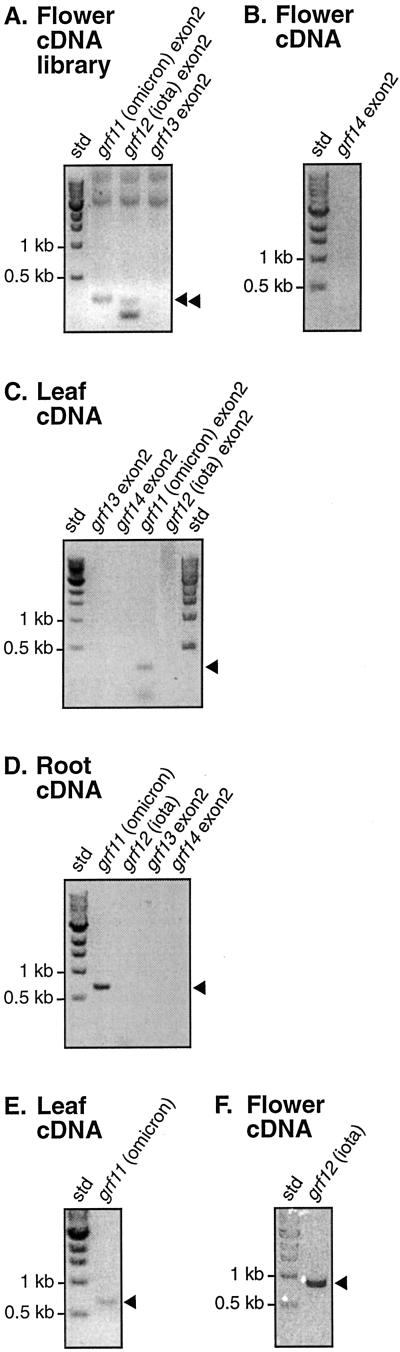
Examining the Arabidopsis genome revealed three additional novel GF14 genes, grf12, grf13, and grf14, in addition to the two recently reported by Rosenquist et al. (2000), grf11 and grf15 (Fig. 1). One of the two previously reported novel genes, grf15 (Table I), only codes for approximately 30% of the length of a GF14 protein (Fig. 1). Furthermore, the 82 amino acids the gene encodes correspond to the C terminus of a functional 14-3-3, and show only 44% identity with its closest relatives, GF14psi and upsilon (data not shown). Grf15 and its putative product were therefore excluded from further analysis. To determine if the four other novel genes are expressed or not, RT-PCR and cDNA library PCR screening were performed. Thus, primers designed for the novel 14-3-3 genes, grfs 11 through 14, found in BAC clones F21H2, T1K7, T16E15, and T11I11 (Table I), were used to PCR screen a cDNA library made from developing flowers, as well as for RT-PCR on mRNA prepared from leaves, roots, and flowers, respectively. Only two of these four novel GF14 genes, grf11 and grf12, were found to be expressed (Fig. 2). No corresponding cDNAs or ESTs were found in the database, which suggests that these mRNA species are present in low abundancy. The two novel isoforms were named GF14omicron, encoded by grf11 (F21H2.3), and GF14iota, encoded by grf12 (T1K7.15; Table I), in accordance with the designations used earlier, using Greek letters for the proteins and Arabic numbers for the genes. Both GF14omicron and GF14iota contain the expected conserved regions found in all other 14-3-3s (Rosenquist et al., 2000), and most divergence is found in the N and C termini (Fig. 3). The annotation of grf12 (T1K7.15) in the Arabidopsis sequencing project did not correspond to the sequence we obtained for the cDNA, suggesting that the predicted splicing in the database is incorrect.
Figure 1.
Gene maps of all Arabidopsis 14-3-3 (GF14) genes. Exons are indicated as black boxes and introns as white boxes. Exon and intron sizes are indicated with the number of bases within in each box. The names of the resulting proteins are within parentheses. The genes can be divided into two groups based on exon patterns: grfs 1 through 8, and grfs 9 through 14, excluding the severely truncated grf15 that does not readily fit into either group. The five novel genes discussed in this paper are grf11, grf12, grf13, grf14, and grf15, of which grf11 and grf15 were already reported by Rosenquist et al. (2000). The abbreviation grf stands for G box regulating factor.
Table I.
Summary of available database information on Arabidopsis 14-3-3 (GF14) genes and proteins
| GF14 Isoform | BAC | Gene | mRNA Accession No. | BAC Accession No. | Protein Accession No. |
|---|---|---|---|---|---|
| Chromosome 1 | |||||
| Putative 14-3-3 | T11I11 | grf13a (T11I11.16) | – | AC012680 | AAG52105b |
| Similar to epsilon | T16E15 | grf14 (T16E15.9c) | – | AC068562 | AAF87262d |
| Iota | T1K7 | grf12 (T1K7.15e) | AF335544 | AC013427 | AAF98570 |
| Omicron | F21H2 | grf11 (F21H2.3) | AF323920 | AC007894 | AAG47840 |
| Epsilon | T16E15 | grf10 (T16E15.8) | U36446 | AC068562 | P48347 |
| Omega | F3F9 | grf2 (F3F9.16) | M96855 | AC013430 | Q01525 |
| Phi | T32G9 | grf4 (T32G9.30) | L09111 | AC079605 | P46077 |
| Chromosome 2 | |||||
| Mu | F14N22 | grf9 (F14N22.14) | U60444 | AC007087 | AAD51784 |
| 14-3-3 Like | F12P23 | grf15 (F12P23.4) | – | AC007264 | AAD28654 |
| Chromosome 3 | |||||
| Nu | F16B3 | grf7 (F16B3.15) | U60445 | AC021640 | AAD51782 |
| Chromosome 4 | |||||
| Chi | Contig. 25 (F23J3) | grf1 | L09112 | AL161513 | P42643 |
| Chromosome 5 | |||||
| Upsilon | F1N13 | grf5 (F1N13_190) | L09109 | AL391145 | AAB62225 |
| Psif | P1 Clone MXI10 + MBB18 | grf3 (MXI10.21) | L09110 | AB005248+AB005231 | P42644 |
| Kappa | P1 Clone MNA5 | grf8 (MNA5.16) | U36447 | AB011479 | AAD51783 |
| Lambda (or AFT1) | F12B17 | grf6 | U68545 | AL353995 | P48349 |
| 14-3-3-Like protein AFT1g | F12B17 | grf6 (F12B17_200) | – | AL353995 | CAB89398 |
| RCI2h | – | RCIIB | X74141 | – | CAA52238 |
The chromosomes and bacteria artificial chromosome (BAC) clones the different genes are found on are indicated, as well as the accession no. of the BAC clone, the gene no. in the BAC, and the accession no. of the mRNA. Accession nos. to putative and expressed gene products are also indicated.
The abbreviation grf stands for G-box regulating factor.
Incorrect annotation resulted in a shortened N terminus (first exon and 5′ part of second exon were not included).
T16E15.9 is located 1,329 bp from grf10 (T16E15.8) in the same BAC clone.
A single nucleotide was counted as a fifth intron when sequence was annotated in the original entry.
Splicing suggested in database is incorrect.
The isoform GF14psi is identical to the entry RCI1 (protein accession no. S47969).
Putative product due to alternative splicing.
An additional thymine at the end of exon 3 of grf6 causes a frame shift resulting in this putative isoform, which is probably the result of erroneous sequence analysis.
Figure 2.
Detection of novel 14-3-3 gene transcripts in flower (A, B, and F), leaf (C and E), and root (D) tissues by RT-PCR and cDNA library PCR. Primary RT-PCR reactions were performed with primers corresponding to exon 2 (see Fig. 1) of grf11 (omicron; A and C), grf12 (iota; A and C), grf13 (A, C, and D), and grf14 (B, C, and D). Subsequent RT-PCR reactions were performed with full-length primers for grf11 on leaf (E) and root (D) cDNA, and for grf12 on flower (F) and root (D) cDNA. Products were run on 1.5% (w/v) agarose gel (inverted image) and bands of interest are indicated with black arrowheads. The identities of these bands were confirmed by sequencing. Sizes of standard (std) markers are indicated to the left. Note that the band grf12 (iota) exon2 (A) only corresponds to 290 bp of the exon due to optimization of primer positions, and therefore is 17 bp shorter than grf11 (omicron) exon2.
Figure 3.
DNA sequences of the two expressed novel Arabidopsis 14-3-3 (GF14) genes, grf11 and grf12, and corresponding amino acid sequences for the proteins, GF14omicron and GF14iota, respectively. Conserved identity between all expressed GF14s is indicated with gray boxes, and conserved homology is indicated with white boxes.
The GF14omicron gene grf11 was expressed in leaves, roots, and flowers, whereas the GF14iota gene grf12 was only expressed in flowers (Fig. 2). The expression of these isoforms in flowers, and particularly the exclusive expression of GF14iota in this tissue, is interesting because when examining binding of the 10 previously identified GF14s to the phosphorylated C terminus of the flower-specific Arabidopsis plasma membrane H+ ATPase isoform 9, AHA9, using surface plasmon resonance, all 10 GF14s were classified as nonbinding (data not shown). The C terminus of AHA9 differs from the one in AHA2 (which showed different degrees of binding to the nine GF14 isoforms tested; Rosenquist et al., 2000) on three out of six amino acid positions, and the 14-3-3 binding motifs of AHA9 and AHA2 thus are sufficiently different to suggest that they are activated by different GF14s. Therefore, GF14omicron and particularly GF14iota are good candidates as regulators of AHA9.
The 14-3-3 genes grf13 (T11I11.16; Table I) and grf14 (T16E15.9; Table I) were not expressed in any of the examined tissues. The coding sequence of gene grf14 contains a single nucleotide insertion in exon 4 (which we have confirmed by additional sequencing [data not shown]) causing a frame shift, which creates a stop codon and loss of the last 45 amino acids (Fig. 1). Given the fact that seven of the 17 conserved target-binding amino acid residues, as well as the nuclear export signal (Rittinger et al., 1999), are located within the missing area, grf14 does not likely produce a functional 14-3-3 isoform, although possible expression cannot be ruled out. The fact that no expression of the gene grf13 was observed is intriguing. The amino acid sequence it encodes is the most divergent of all GF14s, suggesting that if it is expressed it might have a very specific function. Therefore, our failure to detect transcription may be due to a rather specific expression of this isoform, either regarding developmental stage, tissue, or stress condition. We have refrained from naming the gene products of grf13 and grf14 as no expression has been shown so far.
When examining the 10 previously characterized grfs, we found that the gene coding for GF14psi, grf3 (MXI10.21), is divided between P1clone MXI10 and MBB18 (Table I). A striking discovery was that the annotation in the Arabidopsis genome project of BAC clone F12B17 corresponds to a putative protein, “14-3-3-like protein AFT1,” and not to the expressed GF14lambda of Wu et al. (1997). Thus, an alternative splicing of the pre-mRNA of the GF14lambda coding gene, grf6, is suggested, resulting in the putative GF14 protein “14-3-3-like protein AFT1” (CAB89398) with a completely different C terminus from the previously reported GF14lambda (P48349). The C terminus of the putative “14-3-3-like protein AFT1” is 25 amino acids longer than the C terminus of GF14lambda. Our attempts to detect mRNA corresponding to “14-3-3-like protein AFT1” have been negative, suggesting that the gene grf6 codes for no other product than GF14lambda, and that the predicted splicing in the database is incorrect. There is, in fact, no experimental evidence to date that alternative splicing of 14-3-3 genes exists. Thus, the annotation in the database of the grf6 gene (F12B17_200) in the BAC F12B17 should be changed to correspond to GF14lambda. The RCIIB gene reported by Jarillo et al. (1994) results in a protein, RCI2 (CAA52238), that only differs from GF14lambda in the last eight amino acids. The RCIIB gene cannot be found among the data from the Arabidopsis genome project, but is due to an additional thymine at the end of exon 3 in grf6 (Fig. 1). Because this is most likely the result of erroneous sequence analysis, RCIIB has been excluded from the present analysis.
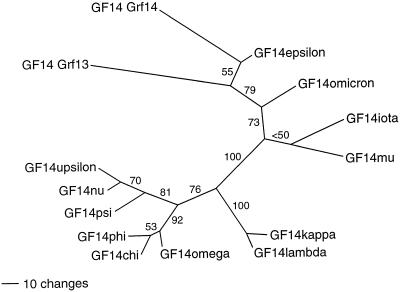
Examining the gene maps of all GF14 genes, one can observe two major groups based on exon patterns; grfs 1 through 8 form one group, where all genes are expressed, and grfs 9 through 14 form a second group (Fig. 1). Unrooted phylogenetic trees based on either the resulting mRNA or amino acid sequences (including the putative products of grf13 and grf14) show that the gene products of grfs 1 through 8 form three branches in one part of the tree, and that the products of grfs 9 through 14 form the other, more divergent, part of the tree (Fig. 4; mRNA data not shown). It is intriguing that a branch-wise specificity was found when the affinity between nine of the GF14s and the phosphorylated C terminus of AHA2 was measured (Rosenquist et al., 2000).
Figure 4.
A phylogenetic tree with topology representative for the Arabidopsis 14-3-3 (GF14) protein family. The putative gene products of grf13 and grf14 are designated Grf13 and Grf14, respectively. A heuristic search using the maximum parsimony method was done on the alignment of GF14 protein sequences using the PAUP 4.0b4 software (Sinauer Associates, Inc. Publishers, Sunderland, MA), with gaps treated as missing data. Bootstrap values for 12,000 replicates are indicated on each branch. The hypothetical 14-3-3-like protein coded by grf15 (Table I; Fig. 1) was excluded from the analysis due to strong deviance in both sequence and length. The 82 amino acids encoded by grf15 show highest homology (44% identity) to GF14psi and upsilon, and correspond to the C terminus of a functional 14-3-3 (data not shown).
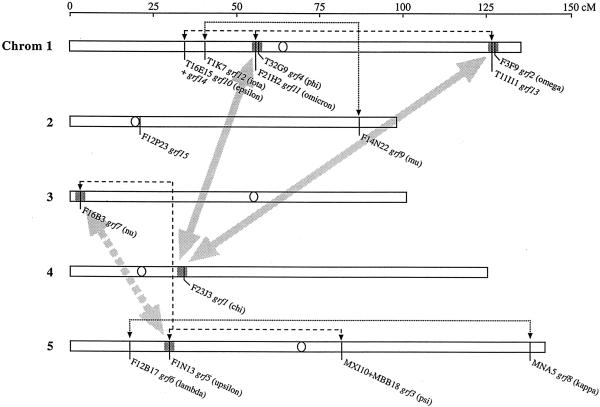
In Figure 5, the locations of the different BAC clones containing GF14 genes are indicated on the chromosomes, and two major duplication events 170 and 50 million years ago (Vision et al., 2000), which may be responsible for the formation of some of the isoforms, are indicated. The genes grf5 (upsilon) and grf7 (nu) are located in regions of chromosome 5 and 3, respectively, that were duplicated 170 million years ago. However, considering the very close relation between GF14 upsilon and nu (Fig. 4), it is more likely that the duplication leading to the formation of grf5 (upsilon) and grf7 (nu) was a much more recent event. The gene grf2 (omega) and grf13 are situated close together on chromosome 1, and this is also the case for grf4 (phi) and grf11 (omicron). These two gene pairs, as well as grf1 (chi) on chromosome 4, are located within areas that were duplicated 50 million years ago. It is notable that no other GF14 gene is located close to grf1 (chi), suggesting that a gene related to grf11 (omicron) and grf13 has been lost. The fact that GF14phi, chi, and omega are much more closely related than GF14Grf13 (the putative product of grf13) is to omicron (Fig. 4) is noteworthy because they all seem to originate from the major duplication event 50 million years ago (Fig. 5). Considering their common evolutionary history, one would expect GF14Grf13 and omicron to show about the same high homology as GF14phi, chi, and omega (Fig. 4). As proteins involved in complex formation, 14-3-3s are subject to a high selection pressure, which acts to conserve these proteins. Thus, if GF14phi, chi, and omega have retained their functional specificity this will have limited their evolution. In a converse manner, if GF14omicron and the putative gene product of grf13 (GF14Grf13) have obtained different functions this may explain their low homology, or if grf13 is not expressed at all, it has of course evolved without any constraints.
Figure 5.
Arabidopsis chromosome map. BAC clones harboring 14-3-3 (GF14) genes are indicated with black lines. Gene names, with corresponding gene products written in parentheses, are indicated by each BAC. Centromeres are indicated with white circles. The gray dashed arrow indicates a major duplication event that took place 170 million years ago, and the two gray arrows indicate another major duplication event that took place 50 million years ago (Vision et al., 2000). Thin dashed and dotted lines connect relatively closely related genes and hence suggest other duplication events. The gene grf14 (T16E15.9) is located 1,329 bases from grf10 (T16E15.8) within the same BAC clone. Given the low homology of the severely truncated 14-3-3 gene grf15 in F12P23, no connection to other genes is indicated.
Previous phylogenetic analyses of plant 14-3-3 isoforms indicate that at the time of the split into eudicotyledons and monocotyledons, about 200 million years ago, two 14-3-3 isoforms were already present (Wang and Shakes, 1996; Rosenquist et al., 2000). This is in accordance with both the gene map in Figure 1 and the phylogenetic tree in Figure 4. The gene map (Fig. 1) divides the Arabidopsis 14-3-3 genes into two major groups; grfs 1 through 8, and grfs 9 through 14, respectively. The phylogenetic tree (Fig. 4) similarly suggests two major branches: the GF14 upsilon/nu/psi, phi/chi/omega, and lambda/kappa branches forming a major branch in the lower part of the tree, and the GF14 mu, iota, omicron, epsilon, Grf14, and Grf13 forming an upper major branch. One of the gene pairs, grf2 (omega)/grf13, grf4 (phi)/grf11 (omicron), or grf1 (chi) and its putative aborted neighbor, is probably the direct descendant of the two ancestral 14-3-3 genes (Fig. 5).
The completion of the sequencing of the Arabidopsis genome finally enabled the determination of the total number of 14-3-3 genes to be 15. The challenge now is to examine the expression pattern of all GF14 isoforms, to identify new target proteins, and to determine the functional specificity of all GF14 isoforms, in order to reveal all the cellular processes involving regulatory 14-3-3 proteins in the model plant Arabidopsis.
MATERIALS AND METHODS
Examination of Database Entries
GF14 database entries were obtained by a simple keyword search at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). A blast search of the Arabidopsis BAC clones at The Arabidopsis Information Resource (http://www.Arabidopsis.org) revealed additional isoforms, and allowed mapping of the genes.
Computational Analysis of GF14 Gene Sequences
DNA sequences corresponding to GF14 genes were obtained from the GenBank database via the National Center for Biotechnology Information. Clustal W multiple alignments of sequences were performed using the MacVector 7.0 software (Oxford Molecular Group Plc, Oxford). The alignments were carried out using the Blossum series matrix, with an open gap penalty of 10 and an extend-gap penalty of 0.05. Alignments were examined and adjusted manually. A heuristic search using the maximum parsimony method was done on the alignment using the PAUP 4.0b4a software (Sinauer Associates, Inc. Publishers), with gaps treated as missing data. The tree bisection reconnection branch-swapping algorithm was used. Tree building and calculation of Bootstrap values, with 12,000 replicates, were also carried out using the PAUP 4.0b4a software. Gene maps were drawn from splicing patterns indicated in the respective BAC entries. The splicing patterns of grf13 (T11I11.16) and grf14 (T16E15.9) suggested in the database were incorrect, and hence adjusted.
Plant Material
Arabidopsis ecotype Columbia-0 was grown on soil at 22°C with a 9-h-light/15-h-dark photoperiod (170 μE) and 70% relative humidity. Root cultures were grown in darkness in flasks with 100 mL of Murashige and Skoog medium, pH 5.7, supplemented with 3% (w/v) Suc (Malamy and Benfey, 1997). Plants for flower RNA preparation were grown under a long-day light regime (16 h light).
mRNA Isolation and RT-PCR
Leaves were harvested after 4 weeks and total RNA was prepared with a conventional phenol/chloroform extraction. Subsequent mRNA isolation was carried out according to the manufacturer's protocol using the QIAGEN Oligotex mRNA Purification Kit (Merck KGaA, Darmstadt, Germany). First-strand synthesis was performed with the First Strand cDNA Synthesis Kit (Boehringer Mannheim, Basel). The same procedures were used on roots harvested after 10 weeks of cultivation. Total RNA was extracted according to the method developed by Chang et al. (1993) from a mix of flowers in all stages of anthesis.
Primary RT-PCR was carried out with Taq polymerase (Roche, Basel) and primers designed against the largest exon of the newly identified GF14 genes (see Fig. 1), as well as for an alternatively spliced grf6. Fragments obtained were run on a 1.5% (w/v) agarose gel and sequenced for identification. RT-PCR subsequently was done with primers corresponding to the full-length genes. Full-length fragments were cloned into the TA vector pGEM (Promega Corp., Madison, WI), and sequenced. Restriction sites were added to full-length primers to facilitate cloning into suitable vectors.
cDNA Library PCR Screening
A cDNA library from Arabidopsis ecotype Landsberg erecta developing flowers (Weigel et al., 1992) was obtained from the Arabidopsis Biological Resource Center. Conventional in vivo excision on the entire library was performed on infected Escherichia coli XL1-Blue with E. coli SOLR (AH Diagnostics, Stockholm). PCR with exon-specific primers was performed on the excised library, for detection purpose only. Full-length clones were amplified from cDNA synthesized from the flower mRNA preparation from Arabidopsis (Columbia-0) described above.
ACKNOWLEDGMENT
We thank Dr. Ove Nilsson (Swedish Agricultural University, Umeå, Sweden) for preparation of total RNA from Arabidopsis flowers.
Footnotes
This work was supported by the Swedish Foundation for Strategic Research (to C.L. and M.S.), by the Swedish Natural Science Research Council (to C.L. and M.S.), by the Kungliga Fysiografiska Sällskapet (to M.R.), and by the Royal Swedish Academy of Sciences (to M.R.).
LITERATURE CITED
- Bachmann M, Huber JL, Athwal GS, Wu K, Ferl RJ, Huber SC. 14-3-3 proteins associate with the regulatory phosphorylation site of spinach leaf nitrate reductase in an isoform-specific manner and reduce dephosphorylation of Ser-543 by endogenous protein phosphatases. FEBS Lett. 1996;398:26–30. doi: 10.1016/s0014-5793(96)01188-x. [DOI] [PubMed] [Google Scholar]
- Brandt J, Thordal-Christensen H, Vad K, Gregersen PL, Collinge DB. A pathogen-induced gene of barley encodes a protein showing high similarity to a protein kinase regulator. Plant J. 1992;2:815–820. [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. [Google Scholar]
- Finnie C, Borch J, Collinge DB. 14-3-3 proteins: eukaryotic regulatory proteins with many functions. Plant Mol Biol. 1999;40:545–554. doi: 10.1023/a:1006211014713. [DOI] [PubMed] [Google Scholar]
- Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Hirsch S, Aitken A, Bertsch U, Soll J. A plant homologue to mammalian brain 14-3-3 protein and protein kinase C inhibitor. FEBS Lett. 1992;296:222–224. doi: 10.1016/0014-5793(92)80384-s. [DOI] [PubMed] [Google Scholar]
- Jahn T, Fuglsang AT, Olsson A, Brüntrup IM, Collinge DB, Volkmann D, Sommarin M, Palmgren MG, Larsson C. The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell. 1997;9:1805–1814. doi: 10.1105/tpc.9.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo JA, Capel J, Leyva A, Martinez-Zapater JM, Salinas J. Two related low-temperature-inducible genes of Arabidopsis encode proteins showing high homology to 14-3-3 proteins, a family of putative kinase regulators. Plant Mol Biol. 1994;25:693–704. doi: 10.1007/BF00029607. [DOI] [PubMed] [Google Scholar]
- Lu G, DeLisle AJ, de Vetten NC, Ferl RJ. Brain proteins in plants: an Arabidopsis homolog to neurotransmitter pathway activators is part of a DNA binding complex. Proc Natl Acad Sci USA. 1992;89:11490–11494. doi: 10.1073/pnas.89.23.11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Rooney MF, Wu K, Ferl RJ. Five cDNAs encoding Arabidopsis GF14 proteins. Plant Physiol. 1994;105:1459–1460. doi: 10.1104/pp.105.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124:33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- Moore B, Perez V. Specific acidic proteins of the nervous system. In: Carlson FD, editor. Physiological and Biochemical Aspects of Nervous Integration. Englewood Cliffs, NJ: Prentice Hall; 1967. pp. 343–359. [Google Scholar]
- Moorhead G, Douglas P, Morrice N, Scarabel M, Aitken A, MacKintosh C. Phosphorylated nitrate reductase from spinach leaves is inhibited by 14-3-3 proteins and activated by fusicoccin. Curr Biol. 1996;1996 6:1104–1113. doi: 10.1016/s0960-9822(02)70677-5. [DOI] [PubMed] [Google Scholar]
- Oecking C, Piotrowski M, Hagemeier J, Hagemann K. Topology and target interaction of the fusicoccin-binding 14-3-3 homologs of Commelina communis. Plant J. 1997;12:441–453. [Google Scholar]
- Rittinger K, Budman J, Xu J, Volinia S, Cantley LC, Smerdon SJ, Gamblin SJ, Yaffe MB. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol Cell. 1999;4:153–166. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- Rosenquist M, Sehnke P, Ferl RJ, Sommarin M, Larsson C. Evolution of the 14-3-3 protein family: does the large number of isoforms in multicellular organisms reflect functional specificity? J Mol Evol. 2000;51:446–458. doi: 10.1007/s002390010107. [DOI] [PubMed] [Google Scholar]
- Toroser D, Athwal GS, Huber SC. Site-specific regulatory interaction between spinach leaf sucrose-phosphate synthase and 14-3-3 proteins. FEBS Lett. 1998;435:110–114. doi: 10.1016/s0014-5793(98)01048-5. [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Wang W, Shakes DC. Molecular evolution of the 14-3-3 protein family. J Mol Evol. 1996;43:384–398. doi: 10.1007/BF02339012. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. Leafy controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- Wu K, Rooney MF, Ferl RJ. The Arabidopsis 14-3-3 multigene family. Plant Physiol. 1997;114:1421–1431. doi: 10.1104/pp.114.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]