Abstract
The specific functions of the light-harvesting proteins Lhca2 and Lhca3 were studied in Arabidopsis ecotype Colombia antisense plants in which the proteins were individually repressed. The antisense effect was specific in each plant, but levels of Lhca proteins other than the targeted products were also affected. The contents of Lhca1 and Lhca4 were unaffected, but Lhca3 (in Lhca2-repressed plants) was almost completely depleted, and Lhca2 decreased to about 30% of wild-type levels in Lhca3-repressed plants. This suggests that the Lhca2 and Lhca3 proteins are in physical contact with each other and that they require each other for stability. Photosystem I fluorescence at 730 nm is thought to emanate from pigments bound to Lhca1 and Lhca4. However, fluorescence emission and excitation spectra suggest that Lhca2 and Lhca3, which fluoresce in vitro at 680 nm, also could contribute to far-red fluorescence in vivo. Spectral forms with absorption maxima at 695 and 715 nm, apparently with emission maxima at 702 and 735 nm, respectively, might be associated with Lhca2 and Lhca3.
During photosynthesis, light energy is captured by pigments in the light-harvesting complex (LHC) proteins and transferred to the reaction centers of the thylakoid membrane in green plants. There are 10 different classes of nuclear-encoded Lhc genes, which encode 10 abundant LHC proteins in higher plants (Jansson, 1994). Of these 10 proteins, Lhca1 through Lhca4 form the LHC I complex, exclusively associated with photosystem I (PS I). Lhcb3 through Lhcb6 are exclusively associated with photosystem II (PS II) and Lhcb1 and Lhcb2 form mixed trimers (LHC II) that can be associated with either photosystem. The gene family also includes some related proteins such as the early light-induced proteins, PsbS, and a few recently identified, small-Mr proteins (Jansson, 1999).
LHC proteins bind chlorophyll (chl) a, chl b, and carotenoids with weak, non-covalent bonds. The ancestor of the LHC proteins was probably a form of the cyanobacterial high-light-inducible protein from which the LHC proteins seem to have evolved as the green algae and plants appeared. The gross architecture of the higher plant light-harvesting antenna appears to have been present more than 350 million years ago because the same LHC proteins are present in all higher plants (Jansson, 1994). This also implies that all 10 LHC proteins must have specific functions, otherwise some of the genes would have been lost through genetic drift during evolution. Much remains, however, to be learned about these specific functions. Besides light harvesting, they also take part in the dissipation of excess light. In light, violaxanthin is photoconverted to zeaxanthin through antheraxanthin. This is believed to facilitate the dissipation of excess light as heat, and the crucial importance of PsbS in this process has recently been demonstrated (Li et al., 2000).
In Arabidopsis, the genes that encode the LHC I proteins (Lhca1–4) are all present as single-copy genes (Jansson, 1999) that, like all other LHC genes, are located in the nuclear genome. The Lhca proteins vary in size from 20 to 24 kD, and are believed to exist as dimers, each independently binding to PS I. A tentative model for the arrangement of the LHC I dimers around PS I has also been published (Jansson et al., 1996). Isolation of native Lhca proteins is a difficult task, but preparation of LHC I on Suc gradients gives two major subfractions which differ in protein composition and protein content (Lam et al., 1984). One fraction has high density and consists of Lhca1 and Lhca4, which have been shown to form a heterodimer (Jansson et al., 1996; Schmid et al., 1997). This fraction has a strong 77-K fluorescence emission with a maximum at 730 nm and consequently has been named LHCI-730 (Lam et al., 1984; Knoetzel et al., 1992). The other fraction has lower density, is composed of Lhca2 and Lhca3, fluoresces maximally at 680 nm in vitro (Lam et al., 1984; LHCI-680) and can sometimes be separated into two fractions, one containing Lhca2, and the other Lhca3 (Knoetzel et al., 1992). The oligomeric state of LHCI-680 preparations has not been established. Although Lhca2 and Lhca3 seem to exist as dimers in the native state (Jansson et al., 1996), the separation of LHCI-680 on Suc gradients indicates that under these in vitro conditions they are in a monomeric state and thus do not represent true functional units. Other biochemical data have also suggested that Lhca2 is not tightly associated with Lhca3 (Jansson et al., 1996).
The 730-nm fluorescence emission of LHCI-730 is remarkable. chl molecules in solution, or bound to most sites in the pigment-binding protein complexes of PS I and PS II (bulk chlorophylls), fluoresce at 670 to 690 nm, but a few fluoresce at longer wavelengths. Because these far-red fluorescing chlorophylls obviously have a lower energy than the PS I reaction center, P700, it has been questioned whether they really could function as antenna pigments or, rather, if they are really sinks for excess excitation energy. Data recently have suggested that the far-red fluorescing chls really are true antenna pigments that at physiological temperatures could transmit their energy to the P700 (Pålsson et al., 1998; Rivadossi et al., 1999), but the direct role of the far-red fluorescing chls in the plant needs to be directly demonstrated. Regardless of these considerations, there seem to be five to 10 far-red fluorescing chlorophylls present in each PS I complex, one or two of which are believed to be associated with the PS I core (Croce et al., 1998). There is heterogeneity among the far-red fluorescing chls. Some emit at 715 to 720 nm, and these are clearly located in the reaction center complex because they also are present in cyanobacteria (which lack LHC I), and the barley (Hordeum vulgare) mutant vir-k23, which is devoid of LHC I (Knoetzel et al., 1998). The other far-red fluorescing chls, emitting at 730 to 740 nm, have been suggested, unsurprisingly, to be associated with LHCI-730 (Croce et al., 1998).
Studies on biochemical preparations give valuable information, but it is known that the in vivo energy transfers and subsequent fluorescence emissions are altered when the complexes are bound to each other and to the reaction center (Bossman et al., 1997; Schmid et al., 1997). Therefore, the best way of resolving the functions of the LHC proteins and the energy transfers in the complex is to analyze mutants lacking individual proteins. Studies have been made on several such mutants, for instance chl b-deficient barley mutants, but the results have been inconclusive because the plants lack many LHC proteins bound both to PS I and PS II (Bossman et al., 1997). In plants where homologous recombination is a rare event, the construction of plants lacking specific proteins is most efficiently performed using antisense inhibition of gene expression. We have previously reported successful antisense inhibition of Lhca4 (Zhang et al., 1997), Lhcb4, and Lhcb5 (Andersson et al., 2001) and here we present the results of an investigation involving the construction and analysis of Lhca2 and Lhca3 antisense Arabidopsis plants.
RESULTS
Highly Efficient Antisense Inhibition
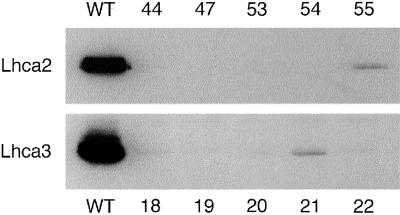
The seeds from the transformed plants were collected and placed on kanamycin agar plates. Of the kanamycin-resistant T1 plants (20 from each transformation), over 80% showed more than 95% depletion of the corresponding protein, demonstrating highly efficient antisense inhibition, and none of the remaining transformants had protein levels higher than 10% of the wild type (Fig. 1). Three Lhca2 and Lhca3 antisense lines with no visible amounts of the Lhca2 or Lhca3 proteins, respectively, were self-pollinated to give the T2 progeny and were analyzed by Southern blots using probes for the antisense genes. The differences in the restriction patterns confirmed that they originated from individual transformation events (data not shown). To ensure that the protein corresponding to the inhibited gene was depleted in all the plants discussed in this paper (i.e. that the antisense effect was stable), all plants were screened by immunoblotting and 77-K fluorescence emission spectroscopy (see below) prior to all subsequent measurements. The results from the three independent lines were always consistent; consequently, in the following presentation, for clarity, we only show the results from one line per transformation.
Figure 1.
Screening for plants deficient in Lhca2 or Lhca3 by immunoanalysis of total leaf membrane preparations from individual antisense lines (44, 47, etc.). Protein corresponding to 3 μg of chl from wild type (WT) and five antisense plants from each transformation was loaded in each lane and separated by SDS-PAGE. The Lhca2 and Lhca3 proteins were detected with monospecific antibodies.
The transgenic plants showed no obvious visible phenotypic deviation from wild type under the growth conditions used in terms of pigmentation or morphology. Even in long-term growth experiments, the growth rates of the antisense plants were similar to wild-type rates (data not shown).
The Antisense Inhibition Is Specific
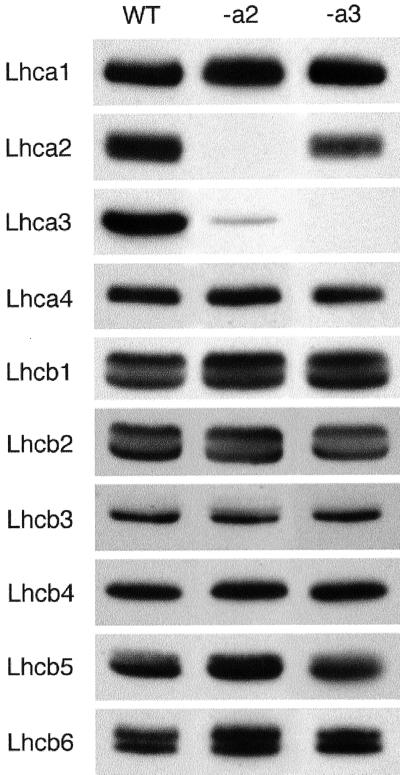
The different Lhca genes show quite a high degree of homology to each other. To elucidate whether the antisense inhibition was specific, transcription of the four Lhca genes was studied using northern-blot analysis (Fig. 2). In both cases (Lhca2 and Lhca3), the transcripts corresponding to the respective inserted antisense gene were depleted below the level of detection by this method (more than 99%), whereas the transcript levels for the other Lhca genes were not changed. Hybridization with a 25S rDNA probe confirmed that equal amounts of RNA were loaded in each lane (not shown). This shows that the antisense inhibition was not only efficient but also specific in the sense that only the mRNA level of the corresponding antisense construct was depleted, leaving the other genes unaffected.
Figure 2.
Verification of specific antisense inhibition by analyzing transcript levels of Lhca genes. Total RNA preparations (2 μg) from wild-type (WT) and Lhca2 (-a2) or Lhca3 (-a3) antisense plants were separated on agarose gels. The corresponding transcripts were detected with homologous probes. A 25S rDNA fragment was used as a control for equal loading (not shown).
Decreased Stability of the Other Lhca Proteins
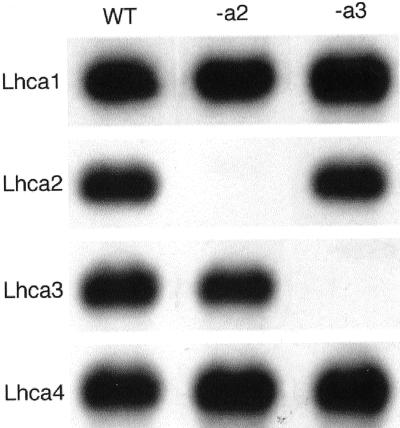
These data prompted us to analyze the quantity of the other Lhca proteins in the antisense lines, to determine if removal of Lhca2 or Lhca3 affected the other LHC proteins of the PS I antenna. The relative levels of the LHC proteins were analyzed by immunoblotting (Fig. 3). In the Lhca2 antisense line, the amounts of Lhca1 and Lhca4 were the same as in the wild type, whereas Lhca3 decreased to less than 10% of the wild-type level. The Lhca3 plants also showed wild-type amounts of Lhca1 and Lhca4, but the Lhca2 levels decreased to about 30%. None of the Lhcb1-Lhcb6 proteins were affected (Fig. 3). Because the Lhca3 mRNA level in the Lhca2 antisense line was the same as in the wild-type plants, our conclusion is that the Lhca3 protein is significantly less stable in the absence of Lhca2. In a converse manner, the low amount of Lhca2 in the Lhca3 antisense plants most probably indicates that Lhca2 is much less stable in the absence of Lhca3. The protein levels simply were not reciprocal to each other because Lhca3 amounts declined more than Lhca2 in the absence of the other protein. This difference was found in three batches of plants grown on three different occasions. These data suggest that the Lhca2 and Lhca3 proteins are in direct contact with each other in PS I, otherwise the interdependence of the protein levels is difficult to explain.
Figure 3.
Analysis of the LHC protein content in total leaf membrane preparations from Lhca2 and Lhca3 antisense plants by immunoblotting. Membrane preparations from wild type (WT) and Lhca2 (-a2) or Lhca3 (-a3) antisense plants were subjected to SDS-PAGE. Protein corresponding to 3 μg of chl was loaded in each lane. The proteins were detected with a collection of antibodies specific for the 10 different LHC proteins.
Both Antisense Lines Have Decreased Long Wavelength Fluorescence
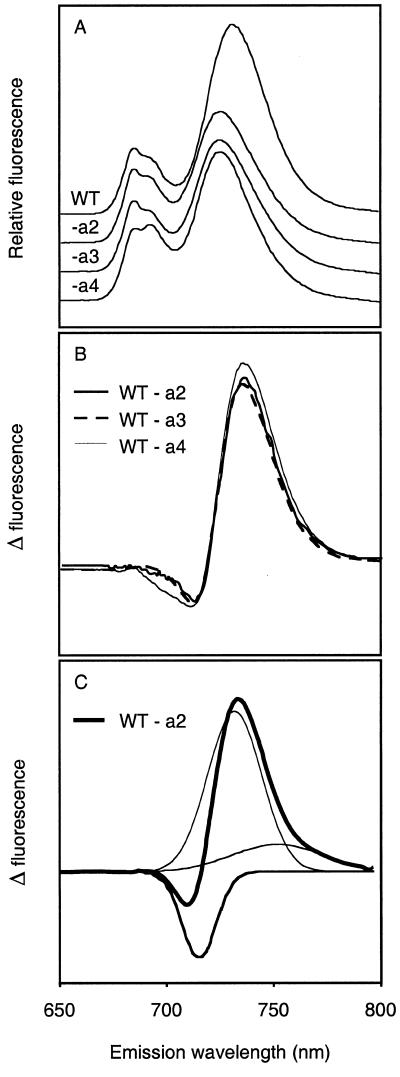
According to previous suggestions, the far-red fluorescing chls in LHC I should be associated with the Lhca1/Lhca4 heterodimer, and not with Lhca2 and Lhca3 (which fluoresce at 680 nm in vitro). Therefore, it was interesting to see that the in vivo 77-K chl fluorescence emission from leaves of the transgenic lines excited at 440 nm (chl a) showed significant differences in the red region of the spectrum as compared with the wild type (Fig. 4A). In both the Lhca2 and Lhca3 transgenic plants, the 732-nm peak is shifted 6 to 726 nm, and the amplitude is dramatically decreased. The difference was not a consequence of different chl content; the antisense lines did not differ significantly from the wild type in this respect. The effect on the fluorescence emission spectrum upon removal of Lhca2 and Lhca3 resembled the effect seen in Lhca4 antisense plants, in which the LHCI-730 complex (Lhca1/Lhca4) is missing (U. Ganeteg and S. Jansson, unpublished data). The Lhca4 antisense gene has been introduced into another genetic background (strain C24), which has a slightly different 77-K fluorescence emission spectrum. Therefore, to compare the fluorescence emission characteristics of the different antisense lines more carefully, difference spectra (wild type-antisense plant) were calculated for the three different antisense lines (Fig. 4B). Gaussian deconvolution of the difference spectra gave identical results with three peaks, one negative at 715 nm, one strongly positive at 733 nm, and a minor (very wide) band at 759 nm (Fig. 4C), which were sufficient to explain the difference spectrum. This shows that removal of Lhca2/Lhca3 or Lhca1/Lhca4 had the same consequences spectroscopically: Fluorescence emission at 733 nm is greatly reduced, but molecules emitting at 715 nm, assumed to be located in the PS I core, fluoresce more strongly in the antisense plants.
Figure 4.
Analysis of 77-K fluorescence emission spectra of wild-type (WT), Lhca2 (-a2), Lhca3 (-a3), and Lhca4 (-a4) antisense plants. A, Seventy-seven-Kelvin fluorescence emission spectra for excitation at 440 nm. The spectra were normalized to the 680-nm peak. B, Difference spectra of corresponding wild-type and antisense plants for Lhca2, Lhca3, and Lhca4. C, Gaussian deconvolution of the Lhca2 difference spectrum shown in Figure 4B and by the thickest solid line. The deconvolution of the Lhca3 and Lhca4 spectra was virtually identical to that of the Lhca2.
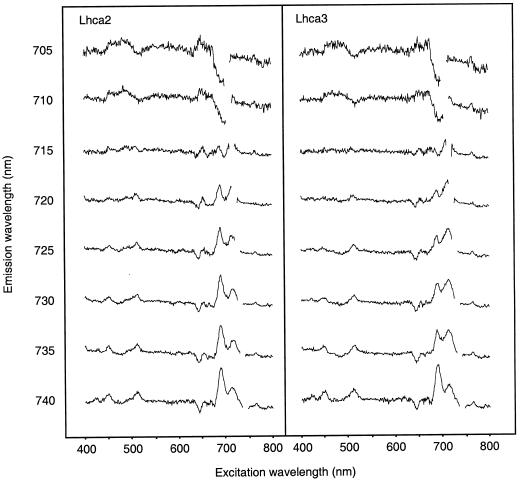
We also analyzed the fluorescence excitation spectra of the antisense lines, monitoring emission at different wavelengths in the far-red region (705–740 nm) and found several interesting features. First of all, we could record differences between the excitation spectra from individual leaves of the same plant (wild type or antisense). We believe this is due to minor differences in environmental conditions experienced by the individual leaves especially, most probably, shading by other leaves. To eliminate such leaf-to-leaf variation, we sampled four non-shaded leaves from different plants, measured their excitation spectra, averaged the data for each line and used the resulting averages for the calculations presented. The spectra were normalized to the 439-nm chl a peak and a difference spectrum was calculated by subtracting the spectra from the Lhca2 or Lhca3 transgenic plants from the wild-type spectra (Fig. 5).
Figure 5.
Difference excitation spectra of Lhca-deficient antisense plants. The fluorescence excitation spectra at 77 K were measured at different emission wavelengths on four non-shaded leaves per antisense line. The spectra were normalized to the 439-nm chl a peak, the four spectra were averaged and the difference spectra were calculated (wild type minus antisense) for Lhca2 and Lhca3 antisense plants.
A decrease in fluorescence in the transgenic plant results in a positive peak and an increase yields a negative peak in the difference spectra. The most prominent differences found were positive peaks in both antisense lines at 695 and 715 nm, when fluorescence was monitored at 720 to 740 nm. Pigments absorbing at 695 and 715 nm were probably missing in the antisense lines. Alternatively, the 695- and 715-nm pigments may not efficiently deliver their excitation energy to another pigment, emitting between 720 and 740 nm. At the same emission wavelengths, positive peaks were found at 510 nm, and negative peaks at 645 nm. A510 and A645 nm is mainly attributed to xanthophylls and chl b, respectively, indicating that xanthophylls were less efficient, and chl b more efficient, in exciting the far-red fluorescing chls in the antisense lines, as compared with the wild type.
No Changes in the Xanthophyll Cycle Pigment
When measuring the chl a/b ratio in the wild type and transgenic plants, no significant differences could be seen. Also, there were no differences in the carotenoid composition of (dark-adapted) wild-type and transgenic plants (data not shown). We also studied the kinetics of the xanthophyll cycle and the maximum level of de-epoxidation by subjecting plants to high light for either 5 min or 1 h. None of the transgenic lines showed any significant differences in the epoxidation state as compared with the wild type (Table I), after either short- or long-term exposure.
Table I.
Analysis of the epoxidation state in Lhca-deficient antisense plants
| Time after High-Light Treatment | 0 Min | 5 Min | 60 Min |
|---|---|---|---|
| WT | 0.99 | 0.89 | 0.55 |
| -a2 | 0.99 | 0.93 | 0.61 |
| -a3 | 0.97 | 0.89 | 0.55 |
Dark-adapted wild-type (WT) and Lhca2 (-a2) and Lhca3 (-a3) antisense plants were illuminated with a PPFD of 700 μmol m−2 s−1. Leaves were taken from four plants, pooled, and frozen in liquid nitrogen 0, 5, and 60 min after high-light treatment. The pigment content was analyzed by HPLC and epoxidation state was calculated as (V + A/2)/(V + A + Z), where A, V, and Z denote concentrations of antheraxanthin, violaxanthin, and zeaxanthin, respectively.
DISCUSSION
Photosynthesis is an extremely complex process in which hundreds (if not thousands) of proteins are involved. PS I of higher plants consists of at least 17 polypeptides, with additional proteins interacting with the complex. To define the functions of all these photosynthetic proteins is an important, but complex, task. However, the standard genetic procedure, of identifying mutants by phenotypic screening, isolating the corresponding gene, and investigating the phenotype by physiological and biochemical methods is not readily applicable due to problems in designing relevant screenings or selections. Moreover, photosynthesis is essential for higher plants, so null mutations in many photosynthetic genes are lethal. Instead, much effort has been spent on biochemical characterization of photosynthetic protein preparations and a large amount of data has been gathered on the components of the photosynthetic machinery. In this situation, reverse genetics is a powerful tool for dissecting the functions of the different proteins. Antisense plants or knockout mutants can potentially provide plant material lacking specific components of multiprotein complexes. In this report, we show two examples of successful reverse genetics where we have specifically repressed the expression of two different LHC I proteins and used the transgenic lines to obtain data related to their structure and function. We show that the antisense effect not only effectively removes the proteins of choice, but it is also specific in the sense that the mRNA levels encoding related proteins are not affected. Under normal controlled growth conditions, no effect of the transformations on growth could be detected, and the absence of Lhca2 or Lhca3 did not affect the xanthophyll cycle.
Structure of LHC I
Electron microscopy of negatively stained PSI-200 particles and pigment stoichiometries have indicated that eight Lhca subunits are associated with each PS I (Boekema et al., 1990). We have, however, recently found that the Lhca protein composition of LHC I is flexible and varies with both the intensity (Bailey et al., 2001) and spectral properties (S. Benson, U. Ganeteg, P. Horton, and S. Jansson, unpublished data) of the light during growth. Under standard laboratory conditions, the mRNA levels of Arabidopsis Lhca1, Lhca2, Lhca3, and Lhca4 are about the same (Jansson, 1999), so we believe that the LHC I in “normal” light could consist of equimolar amounts of the four polypeptides but that this is certainly not true for all light conditions. Lhca1 and Lhca4 appear to exist mainly as heterodimers. This has been corroborated by studies involving cross-linking, reconstitution of the LHCI-730 complex, and subsequent fluorescence measurements (Schmid et al., 1997). Our studies of antisense plants (Zhang et al., 1997; S. Benson, U. Ganeteg, P. Horton, and S. Jansson, unpublished data), barley mutants (Bossman et al., 1997), and Arabidopsis grown in different light intensities (Bailey et al., 2001) have shown that Lhca1/Lhca4 stoichiometries may vary considerably in vivo. This means that under some conditions these proteins can appear without each other, perhaps as homodimers or monomers.
We found that the contents of the Lhca2 and Lhca3 proteins were interdependent. In our opinion, this indicates that they are in physical contact with each other in the PS I holocomplex. This could suggest that Lhca2 and Lhca3 form a heterodimer, but cross-linking and other data (Jansson et al., 1996) indicate that Lhca2 and Lhca3 form homodimers instead. We believe that the interdependence implies that Lhca2 and Lhca3 dimers are in contact with each other, as also indicated by cross-linking data. Because both Lhca2 and Lhca3 could be affected by removal of Lhca1/Lhca4 (U. Ganeteg and S. Jansson, unpublished data), all Lhca dimers (two Lhca1/4, one Lhca2, and one Lhca3) should be located adjacent to each other in PS I.
The Far-Red Fluorescing chls
It is known that Lhca2 and Lhca3 preparations have an in vitro fluorescence at 680 nm (Knoetzel et al., 1992). We have shown that Lhca2 and Lhca3 antisense plants have drastically reduced fluorescence at long wavelengths, although the LHCI-730 dimer, which is thought to be responsible for this fluorescence, is present in wild-type amounts. Interpretations of fluorescence data are complicated because changes in fluorescence do not necessarily mean that emitting chl molecules are absent; for example, changes in reabsorption, excitation transfer, and fluorescence quenching could also affect fluorescence yield, especially in a mutant where rearrangements of the system could occur. Taken together, we believe that the most probable explanation of our data nevertheless is that Lhca2 and/or Lhca3 also bind far-red fluorescing chls, with emission peaks at 733 nm. However, other explanations, for example that chls bound to Lhca2 and/or Lhca3 are crucial for energy transfer to LHCI-730, cannot be ruled out. A few far-red fluorescing chl molecules have been shown to be present in each PS I complex, and these are characterized by unusually large Stokes shifts (the difference between the excitation and fluorescence emission wavelengths). For normal chl molecules, Stokes shifts are normally around 3 to 4 nm, but the far-red fluorescing chls have been suggested to have 10 to 25 nm (Wittmershaus, 1987; Gobets et al., 1994), 6–11 nm (Croce et al., 1998), and, most recently, 22-nm shifts (Ihalainen et al., 2000). When our Lhca4 antisense lines are grown under conditions in which they completely lack Lhca1 and Lhca4, 733-nm chls are obviously missing, but long wavelength fluorescence is reduced by about 50% rather than being completely removed. The same holds true for the Lhca2 and Lhca3 antisense lines. A preliminary analysis of the 77-K fluorescence excitation spectra of the Lhca4 antisense plants also gave results very similar to those of the Lhca2 and Lhca3 antisense plants, i.e. strong positive peaks at 695 and 715 nm are found in the difference spectra. This could easily be explained by assuming that chls absorbing at 715 nm and emitting at 733 nm have been removed in all three types of antisense plants, but less readily by assuming that energy transfer to these pigments has been reduced by removal of Lhca2 and/or Lhca3. Thus, our data indicate that chls absorbing at 715 nm and emitting at 733 nm with extremely large (18 nm) Stokes shifts, probably due to an unusually strong coupling, are physically associated with both Lhca1/4 and Lhca2/Lhca3. Ihalainen et al. (2000) also identified chls associated with Lhca2 and Lhca3 that emit at 702 nm in isolated LHC I preparations, although this fluorescence emission is not detectable in PS I preparations. They conclude that these pigments in the native PS I complex efficiently transfer their energy to the chlorophylls emitting at 720 nm, 730 nm, and perhaps even to P700 itself. The pigments absorbing at 695 nm and giving rise to fluorescence at long wavelengths that are missing in our Lhca2/Lhca3 deficient plants might be identical to these 702-nm chl. The fact that we see no reduction in fluorescence emission at 702 nm in our plants corroborate the suggestion by Ihalainen et al. (2000) that this fluorescence is efficiently quenched in vivo.
If we accept the view that Lhca2 and/or Lhca3, in addition to Lhca1/Lhca4, bind far-red fluorescing chl, the similar effect on fluorescence upon removal of Lhca1/4 or Lhca2 (or Lhca3) suggests that there are similar amounts of far-red fluorescing chls associated with all Lhca dimers, perhaps one per dimer. This is also consistent with the blue shift of Arabidopsis PsaK antisense plants, which have slightly decreased amounts of Lhca2 and Lhca3 but wild-type levels of Lhca1 and Lhca4 (Jensen et al., 2000), and the 77-K emission spectrum of barley chlorina mutants (Bossman et al., 1997; Knoetzel et al., 1998). This also implies that Lhca2 and Lhca3 preparations differ from the native complexes, and that the 680-nm fluorescence (used to designate the complex) is an artifact, probably due to the monomerization of dimeric complexes during preparation.
We also observed a reduction in the relative efficiency of the xanthophylls, but increased efficiency of chl b in exciting the 735-nm emitting forms in the antisense lines. Our interpretation of this is that more LHC II is associated with PS I in the antisense lines, compensating for the loss of LHC I. LHC II binds more chl b than LHC I and, as a consequence, less chl a and xanthophylls on a total pigment basis (Jansson, 1994). The antisense plants should also show an increased reduction of their plastoquinone pools if the PS I antenna size is decreased. Therefore, LHC II phosphorylation will increase and the plant will be driven toward state II, with a higher proportion of the mobile LHC II associating with PS I. Again, other interpretations cannot be excluded.
In conclusion, we have found that Lhca2 and Lhca3, the polypeptides previously collectively named LHCI-680, contribute to the long wavelength fluorescence and thus we think that this name is inappropriate and that Lhca2 and Lhca3 should be used to designate the polypeptides instead. Our study also demonstrates the power of reverse genetics because the different polypeptides can be studied in vivo, where sometimes features such as fluorescence properties can be strikingly different from those recorded in vitro. We finally want to point out that the functions of the far-red fluorescing chl forms are still unclear, but studies on these aspects are under way using the different Lhca antisense lines.
MATERIALS AND METHODS
Plant Growth Conditions
Wild-type Arabidopsis ecotype Colombia and the transgenic plants were germinated and grown on soil/perlite with a day/night temperature regime of 23°C/18°C, a photoperiod of 8 h with a PPFD of 150 μmol m−2 s−1 (fluorescent lamps), and 75% humidity. For screening seeds for positive transformants, plants were germinated on plates with 1× Murashige and Skoog nutrients (Sigma, St. Louis) and 50 μg mL−1 kanamycin, with a photoperiod of 16-h light.
Vector Construction
The binary vector pPCV702 (Koncz and Schell, 1986) was first converted to a more versatile plant expression vector. pPCV702 DNA was digested with EcoRI, the protruding ends were repaired using Klenow enzyme, and the vector was religated. After digestion with BamHI, oligonucleotides (sequence: 5′ GAGCTCGAATTCGTCGACCCGGGAGATCCCC 3′) were added and ligated into the BamHI site. Thus, the new cloning vector, designated pSJ10, had the single BamHI cloning site of pPCV702 (flanked by the cauliflower mosaic virus 35S promoter and the polyadenylation signal) replaced with a SacI/EcoRI/SalI/SmaI cloning cassette. The whole cDNA clone of Lhca2, amplified by PCR from the full-length Arabidopsis expressed sequence tag (EST) clone 32F4T7 using the primers T7 (Life Technologies, Grand Island, NY) and FWE (with the sequence 5′ TCGCGAATTCGCGTACGTAAGCTTGGATCC 3′), was EcoRI digested and cloned into the pSJ10 vector digested with EcoRI. The Lhca3 fragment was digested from the full-length Lhca3 EST clone 40G8T7 with SalI and NotI. The protruding ends were repaired and the fragment was subcloned into pUC19 digested with SmaI. The subclone was digested with SacI and SalI, and the fragment was directionally cloned into SacI/SalI digested pSJ10. The antisense orientation of the Lhca2 and Lhca3 inserts was verified by DNA sequencing.
Arabidopsis Transformation
Adult Arabidopsis plants were transformed by in planta vacuum infiltration with a solution containing Agrobacterium tumefaciens, as follows. Arabidopsis seeds were sprinkled over soil-filled pots covered with a piece of fly screen. The seedlings grew through the fly screen and the plants were grown under controlled-environment conditions with a photoperiod of 8 h. After about 6 weeks, plants were shifted to a photoperiod of 16 h to induce flowering. When the plants had developed a short inflorescence, the pots with Arabidopsis plants were submerged in a solution containing the A. tumefaciens (resuspended in 1× Murashige and Skoog salts, 1% [w/v] Suc, and 0.044 μm benzylamino purine), a vacuum was applied and held for 5 min, before being rapidly released. After the transformation the plants were placed in the growth chamber and seeds were collected. The seeds were screened on Murashige and Skoog plates containing 50 μg mL−1 kanamycin. Kanamycin-resistant plants were retained, screened by immunoblotting to verify the efficiency of the antisense inhibition, allowed to self-pollinate, and the T2 (or T3) generation was used for the following experiments. Seeds from the antisense lines have (stock nos. CS3889, CS3890, CS3891, CS3892, CS3893, and CS3894) have been deposited at the Arabidopsis Biological Resource Stock Center (Ohio State University, Columbus).
Thylakoid Protein Preparation and Immunoblotting
Arabidopsis thylakoid proteins were prepared as described previously (Zhang et al., 1997) from leaves of 6- to 8-week-old plants taken 3 h into the photoperiod. The proteins were separated according to Jansson et al. (1997), except that the preparations were solubilized at 95°C. After electrophoresis the proteins were transferred to 0.2-μm nitrocellulose membranes (Micron Separations Inc., Westborough, MA) using a minitrans blot system (Bio-Rad, Hercules, CA) as recommended by the vendor. The antibody collection used to detect the different LHC proteins was essentially identical to the one described before (Jansson et al., 1997), although new batches of Lhca1 and Lhca4 antibodies were prepared using the same antigens as described earlier (Król et al., 1995).
RNA Preparation and Northern-Blot Analysis
Leaf samples for RNA preparation were taken 3 h into the photoperiod, frozen in liquid nitrogen, and stored at −80°C. RNA was prepared from 100 mg of leaf tissue from five plants using TRIZOL Reagent (Molecular Research, Inc., Cincinnati) as recommended by the vendor. The purity and concentration of the preparations were determined spectrophotometrically (GeneQuant, Pharmacia, Uppsala). Northern-blot analyses were performed on 2-μg RNA samples as described earlier (Strand et al., 1997). Homologous Arabidopsis EST clones (Jansson, 1999) were used as probes for transcripts of the different Lhca genes. A 25S rDNA probe was used as a control to check that loading was equal.
Fluorescence Spectrum Measurements
The chl emission and excitation spectra at 77 K were measured, using a fluorescence spectrophotometer (Fluoro-Max-2, ISA Inc., Edison, NJ), in dark-adapted intact leaves directly exposed to the growth light of 6- to 8-week-old plants. Fluorescence emission spectra were measured using an excitation light with a wavelength of 440 nm, and the spectra were normalized to the 680-nm peak. The fluorescence excitation spectra were measured at different emission wavelengths with an integration time of 0.5 s, with a 3-nm slit width, and the spectra were normalized to the 439-nm chl a peak.
Pigment Preparation and Analysis
For pigment measurements, four plants from each line were dark adapted for 16 h and subsequently illuminated with a PPFD of 700 μmol m−2 s−1. Leaves from the four plants were taken, pooled, and frozen in liquid nitrogen after 0, 5, and 60 min. The pigments were extracted in 80% (v/v) acetone, and after centrifugation the pellet was re-extracted with 100% (v/v) acetone. The two extracts were pooled and the pigment composition was analyzed by HPLC as described previously (Król et al., 1995). The epoxidation states of the xanthophyll pigments were calculated according to the formula: [(V + A/2)/(V + A + Z)], where V, A, and Z are the concentrations of violaxanthin, antheraxanthin, and zeaxanthin, respectively.
ACKNOWLEDGMENTS
We are grateful to the Ohio Stock Center for EST clones, Olof Olsson for providing pPCV702 and sequence data, Peter Horton for useful comments, Igor Rojdestvenski for help with Gaussian deconvolution, and Jan Dekker for sharing unpublished data.
Footnotes
This work was supported by grants from the Swedish Forestry and Agricultural Research Council.
LITERATURE CITED
- Andersson J, Walters R, Horton P, Jansson S. Antisense inhibition of the photosynthetic antenna proteins CP29 and CP26: implications for the mechanism of protective energy dissipation. Plant Cell. 2001;13:1193–1204. doi: 10.1105/tpc.13.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Walters R, Jansson S, Horton P (2001) Acclimation of Arabidopsis thaliana to the light environment: the existence of separate low light and high light responses. Planta (in press) [DOI] [PubMed]
- Boekema E, Wynn R, Malkin R. The structure of spinach photosystem I studied by electron microscopy. Biochim Biophys Acta. 1990;1017:49–56. [Google Scholar]
- Bossman B, Knoetzel J, Jansson S. Screening of chlorina mutants of barley (Hordeum vulgare L.) with antibodies against light-harvesting proteins of PS I and PS II: absence of specific antenna proteins. Photosynth Res. 1997;52:127–136. [Google Scholar]
- Croce R, Zucchelli G, Garlaschi FM, Jennings RC. A thermal broadening study of the antenna chlorophylls in PSI-200, LHCI, and PSI core. Biochemistry. 1998;37:17355–17360. doi: 10.1021/bi9813227. [DOI] [PubMed] [Google Scholar]
- Gobets B, van Amerongen H, Monshouwer R, Kruip J, Rogner M, van Grondelle R, Dekker J. Polarized site-selected fluorescence spectroscopy of isolated photosystem I particles. Biochim Biophys Acta. 1994;1188:75–85. [Google Scholar]
- Ihalainen JA, Gobets B, Sznee K, Brazzoli M, Croce R, Bassi R, van Grondelle R, Korppi-Tommola JEI, Dekker JP. Evidence for two spectroscopically different dimers of light-harvesting complex I from green plants. Biochemistry. 2000;39:8625–8631. doi: 10.1021/bi0007369. [DOI] [PubMed] [Google Scholar]
- Jansson S. The light-harvesting chlorophyll a/b binding-proteins. Biochim Biophys Acta. 1994;1184:1–19. doi: 10.1016/0005-2728(94)90148-1. [DOI] [PubMed] [Google Scholar]
- Jansson S. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 1999;4:236–240. doi: 10.1016/s1360-1385(99)01419-3. [DOI] [PubMed] [Google Scholar]
- Jansson S, Andersen B, Scheller HV. Nearest-neighbor analysis of higher-plant photosystem I holocomplex. Plant Physiol. 1996;112:409–420. doi: 10.1104/pp.112.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson S, Stefánsson H, Nyström U, Gustafsson P, Albertsson P-Å. Antenna protein composition of PS I and PS II in thylakoid sub-domains. Biochim Biophys Acta. 1997;1320:297–309. [Google Scholar]
- Jensen PE, Gilpin M, Knoetzel J, Scheller HV. The PSI-K subunit of photosystem I is involved in the interaction between light-harvesting complex I and the photosystem I reaction center core. J Biol Chem. 2000;275:24701–24708. doi: 10.1074/jbc.M000550200. [DOI] [PubMed] [Google Scholar]
- Knoetzel J, Bossmann B, Grimme LH. Chlorina and viridis mutants of barley (Hordeum vulgare L.) allow assignment of long-wavelength chlorophyll forms to individual Lhca proteins of photosystem I in vivo. FEBS Lett. 1998;436:339–342. doi: 10.1016/s0014-5793(98)01158-2. [DOI] [PubMed] [Google Scholar]
- Knoetzel J, Svendsen I, Simpson DJ. Identification of the Photosystem I antenna polypeptides in barley: isolation of three pigment-binding antenna complexes. Eur J Biochem. 1992;206:209–215. doi: 10.1111/j.1432-1033.1992.tb16918.x. [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of the TL-DNA gene 5 controls the tissue specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Król M, Spangfort MD, Huner NP, Öquist G, Gustafsson P, Jansson S. Chlorophyll a/b-binding proteins, pigment conversions, and early light-induced proteins in a chlorophyll b-less barley mutant. Plant Physiol. 1995;107:873–883. doi: 10.1104/pp.107.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E, Ortiz W, Malkin R. Chlorophyll a/b proteins of photosystem I. FEBS Lett. 1984;168:10–14. [Google Scholar]
- Li X-P, Björkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK. A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature. 2000;403:391–395. doi: 10.1038/35000131. [DOI] [PubMed] [Google Scholar]
- Pålsson LO, Flemming C, Gobets B, van Groendelle R, Dekker JP, Schlodder E. Energy transfer and charge separation in photosystem I: P700 oxidation upon selective excitation of the long-wavelength antenna chlorophylls of Synechococcus elongatus. Biophys J. 1998;74:2611–2622. doi: 10.1016/S0006-3495(98)77967-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivadossi A, Zucchelli G, Garlaschi FM, Jennings RC. The importance of PS I chlorophyll red forms in light-harvesting by leaves. Photosynth Res. 1999;60:209–215. [Google Scholar]
- Schmid VH, Cammarata KV, Bruns BU, Schmidt GW. In vitro reconstitution of the photosystem I light-harvesting complex LHCI-730: heterodimerization is required for antenna pigment organization. Proc Natl Acad Sci USA. 1997;94:7667–7672. doi: 10.1073/pnas.94.14.7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand Å, Hurry V, Gustafsson P, Gardeström P. Development of Arabidopsis thaliana leaves at low temperatures releases the suppression of photosynthesis and photosynthetic gene expression despite the accumulation of soluble carbohydrates. Plant J. 1997;12:605–614. doi: 10.1046/j.1365-313x.1997.00605.x. [DOI] [PubMed] [Google Scholar]
- Wittmershaus B. Measurements and kinetic modeling of picosecond time resolved fluorescence from photosystem I and chloroplasts. In: Biggins J, editor. Progress in Photosynthesis Research 1. Dordrecht, The Netherlands: Martinus Nijhoff Publisher; 1987. pp. 75–82. [Google Scholar]
- Zhang H, Goodman HM, Jansson S. Antisense inhibition of the photosystem I antenna protein Lhca4 in Arabidopsis thaliana. Plant Physiol. 1997;115:1525–1531. doi: 10.1104/pp.115.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]