Abstract
We report here the finding of a highly significant inverse correlation of the uracil content of 16S rRNA and the optimum growth temperature (Topt) of cultured thermophilic and psychrophilic prokaryotes. This correlation was significantly different from the weaker correlations between the contents of other nucleotides and Topt. Analysis of the 16S rRNA secondary structure regions revealed a fall in the A:U base-pair content in step with the increase in Topt that was much steeper than that of mismatched base-pairs, which are thermodynamically less stable. These findings indicate that the 16S rRNA sequences of thermophiles and psychrophiles are under a strong thermo-adaptive pressure, and that structure–function constraints play a crucial role in determining their 16S rRNA nucleotide composition. The derived relationship between uracil content and Topt was used to develop an algorithm to predict the Topt values of uncultured prokaryotes lacking cultured close relatives and belonging to the phyla predominantly containing thermophiles. This algorithm may be useful in guiding the design of cultivation conditions for hitherto uncultured microbes.
INTRODUCTION
Ecological success reflects the progressive evolution of organismal genomes towards optimal interactions with prevailing physico-chemical and biotic conditions of the habitat, including habitat-specific stresses, and exploitation of available resources. Characterization of the patterns of genetic changes underlying these processes, and their mechanisms, will not only advance understanding the evolutionary processes, but also aid in identifying sequence-based markers specific to a particular adaptative process and that thus form a habitat-specific genomic signature.
Because of their great interest for applications in industrial biotechnology, thermophiles and psychrophiles have been intensively studied and a number of primary sequence and tertiary structure-based markers of thermophily have been proposed (1–7). We report here the finding of an unexpected, highly significant inverse relationship between the uracil contents of 16S ribosomal RNA sequences and the optimal growth temperatures (Topt) of thermophilic and psychrophilic prokaryotes, which is much stronger than correlations between Topt and the contents of other nucleotides.
MATERIALS AND METHODS
16S rDNA sequences of prokaryotes were downloaded from the NCBI website. Sequences with >2% of unknown nucleotides were not considered. For the purpose of estimating the various base-pair contents in the stems, reconstructed structural models of 16S rRNA were obtained from ‘The Comparative RNA Web (CRW) Site’ (http://www.rna.icmb.utexas.edu/). Topt values were taken from the DSMZ German Collection of Microorganisms and Cell Cultures website (http://www.dsmz.de/), and from the Galtier and Lobry dataset (1). We considered prokaryotes having Topt values of >45°C as thermophiles, 45–21°C as mesophiles, and <21°C as psychrophiles. Nucleotide contents of 16S rDNA, i.e. %A, T(U), G, C contents were calculated using in-house written ‘Perl’ scripts.
We divided the compiled list of 16 rDNA sequences in two groups, the first from cultured microbes whose Topt values were known, and the second from uncultured microbes. Group 1 comprised sequences from 70 mesophiles, 117 thermophiles and 19 psychrophiles that were analyzed for identification of a marker that correlated with Topt values (Supplementary Table S1). Only one strain per species was included in the analysis to avoid statistical bias. The members of Group 1 were selected to represent a wide phylogenetic and genomes G+C content range (30–70% in this study). Group 2 contained members belonging to the Aquificae, Deinococcus–Thermus, Thermomicrobia, Thermotogae and Thermodesulfobacteria phyla which are known to be mostly thermophilic (8,9) (http://141.150.157.117:8080/prokPUB/index.htm), comprising of sequences of uncultured bacteria that do not have a cultivated close relative, and was used to make predictions of the Topt value. The Group 2 16S rRNA sequences of uncultured prokaryotes were obtained from the ‘Ribosomal Database project—II release 9.0’ database (RDP), using the hierarchy browser tool available at the following URL: http://rdp.cme.msu.edu/hierarchy/hb_intro.jsp. Only sequences >1200 bp were considered. For each 16S rDNA sequence from an uncultured bacterium, a search was made using the ‘Bl2seq’ program available at NCBI website (http://www.ncbi.nlm.nih.gov) for a close relative (≥97% identity) amongst the sequences of cultivated bacteria.
Phylogenetic analyses were performed using PHYLIP package version 3.6 (http://evolution.gs.washington.edu/phylip.html) and ‘ClustalW’ online tool (http://www.ebi.ac.uk/clustalw). DNADIST and NEIGHBOR programs of the PHYLIP package were used to construct the phylogenetic tree. The consensus tree was visualized using ‘TreeExplorer’ software written by Koichiro Tamura (http://evolgen.biol.metro-u.ac.jp/TE/TE_man.html).
The statistical methods included estimation of Pearson's correlation coefficient ‘r’ between two variables and Z-statistic for pair-wise comparison of correlation coefficients using statistical analyses software MedCalc 7.4 (Medcalc Software, Mariakerke, Belgium) and MS-EXCEL (Microsoft Corp.).
RESULTS AND DISCUSSION
Topt and 16S rRNA uracil content relationship
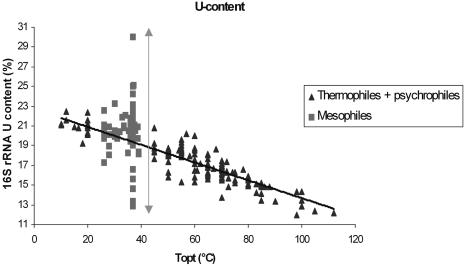
The nucleotide composition of the 16S rRNA sequences of a phylogenetically diverse group of cultured organisms (Group 1; Materials and Methods) was analyzed for possible correlation with their cognate optimum growth temperatures. Interestingly, the uracil content of 16S rRNA showed a strong negative linear correlation with the Topt in thermophiles and psychrophiles. In contrast, the U-content of 16S rRNA molecules of mesophiles varied widely, particularly in those organisms with Topt values close to that of the mammalian body temperature (Figure 1). This points to variables other than temperature shaping the nucleotide composition in mesophiles. Mesophiles having extremely high uracil contents were predominantly host-dependent parasites/symbionts (which is consistent with the fact that the parasites/symbionts are AT-rich), whereas those having low U content reflected their low genome AT contents.
Figure 1.
Relationship of uracil content of 16S rRNA and Topt of cultivated prokaryotes. The bidirectional arrow depicts the extent of variation in the uracil content of 16S rRNA of the mesophiles within a narrow Topt range.
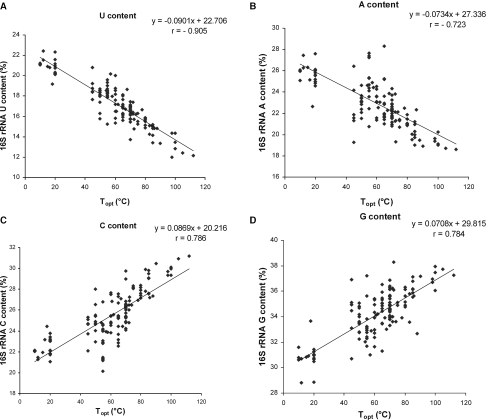
Figure 2 shows the plots of 16S rRNA nucleotide contents as a function of Topt values for thermophiles and psychrophiles (but excluding mesophiles). The correlation coefficient (r) of Topt values to U, A, C, G and G+C contents were −0.905, −0.723, +0.786, +0.784, +0.833, respectively; all having significant P-value of <0.0001. Only uracil content showed a high degree of correlation with Topt values, and there was a statistically significant difference in absolute correlation coefficient values of U content and Topt with respect to other mononucleotides having P-value ≤ 0.001 (Table 1). Thus, the uracil content of 16S rRNA seems to represent a useful predictor of Topt in thermophiles and psychrophiles. The global standard error (SE) for the Topt predictions was ±9.4°C. In 90% of the cases, the SE was ±7.0°C, and 67% had an SE of ±4.7°C.
Figure 2.
Relationships of 16S rRNA contents of different nucleotide and Topt of cultured prokaryotes: (A) U content; (B) A content; (C) C content; (D) G content.
Table 1.
Pair-wise comparison of absolute correlation coefficients
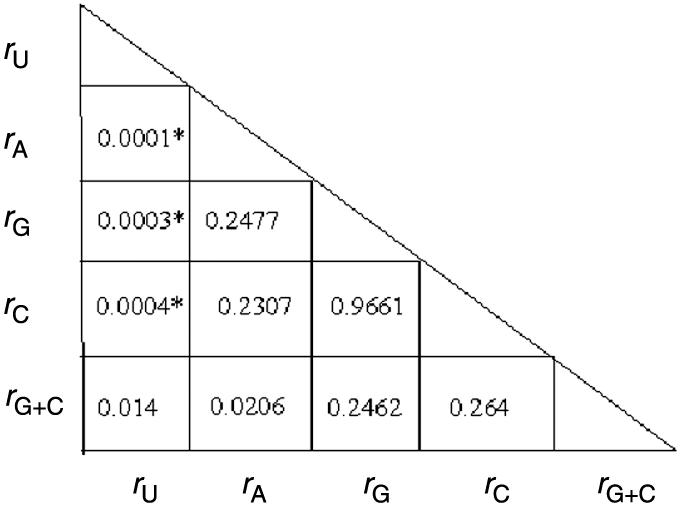 |
The P-values shown indicate the probabilities for the null hypothesis that there is no significant difference between the two absolute values of correlation coefficients (of Topt and rA, rU, rG, rC, rG+C).
Asterisk indicates a very significant difference between the absolute correlation coefficient values being compared (P-value ≤ 0.001).
The uracil deficiency in 16S rRNA of thermophiles: loops and base-paired double-stranded stems
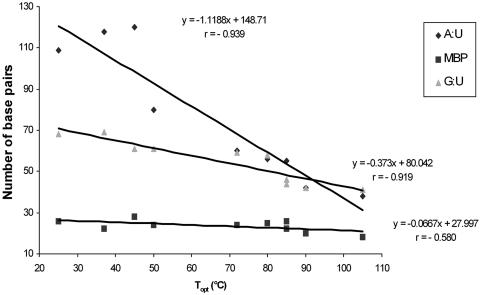
The G+C content of rRNA base-paired stems has been previously shown to correlate positively with Topt (1), and assumed to reflect selection pressure for the more thermo-stable secondary structure provided by triple-hydrogen bonded G:C base pair, and counter selection of the less stable A:U base pair. rRNA stems also contain mismatched base pairs, the majority of which are G:U pairs (10). Since G:U mismatches are less stable than A:U matches, they would be expected, on thermodynamic grounds, to be counter selected over A:U pairs as Topt increases. We assessed this prediction by performing a detailed comparative analysis of the 16S rRNA secondary structural regions, using the re-constructed models of Cannone et al. (11) and manually counting the A:U, G:U and total mismatched base-pair contents in the stems. As can be seen in Figure 3, there was, surprisingly, a relatively constant mismatched base-pair content in the stems. Perhaps, equally surprising was a much sharper drop in the more stable A:U pairs with Topt than G:U pairs. Thus, thermodynamic stability is not the overriding selective force in stem sequence determination, and structure:function constraints maintain less stable mismatched base pairs (including G:U) at key locations, either as recognition sites, or as tertiary structure determinants, as has been previously suggested for G:U base-pairs (10). The structure:function constraint determines the extent of evolutionary compulsion on depletion of the uracil-containing base pairs (A:U > G:U) to raise the stem G:C content, which explains the observed inverse relationship between uracil content and Topt.
Figure 3.
A:U, G:U and mismatched base pair (MBP) content of 16S rRNA stems as a function of Topt. Data was obtained from analysis of the re-constructed 16S rRNA secondary structure models of the following prokaryotes (with NCBI accession Id and Topt within brackets): Aeropyrum pernix (str. K1) AP000062 (90°C), Aquifex aeolicus AE000709 (85°C), Aquifex pyrophilus M83548 (85°C), Clostridium perfringens M69264 (45°C), Escherichia coli J01695 (37°C), Psychrobacter pacificensis AB016054 (25°C), Pyrodictium occultum M21087 (105°C), Streptomyces thermoviolaceus Z68096 (50°C), Thermus aquaticus L09663 (72°C), Thermos thermophilus X07998 (80°C).
We further analyzed for the mononucleotides variation pattern in the 16S rRNA loops of certain of the Group 1 extremophiles (Supplementary Table S2) by collecting their secondary structure data from ‘The European ribosomal RNA database’ (http://www.psb.ugent.be/rRNA/). In these regions, the only nucleotide showing a negative correlation with Topt with a statistically significant P-value of <0.001 is uracil, however with a moderate correlation coefficient of −0.436 (Table 2). It is therefore tempting to speculate that a subtle modification in loop uracil-content may alter the loop energies in a way so as to have an effect on the determination of the tertiary rRNA fold structure, given the widely known fact that there are a large number of ways in which rRNA hairpins and loops can be formed. Furthermore, RNAs are increasingly sensitive to chemical hydrolysis at temperatures greater than 50°C (12), hence, it will be of interest to know whether altering the uracil content has any temperature-dependent effect on 16S rRNA and pre-rRNA stability, especially with regard to its half-life.
Table 2.
Correlation analysis of nucleotide composition of 16S rRNA loops and Topt in thermophiles and psychrophiles
| Correlation coefficient (r) | P-value | |
|---|---|---|
| A | −0.023 | 0.864 |
| C | 0.132 | 0.322 |
| G | 0.257 | 0.051 |
| T (U) | −0.436 | <0.001 |
P-value ≤ 0.001 was considered to be significant.
Topt prediction for uncultured thermophiles
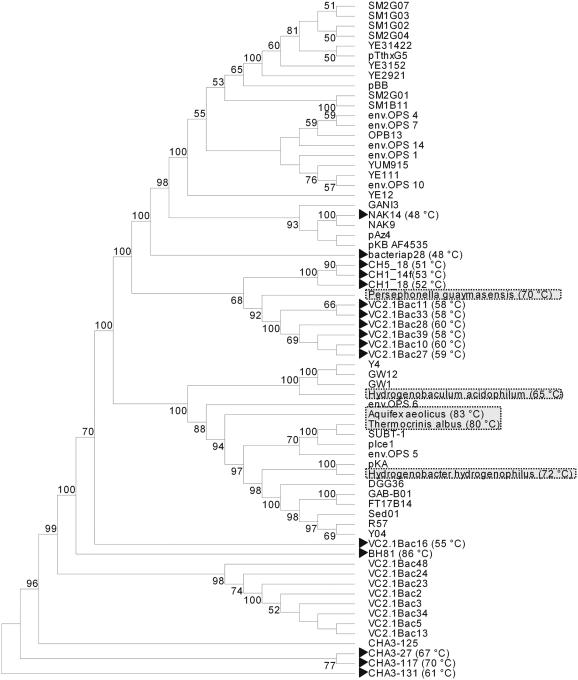
Having found a strong relationship between uracil content of 16S rRNA and Topt of extremophiles, we applied the finding as a method to predict the Topt values of prokaryotes. We know that organisms can only grow within certain windows of physico-chemical conditions, like temperature, pH, redox, etc., so getting these parameters right lies at the root of cultivation. In certain cases, it is possible to infer the growth requirements of an organism from the physiology of one or more already cultivated and characterized close relatives, if available. Unfortunately, many uncultured microbes do not have cultivated close relatives, and, even if they do, phylogenetic relatedness is no guarantee of physiological similarity [e.g. although Prochlorococcus strain MED4 is phylogenetically related to strain MIT9313, the former is high-light adapted whereas the latter is low-light adapted; (13), and in the order Thermotogales, hyperthermophiles such as Thermotoga maritima and Thermotoga neapolitana have optimal growth temperatures (Topt) of ∼80°C, whereas Geotoga petraea, Geotoga subterranea and Petrotoga miotherma have Topt values ranging between 45 and 55°C (14)]. In other cases, it may be possible to infer the growth requirements of microbes from a knowledge of the physico-chemical conditions of their habitats, but such information can be misleading: for example, extremophiles have been found in non-extreme habitats (15–17). Moreover, physico-chemical conditions can vary steeply across gradients in many environments (e.g. a hydrothermal vent chimney with temperatures ranging from 25 to 200°C). Since it is known that large differences in Topt can exist for microbes belonging to the same phylogenetic clade, the value of the approach we have developed is in predicting Topt values of uncultured microbes, particularly those lacking cultured close relatives or those having cultured close relatives whose Topt values are, for the purposes of prediction, misleadingly different. We therefore predicted Topt values of uncultured and cultured members of the following phyla: Aquificae, Deinococcus-Thermus, Thermomicrobia, Thermotogae and Thermodesulfobacteria (Group 2, Materials and Methods) using the mathematical equation, Topt = (22.706 − %U content)/0.0901. Consistent with the fact that most of the cultured members of these phyla are known to be thermophilic, our algorithm predicts the majority of the uncultured members (∼85%) also to be thermophiles. We additionally included Cyanobacteria in our analysis, as this phylum is also known to contain thermophiles (Table 3). The predicted Topt values of the uncultured microbes for the studied phyla ranged from 45 to 105°C. Figure 4 shows an example of the phylogenetic tree of uncultured members of the Aquificae, based on 16S rRNA sequence homology analysis with the Bl2seq program, combined with predicted Topt values. There are clearly some unanticipated predicted Topt values for this phylum: the uncultured environmental clones VC2.1 Bac10, Bac11, Bac27, Bac28, Bac33, Bac39; and CH1_14f, CH5_18, CH1_18 have Topt ranging from 50 to 60°C, while the closest available isolated counterparts, i.e. Persephonella species have a Topt of 70°C, some 10–20°C higher. This shows that being guided by the physico-chemical properties appropriate for cultivation of the nearest cultured relative can be misleading and supports the utility of the approach described here for predicting Topt values of uncultured microbes.
Table 3.
Percentage of predicted thermophilic uncultivated prokaryotes without cultivated close relative, and range of predicted Topt values, in various phyla considered in this study
| Phylum | Total sequences of uncultured organisms in RDP database | Percentage of uncultured thermophiles with no close cultured relative | Examples of diverse predicted Topt values of uncultured members |
|---|---|---|---|
| Aquificae | 63 | 25.4 | Clone BH81 (AF352537), Near Boiling Silica-Depositing Thermal Springs: 86°C. VC2. 1 Bac16 (AF068794), Mid-Atlantic Ridge hydrothermal vent: 55°C |
| Cyanobacteria | 196 | 5.6 | Clone S M2A08 (AY293403), Travertine Terraces at Yellowstone Hot Springs: 60°C. SM2A09 (AY195603), Travertine depositional system at Mammoth Hot Springs: 48°C |
| Deinococcus-Thermus | 32 | 12.5 | Thermus sp. clone bacteriap1 (AF402971), Kuirau Park: 71°C. Clone PK19 (AY555779), Bor Khlueng Hot Spring in Thailand: 58°C |
| Thermomicrobia | 11 | 100.0 | Clone F BP267 (AY250872), McMurdo Dry Valleys, Antarctica: 79°C. a2b029 (AF419669), hydrothermal sediments in the guaymas Basin: 50°C |
| Thermotogae | 8 | 75.0 | Clone R5p5 (AF 482446), granular sludge: 48°C. TTA_B141 (AY297987), thermophilic anaerobic reactor degrading terephthalate: 59°C |
| Thermodesulfobacteria | 3 | 67.0 | Thermodesulfobacterium group OP B45 (AF027096), Yellowstone hot spring: 71°C |
Figure 4.
A neighbor-joining 16S rRNA-based phylogenetic tree constructed from the uncultured environmental bacterial sequences of Aquificae phylum. The topology shown is a consensus tree obtained after 100 bootstrap replicates. Bootstrap values >50% are shown at branch nodes. Arrows point to those thermophilic uncultured bacteria that do not have cultivated counterparts. Predicted Topt of such bacteria range from 50 to 90°C (brackets). Cultivated representatives of the Aquificae phylum, highlighted in grey, are shown as phylogenetic reference points in the tree. [GenBank accession numbers of the uncultured bacterial sequences is provided in the bracket: BH81 (AF352537); VC2.1 Bac48 (AF068811); VC2.1 Bac2 (AF068784); SUBT-1 (AF361217); VC2.1 Bac23 (AF068799); VC2.1 Bac5 (AF068787); VC2.1 Bac3 (AF068785); pIce1 (AF301907); VC2.1 Bac34 (AF068808); CHA3-125 (AJ132734); pKA (AF453505); CHA3-117 (AJ132733); VC2.1 Bac13 (AF068793); env.OPS 5 (AF018190); VC2.1 Bac24 (AF068800); CHA3-27 (AJ132736); DGG36 (AY082371); FT17B14 (AY251061); Y04 (AF407675); R57 (AF407692); GAB-B01 (AB183857); CHA3-131 (AJ132735); VC2.1 Bac28 (AF068802); VC2.1 Bac10 (AF068791); env.OPS 6 (AF018191); Sed01 (AF407673); VC2.1 Bac27 (AF068801); VC2.1 Bac39 (AF068782); VC2.1 Bac33 (AF068807); VC2.1 Bac11 (AF068792); VC2.1 Bac16 (AF068794); pKB (AF453506); CH1_14f (AY672500); CH1_18 (AY672501); CH5_18 (AY672528); NAK9 (AB005737); NAK14 (AB005738); bacteriap28 (AF402979); pAz4 (AF453508); OPB13 (AF027098); env.OPS 14 (AF018196); env.OPS 4 (AF018189); env.OPS 7 (AF018192); YE12 (AB073124); env.OPS 1 (AF018186); pBB (AF113542); GANI 3 (AB005735); SM2G01 (AF445734); SM1B11 (AF445658); YUM915 (AB073130); env.OPS 10 (AF018194); YE111 (AB073123); SM2G04 (AF445736); pTthxG5 (AF453509); SM1G02 (AF445696); SM2G07 (AF445739); YE31422 (AB073127); SM1G03 (AF445697); YE3152 (AB073128); GW12 (AY559413); YE2921 (AB073126); GW1 (AY559410); Y4 (AY559409); Hydrogenobaculum acidophilum (D16296); Hydrogenobacter hydrogenophilus (Z30242); Thermocrinis albus (AJ278895); Aquifex aeolicus VF5 (AJ309733); Persephonella guaymasensis (AF385630).]
Although the predictions made here were only for uncultured members of known thermophilic clades, the approach may also be useful for predicting the Topt of thermophilic members of clades, such as Firmicutes, Proteobacteria, Bacteroidetes, etc., typically containing mesophilic organisms.
CONCLUSIONS
The observed inverse relationship between the uracil content of 16S rRNA and Topt points to the presence of a strong thermo-adaptive mechanism playing a role in shaping the 16S rRNA nucleotide composition in prokaryotes. This adaptive response may take the form of decreased potential for chemical hydrolysis or to adopt a stable tertiary structure. The results further highlight the role of mismatched base pairs in having a biological relevance, for example, acting as protein recognition sites.
The advantages of our uracil content-based approach in predicting the Topt are that it is entirely primary structure based and provides a better estimate of optimum growth temperatures of thermophiles and psychrophiles that can be used to guide the design of cultivation conditions for uncultured microbes.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
A.N.K. and V.A.P.M.d.S. gratefully acknowledge financial support from the BMBF (project Intergenomics). K.N.T. thanks the Fonds der Chemischen Industrie for generous support. Funding to pay the Open Access publication charges for this article was provided by The German National Research Centre for Biotechnology (GBF).
Conflict of interest statement. None declared.
REFERENCES
- 1.Galtier N., Lobry J.R. Relationships between genomic G+C content, RNA secondary structures and optimal growth temperature in prokaryotes. J. Mol. Evol. 1997;44:632–636. doi: 10.1007/pl00006186. [DOI] [PubMed] [Google Scholar]
- 2.Nakashima H., Fukuchi S., Nishikawa K. Compositional changes in RNA, DNA and proteins for bacterial adaptation to higher and lower temperatures. J. Biochem. (Tokyo) 2003;133:507–513. doi: 10.1093/jb/mvg067. [DOI] [PubMed] [Google Scholar]
- 3.Kreil D.P., Ouzounis C.A. Identification of thermophilic species by the amino acid compositions deduced from their genomes. Nucleic Acids Res. 2001;29:1608–1615. doi: 10.1093/nar/29.7.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paz A., Mester D., Baca I., Nevo E., Korol A. Adaptive role of increased frequency of polypurine tracts in mRNA sequences of thermophilic prokaryotes. Proc. Natl Acad. Sci. USA. 2004;101:2951–2956. doi: 10.1073/pnas.0308594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brocchieri L. Environmental signatures in proteome properties. Proc. Natl Acad. Sci. USA. 2004;101:8257–8258. doi: 10.1073/pnas.0402797101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambros R.J., Mortimer J.R., Forsdyke D.R. Optimum growth temperature and the base composition of open reading frames in prokaryotes. Extremophiles. 2003;7:443–450. doi: 10.1007/s00792-003-0353-4. [DOI] [PubMed] [Google Scholar]
- 7.Wang H.C., Hickey D.A. Evidence for strong selective constraint acting on the nucleotide composition of 16S ribosomal RNA genes. Nucleic Acids Res. 2002;30:2501–2507. doi: 10.1093/nar/30.11.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reysenback A.-L., Götz D., Yernool D. Microbial diversity of marine and terrestrial thermal springs. In: Staley J.T., Reysenbach A.-L., editors. Biodiversity of Microbial Life: Foundation of Earth's Biosphere. New York: Wiley-Liss, Inc.; 2000. pp. 345–398. [Google Scholar]
- 9.Rappe M.S., Giovannoni S.J. The uncultured microbial majority. Annu. Rev. Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. Review. [DOI] [PubMed] [Google Scholar]
- 10.Varani G., McClain W.H. The G × U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Muller K.M., et al. The Comparative RNA Web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:15. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poole A.M., Jeffares D.C., Penny D. The path from the RNA world. J. Mol. Evol. 1998;46:1–17. doi: 10.1007/pl00006275. Review. [DOI] [PubMed] [Google Scholar]
- 13.Rocap G., Larimer F.W., Lamerdin J., Malfatti S., Chain P., Ahlgren N.A., Arellano A., Coleman M., Hauser L., Hess W.R., et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 14.Martins L.O., Carreto L.S., Da Costa M.S., Santos H. New compatible solutes related to di-myo-inositol-phosphate in members of the order Thermotogales. J. Bacteriol. 1996;178:5644–5651. doi: 10.1128/jb.178.19.5644-5651.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purdy K.J., Nedwell D.B., Embley T.M. Analysis of the sulfate-reducing bacterial and methanogenic archaeal populations in contrasting Antarctic sediments. Appl. Environ. Microbiol. 2003;69:3181–3191. doi: 10.1128/AEM.69.6.3181-3191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahman T.J., Marchant R., Banat I.M. Distribution and molecular investigation of highly thermophilic bacteria associated with cool soil environments. Biochem. Soc. Trans. 2004;32:209–213. doi: 10.1042/bst0320209. [DOI] [PubMed] [Google Scholar]
- 17.Summit M., Baross J.A. A novel microbial habitat in the mid-ocean ridge subseafloor. Proc. Natl Acad. Sci. USA. 2001;98:2158–2163. doi: 10.1073/pnas.051516098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.