Abstract
Trinucleotide repeats are involved in a number of debilitating diseases such as myotonic dystrophy. Twelve to seventy-five base-long (CTG)n oligodeoxynucleotides were analysed using a combination of biophysical [UV-absorbance, circular dichroism and differential scanning calorimetry (DSC)] and biochemical methods (non-denaturing gel electrophoresis and enzymatic footprinting). All oligomers formed stable intramolecular structures under near physiological conditions with a melting temperature that was only weakly dependent on oligomer length. Thermodynamic analysis of the denaturation process by UV-melting and calorimetric experiments revealed an unprecedented length-dependent discrepancy between the enthalpy values deduced from model-dependent (UV-melting) and model-independent (calorimetry) experiments. Evidence for non-zero molar heat capacity changes was also derived from the analysis of the Arrhenius plots and DSC profiles. Such behaviour is analysed in the framework of an intramolecular ‘branched-hairpin’ model, in which long CTG oligomers do not fold into a simple long hairpin–stem intramolecular structure, but allow the formation of several independent folding units of unequal stability. We demonstrate that, for sequences ranging from 12 to 25 CTG repeats, an intramolecular structure with two loops is formed which we will call ‘bis-hairpin’. Similar results were also found for CAG oligomers, suggesting that this observation may be extended to various trinucleotide repeats-containing sequences.
INTRODUCTION
Recent molecular genetic studies have revealed a correlation between spontaneous expansion of several DNA trinucleotide repeats and a variety of debilitating human diseases [for reviews see (1–4)]. This class of diseases was first characterized in Fragile-X syndrome (5) and later in myotonic dystrophy and other disorders. Myotonic dystrophy type 1 (DM1) is caused by the expansion of a (CTG)–(CAG) repeat in the DMPK gene (6). To date, at least nine distinct loci show instability with the same (CAG)–(CTG) repeat. These diseases increase in severity with earlier onset in successive generations and have no cure. Although the pathological states show different characteristics, one common feature among them is that the affected repetitive DNA unit has expanded beyond the number of repeats found in the healthy population. Therefore, the expansion of triplet repeats represents a novel mutational mechanism. More recently, other disorders have been associated with the expansion of non-trinucleotide motifs, such as the CCTG tetranucleotide in myotonic dystrophy type 2 (7).
DNA secondary structures may be considered as a common and causative factor for triplet expansion (8–10) but the molecular mechanisms causing the instability are unknown and remain a subject of intensive study. Even if no therapeutic approach is currently available to prevent or revert repeat expansion, in vitro studies suggest that repeat deletion could be induced by various chemotherapeutic agents (11,12); thus, opening a new field of study aimed at the design of trinucleotide repeat-specific ligands. Preliminary results suggest that selective recognition of trinucleotide repeat structures is possible (S. Amrane, unpublished data), but the rational design of such ligands should be facilitated by knowledge of the structure and energetics of their nucleic acid target.
Repetitive CNG sequences are susceptible to the formation of duplexes by self-folding, forming two Watson–Crick G–C pairs and one mismatch pair (13–16). Long hairpins have long lifetimes and inhibit duplex reannealing (17,18). A controversy remains on how different are these structures from classical B-DNA. Besides forming ‘slipped duplexes’ (19), CGG repeats have been reported to form quadruplexes (20–23). However, a recent study demonstrated that CGG repeats are reluctant to form tetraplexes under physiological conditions and this structure is unlikely to be involved in the disease (24): these sequences preferentially fold into antiparallel homoduplexes or hairpins in a length-dependent manner.
Concerning the structure of the bimolecular (CTG–CAG)n duplex, several results revealed a ‘polyhairpin concept’. Studies on (CAG–CTG)30,50 showed that these duplexes formed alternative DNA duplex structures named SDNA and SiDNA (9,19,25–27). This conclusion was reached using atomic force microscopy, electron microscopy, native gel electrophoresis, Mung Bean (MB) and T7 endonuclease cleavage assays. SDNA is a well-described bimolecular polyhairpin structure composed of multiple short (CTG)n and (CAG)n (n = 1–10) slipped-out structures with a hairpin-like single-stranded character. SDNA is formed in the absence of replication when the two complementary strands have the same length. Instead, SiDNA is formed during replication between two complementary strands with a different number of repetitions and results in repeat expansion or deletion. SiDNA is composed of one major slipped-out structure containing 20–30 repeats of the longest (CTG)n or (CAG)n strand.
To date, only the short hairpin structures (containing <10 repeats) involved in the formation of SDNA have been clearly established (15,28) revealing an intramolecular stem with several repetitions of a T·T or A·A mismatch sandwiched between 2 G–C base pairs. In contrast, the nature of the structures observed for longer repeats involved in the formation of SiDNA is still unclear. Even if several studies have previously demonstrated that long trinucleotide repeats adopt more compact structures than the short ones, suggesting the hypothesis of a multi-folded structure (16,29), but little is known about their exact conformation. The elucidation of this point is the purpose of our study; i.e. the analysis of the conformational properties of individual (CTG)n or (CAG)n strands, which may constitute the single slipped-out structures of particular DNA regions.
Initial thermodynamic studies showed the stabilities of CAG and CTG hairpins to be nearly identical under physiological salt concentrations in vitro (17). In contrast, Völker et al. (30) showed that, within a conformationally confined system, (CAG)6 and (CTG)6 form stable, ordered structures with the former triplet less stable than the latter, as also recently confirmed by another group (31). In this study, we analysed the folding of (CTG)n and (CAG)n individual sequences by using biophysical [UV-absorbance, circular dichroism (CD) and microcalorimetry] and biochemical techniques (native gel electrophoresis and enzymatic footprinting). In agreement with the previous results, we found that the folding of these sequences is intramolecular rather than bimolecular and that these structures are stable in physiological conditions. The study of the thermodynamic data obtained by thermal denaturation and microcalorimetry led us to propose an intramolecular ‘bis-hairpin’ like model for (CTG)n and (CAG)n individual strands (n = 12–25), with longer strands possibly giving rise to the formation of multi-branched hairpins. These structures are actually distinct from the multiple hairpins found in SDNA and SiDNA, in which several short hairpins protrude from a DNA duplex at different positions. Our model suggests that each slipped-out single-stranded region found in SDNA and SiDNA can actually fold into structures more complex than previously thought, as soon as 10–12 repeats are present in a single protruding loop. Similar results were also found for (CAG)n repeats.
MATERIALS AND METHODS
Oligodeoxynucleotides
Oligodeoxynucleotide probes were synthesized by Eurogentec (Belgium) on the 0.2 or 1 μmol scale. As all oligomers studied here correspond to DNA, the ‘d-’ prefix was omitted from most sequences. Purity was checked by gel electrophoresis. All concentrations were expressed in strand molarity using a nearest-neighbour approximation for the absorption coefficients of the unfolded species (32).
Thermal difference spectra for CTG and CAG trinucleotide repeats
The thermal difference spectrum (TDS) of a nucleic acid is obtained by simply recording the UV-absorbance spectra of the unfolded and the folded states at temperatures, respectively, above and below its melting temperature (Tm). The difference between these two spectra is defined as the TDS. The TDS has a specific shape that is unique for most structures (33,34) (J.L. Mergny et al., manuscript in preparation); thus, providing a simple, inexpensive and rapid method to gain structural insight into nucleic acid structures, both DNA and RNA, ranging from short oligomers to polynucleotides. Spectra were recorded between 220 and 335 nm with a Kontron Uvikon 940 UV/Vis spectrophotometer using quartz cuvettes with an optical pathlength of 0.2 or 1 cm. The differential spectrum of a CTG repeat structure gives a maximum differential absorbance at an unusually high wavelength (∼277 nm) (34).
UV-melting experiments
The thermal stability of the different trinucleotide repeat structures was estimated by heating/cooling experiments, recording the UV-absorbance at several wavelengths as a function of temperature using a Kontron Uvikon 940 spectrophotometer thermostated with an external ThermoNeslab RTE111 or ThermoHaake Phoenix C25P1 waterbath. The temperature of the bath was typically increased or decreased at a rate of 0.2°C/min, using 0.2 or 1 cm pathlength quartz cuvettes. All experiments were carried out in 10 mM sodium cacodylate buffer (pH 7.0) containing 30–500 mM KCl. Taking into consideration the TDSs and the high strand concentrations used for some UV-melting experiments, we chose to record the denaturation process at 290 nm (using cuvettes of 0.2 cm optical pathlength for the highest concentrations) in order to obtain an absorbance between 0.1 and 1.5. On the contrary, for the thermodynamic analysis (see below), lower strand concentrations were used and the denaturation process was followed at 275 nm where the signal is maximal. However, the observed melting temperatures were in excellent agreement (usually within 0.5°C, data not shown) with the ones determined at other wavelengths (e.g. 260 nm). All melting profiles were perfectly reversible at the chosen temperature gradient, demonstrating that these curves correspond to true equilibrium curves (35).
Thermodynamic analysis
For all parameters listed below, the assumed direction of the thermal process is the single-strand-to-hairpin transition. The thermal reversibility and the known molecularity of the process allow one to calculate the value of the equilibrium constant (Ka) assuming a simple two-state transition model. One must first convert absorbance measurements into folded fraction by manually selecting two baselines corresponding to the completely folded and unfolded form. An uncertainty may therefore arise because of the subjectivity in baseline determination. Starting from the classical Gibbs enthalpy equation,
one can write, for a reversible reaction [in which ΔG° = −RT ln(Ka)], the following van't Hoff equation:
where T is the temperature in Kelvin, while and are, respectively, the standard enthalpy and the entropy change of the reaction. In other words, provided that and are temperature-independent (see below), the so-called van't Hoff representation or Arrhenius plot [ln(Ka) versus 1/T] should give a straight line, with a slope of and a y-axis intercept of . The of this reversible reaction is called the van't Hoff enthalpy and is defined by
In the case of temperature-dependent enthalpies and entropies, one will obtain a significant deviation from linearity. Nevertheless, the above equation is still valid, but the slope at each point may be different, leading to temperature-dependent and values. van't Hoff enthalpies and entropies are said to be ‘model-dependent’: they rely on a two-state equilibrium hypothesis. They are, therefore, less robust than the ‘model-independent’ thermodynamic values provided by calorimetry (see below).
Determination of
The linear fit of an Arrhenius plot assumes that ΔH° is temperature-independent, which in turn means that . is the heat capacity change at constant pressure which is occurring during the thermal process (considered in the single-strand-to-hairpin direction). and are linked to by the following relations:
The hypothesis (widely assumed for nucleic acids) is now challenged. Several experiments have demonstrated that the of a duplex (36–40) or a triplex (41) may significantly depend on the temperature. In the case of UV-melting curves it is often difficult to provide an estimate of (35) because of baseline assumption problems. A small experimental error in ln(Ka) may obscure the temperature dependence of (42). Nevertheless, the Arrhenius curves analysed here were so deviant from linearity that reliable non-zero values could be extracted by fitting the plot of ln(Ka) versus 1/T with Kaleidagraph 3.5, according to the following equation:
where Tm is the melting temperature (in Kelvin) of the single-strand-to-hairpin renaturation process, while and are, respectively, the enthalpy and entropy change of the thermal transition at the melting temperature. In our case, they can be reasonably considered equivalent to the and , respectively. The heat capacity change obtained by non-linear fitting of the Arrhenius plot will be referred to in the text as .
Other methods may be used to measure the heat capacity change such as differential scanning calorimetry (DSC), which uses the difference between the pre- and the post-transition baselines (37), or even better, isothermal titration calorimetry (ITC). However, the DSC method is not always straightforward and requires an optimized experimental setting (40) whereas ITC is not simply applicable to intramolecular reactions.
Differential scanning calorimetry
Microcalorimetry experiments were performed on the (CTG)n oligomers using a Nano DSC-II microcalorimeter (CSC) driven by a DSC-run software. The oligonucleotides were dissolved at concentrations ranging from 40 to 200 μM in 10 mM sodium cacodylate buffer at pH 7 containing 30 mM KCl. Buffer and oligo solutions were carefully degassed prior to their utilization and their thermal profiles were analysed in the 0–95°C temperature range at a scan rate of 1°C/min. An initial calibration of the instrument was performed by filling both cells with the buffer solution and accumulating several scans until the balance was reached. Then, the DSC profile of the oligo was obtained by loading the solution of the oligonucleotide into the sample cell during the last cooling scan of the calibration experiment, leaving the buffer solution into the reference cell (load-on-the-fly method). With this method it was possible to execute a preliminary setting up of the instrument and to perform a real experiment in the same set of scans, without compromising the shape and the calorimetric values of the sample curve. A minimum of six scans was collected for each experiment. Substraction of the baseline and calculation of the thermodynamic parameters were carried out using the Cp-Calc software (Applied Thermodynamics). We observed that for all sequences tested here, the Tm values found by DSC and UV-melting analysis were in good agreement. The systematic small (≈2°C) difference in favour of the DSC Tm is the result of the calorimetric definition of the melting temperature, which corresponds to the temperature of maximum heat release/uptake rather than to the temperature of half association/dissociation. The real Tm (half association/dissociation temperature) is therefore ∼2°C below the DSC Tm, referred to in the text as ‘’. The calorimetric enthalpy () and entropy () for the transition process were determined in a model-independent way from the DSC curve. Comparison of the with the van't Hoff value obtained by the UV-thermal curve () allowed us to confirm or infirm the correctness of the previously assumed two-state model used to describe the entire thermal process (). Additional fitting of the experimental DSC curve with a more general model equation (provided by the Cp-Calc software) and its resolution by deconvolution analysis allowed us to obtain the value of for the whole process, as well as the thermal profile of the ‘daughter’ subcurves corresponding to the intermediate transitions. Deconvolution of the DSC curves represents one of the major advantages of the calorimetric analysis over the spectroscopic one. In fact, while by UV analysis small differences in melting temperature between independent subunits may be masked by an even smaller conformational change in the overall structure (giving rise to an apparent two-state transition profile of the UV-thermal curve), deconvolution of the calorimetric curve into its components can allow for unravelling intermediate states.
Nuclease susceptibility assays
Prior to structural probing reactions, 5′ end-labelled oligonucleotides were subjected to denaturation by heating the samples at 90°C for 3 min and then slowly cooled to room temperature. (CTG)n sequences (with n = 15, 16, 20 or 25) were analysed by enzymatic probing in the presence of increasing concentrations of S1 nuclease (0.1–4 U/μl) and MB nuclease (1–10 U/μl). For both nucleases (Promega), limited digestions of 5′ end-labelled oligonucleotides were performed at 30°C for 5 min in a buffer composed of 10 mM Tris–HCl (pH 7.2), 10 mM MgCl2 and 30 mM KCl. All the reactions were stopped by adding an equal volume of 100% formamide solution and heating for 3 min at 90°C. The nuclease digestion products were subjected to electrophoretic runs (70 W, 1500 V, 90 min) on denaturing polyacrylamide gels (15%) containing 7 M urea/TBE 1× and revealed on a phosphorimager screen (Typhoon; Molecular Dynamics). The cleavage sites for the two nucleases were determined by comparison of the enzymatic digestion products with size markers corresponding to (CAG)n treated with formic acid, (CTG)n treated with DMS and seven non-treated size markers: (CTG)12, (CTG)10, (CTG)8, (CTG)7, (CTG)6, (CTG)4 and (CTG)2. This determination was not straightforward because of the differences in the cleavage sites between chemical reactions (HCOOH or DMS) and enzymatic reactions (S1 and MB nucleases). On the one hand, the chemical reactions produce oligonucleotides with a 3′-phosphate end, whereas on the other hand, the enzymatic reactions produce sequences with a 3′-OH end. Therefore, an untreated size marker and its corresponding one obtained upon chemical cleavage of the parent oligo may not be equally accelerated by the electric field, although they have the same sequence [e.g. the (CTG)7 untreated size marker and its corresponding one obtained upon treatment of (CTG)20 with formic acid].
RESULTS
Confirmation of the folded form and intramolecular folding of CTG repeats
Several groups have previously demonstrated that repeats adopt an intramolecular duplex structure [for a recent example see (31)]. For this reason, we will very briefly describe the experiments that were carried out to confirm this model in our system. As shown in Supplementary Figures S1A and S1B, DNA composed of pure (CTG)n repeats (n = 4–10) exhibit fast, strand concentration-independent, mobility on polyacrylamide gels (29,43–45) in good agreement with an intramolecular structure. This increased mobility is almost completely lost under denaturing conditions (Supplementary Figures S1C and S1D). CD spectra of CTG oligomers are similar to the CD spectral signature of B-DNA with a negative peak ∼255 nm and a positive peak ∼285 nm (Supplementary Figure S2) (31). The concentration dependence of the melting temperature, Tm, was measured to clarify the type of structure formed (Figure 1). Plots of Tm versus C0 (where C0 is the total strand concentration) for a (CTG)n series of oligomers, where n = 8, 15 or 25, produced near-zero slopes (Figure 1B), indicating that unimolecular denaturations as also reported previously (31,46).
Figure 1.
UV-melting curves. (A) Example of UV-absorbance denaturation profile (recorded at 290 nm) at two different concentrations (50 μM, left y-axis and 1 μM, right y-axis) of the (CTG)25 sequence in a 10 mM sodium cacodylate (pH 7.0) buffer containing 30 mM KCl. (B) Concentration dependence of the Tm for three different oligonucleotides: (CTG)8, squares; (CTG)15, circles; and (CTG)25, diamonds.
Thermal absorbance difference spectra
Another method, we recently proposed, to study the conformation of a nucleic acid is to record the ‘thermal absorbance difference spectrum’ between its high and low temperature UV-absorbance spectrum (33,34). The normalized TDS has a shape which is specific for each nucleic acid conformation studied so far, ranging from duplexes to quadruplexes: as shown in Figure 2A, the shapes of TDS for (CTG)n (n = 8–20) were all very close (red curves) suggesting that these oligomers adopt a similar folded conformation. Furthermore, these spectra with two maxima ∼275 and 235 nm are highly reminiscent of pure GC-rich B-DNA TDS [blue curve and (34)]. A similar analysis was performed for CAG repeats (Figure 2B); although the TDS were not superimposable (compare panels A and B), these spectra were also reminiscent of GC-rich B-DNA TDS, but completely different from the ones of other structures such as quadruplexes (33). It is interesting to note that the CTG spectra are very close to the 100% GC spectra (47) (Figure 2A, in blue; these duplexes correspond to sequences where only GC base pairs are formed), whereas CAG repeats resemble mixed duplexes (Figure 2B, in black; these duplexes involve 67% GC/33% TA base pairs).
Figure 2.
Thermal differential absorbance data. Normalized absorbance thermal difference spectra (TDS) of (CTG)8–20 (A) and (CAG)8–20 (B) (TDS were obtained by high temperature minus low temperature absorbance spectra in a 10 mM sodium cacodylate buffer at pH 7 containing 30 mM KCl). The trinucleotide spectra are shown in red. Three representative TDS of pure GC duplexes (in blue) or 67% GC duplexes (in black) are also presented for comparison with the trinucleotide sequences.
Analysis of the melting profiles
Careful analysis of the UV-melting profiles revealed several observations. First, the melting temperature of (CTG)n and (CAG)n was weakly dependent on oligonucleotide length (example provided in Supplementary Figure S3; see also Figure 1B). For the CTG oligomers tested here, the Tms varied between 54 (n = 6) and 58.7°C (n = 25) (Table 1), while for (CAG)n the Tms varied between 51.6 (n = 6) and 54°C (n = 25) (Supplementary Table S1).
Table 1.
Thermodynamic parameters for (CTG)n repeats
| na | (°C)b | (°C)c | (kcal/mol)d | (kcal/mol)e | (cal/mol/K)d | (cal/mol/K)e | (cal/mol/K)f | (cal/mol/K)g | |
|---|---|---|---|---|---|---|---|---|---|
| 6 | 54 | 55.1 | −33.0 | −22.6 | 1.28 | −101 | −69 | −450 | nd |
| 8 | 56.8 | 58.1 | −44.0 | −40.5 | 1.09 | −133 | −122 | −1030 | −380 |
| 10 | 57 | 60.1 | −51.0 | −45.9 | 1.11 | −155 | −138 | −710 | −720 |
| 11 | 57.1 | 59.4 | −53.9 | −51.0 | 1.06 | −165 | −153 | −1360 | −1040 |
| 12 | 57.8 | 60.0 | −56.6 | −62.8 | 0.90 | −171 | −188 | −1410 | −1210 |
| 13 | 57.7 | 60.1 | −52.7 | −62.6 | 0.77 | −159 | −188 | −1700 | nd |
| 14 | 57.6 | 60.8 | −52.6 | −78.7 | 0.67 | −159 | −235 | −1510 | nd |
| 15 | 57.7 | 59.6 | −51.9 | −82.1 | 0.63 | −156 | −293 | −1500 | nd |
| 16 | 57.7 | 61.4 | −51.5 | −95.2 | 0.54 | −156 | −296 | −1500 | nd |
| 20 | 57 | 59.3 | −56.5 | −125.7 | 0.45 | −171 | −385 | −2060 | −1260 |
| 25 | 58.7 | 61.3 | −61.7 | −147.2 | 0.42 | −186 | −440 | −2120 | −1880 |
For all parameters listed, the assumed direction is the single-strand-to-hairpin transition. nd, not determined.
aNumber of (CTG) repeats.
bMelting temperature deduced from the UV-melting curve.
c‘Melting’ temperature deduced from the DSC profile.
dThermodynamic parameters deduced from the UV-melting curves, using a non-linear fit of the Arrhenius plots, where the is close to the ΔH° determined at the . (Error bars for ΔH° values are shown in Figure 4C; highest relative error of 6.4%.)
eThermodynamic parameters deduced from the DSC profiles. (Error bars for ΔH° values are shown in Figure 4C; highest relative error of 5.1%.)
fHeat capacity change deduced from non-linear fitting of the Arrhenius plots.
gHeat capacity change deduced from general model fitting of the DSC profiles.
Second, the Arrhenius representation of the melting curves also revealed a more complex behaviour (35). Determination of the folded fraction for the two samples analysed in Supplementary Figure S3 led to the Arrhenius plots are shown in Figure 3. A significant and reproducible deviation from linearity is seen in both cases. As described previously (Materials and Methods), it is possible to fit these curves to obtain the values (Table 1). Assuming a reaction in the single-strand-to-hairpin direction, these values were found to be always negative, in agreement with the studies on different DNA structures but with surprisingly high absolute values for long sequences [e.g. (CTG)20, (CTG)25 and (CAG)25]. Additionally, even if some discrepancies may be found, the values deduced from van't Hoff plots () were generally in good agreement with those deduced from calorimetry (; Tables 1 and S1).
Figure 3.
Arrhenius plots [(ln(Ka) versus 1/T)] for (CTG)n sequences. These plots were deduced from the UV-absorbance denaturation profiles recorded at 275 nm of two different oligonucleotides: 5 μM (CTG)10 (open circles) and 5 μM (CTG)25 (closed circles).
Third, the thermal stability of these trinucleotide repeats was analysed as a function of KCl concentration. As expected, the Tm values for the (CTG)8 oligomer were found to be indeed dependent on KCl concentration (Supplementary Figure S4): a 10-fold increase in potassium concentration led to a 7°C increase in Tm (from 50°C at 10 mM KCl to 57°C at 100 mM KCl). This ionic strength dependence is somewhat lower than previously reported (16) and lower than expected for regular double-stranded hairpins.
Calorimetric analysis of (CTG)n
The calorimetric values of the enthalpy () and entropy () of renaturation for the CTG oligomers are reported in Table 1 and an example of a DSC denaturation run of (CTG)25 is provided in Figure 4A. As in the UV-thermal analysis, the determined by DSC was almost sequence-length independent (Figure 4B). Next, the van't Hoff enthalpy determined by the analysis of the UV-thermal transition curve () was compared to the calorimetric value obtained by DSC () (Figure 4C and D). For relatively short sequences (n = 4–12) the ratio was close to 1 (1.08 ± 0.13; Table 1). The similarity of the two values validated the hypothesis of the two-state model used in the van't Hoff analysis to describe the thermal transition of the intramolecular trinucleotide repeat. However, in the case of longer sequences, the ratio dramatically dropped to values ∼0.4 (Figure 4D). The failed to become more negative with increased repeat number, whereas continued to increase (in absolute terms) explaining why the agreement was lost (Figure 4C and Table 1). The difference was highly significant as the errors bars (Figure 4C) on and were 6% or lower.
Figure 4.
Enthalpy determination for (CTG)n. (A) Typical differential scanning calorimetric profile for (CTG)25 denaturation. The solid line is the thermogram and the dashed line is the determined baseline. Integration of the area between the thermogram and the baseline yields to the model-independent calorimetric of the transition (in this case, in the hairpin-to-single-strand direction; hence, a positive ; the opposite reaction—formation of the hairpin—is associated with a negative ). Thermograms were obtained in a 10 mM sodium cacodylate buffer at pH 7 containing 30 mM KCl. (B) Comparison between the and the Tm found by DSC (closed circles) and by UV-melting profiles (open circles), respectively, as a function of sequence-length (number of CTG repeats). Linear fits (which are poor, R = 0.65, solid line and 0.69, dashed line for DSC and UV data, respectively) are shown. (C) Comparison between the calorimetric model-independent (closed circles) and the deduced from a van't Hoff analysis of the UV-melting profiles (open circles) as a function of sequence-length (number of CTG repeats). The renaturation process is considered in the single-strand-to-hairpin direction; hence, negative values of ΔH°. Errors bars on and were 6% or lower. (D) Ratio between the and the as a function of sequence-length (number of repeats).
Calorimetric analysis of (CAG)n
The thermal behaviour observed for the CTG repeats was compared with that obtained from CAG repeats of identical length under the same experimental conditions. An example of a DSC denaturation run for a (CAG)n oligo is provided in Figure 5A. Again, we observed that, for all sequences tested, the and the Tm values found by DSC and UV-melting analysis were in good agreement and almost independent on sequence-length (Figure 5B). As shown in Figure 5C and D, the values of and for the (CAG)n thermal denaturations were compared and plotted against the number of triplet repeats, leading to the same conclusions drawn for the (CTG)n analogs (see also Supplementary Table S1).
Figure 5.
Enthalpy determination for (CAG)n. (A) Typical differential scanning calorimetric profile for (CAG)25 denaturation. Identical legend as in Figure 4. Thermograms were obtained in a 10 mM sodium cacodylate buffer at pH 7containing 30 mM KCl. (B) Comparison between the and the Tm found by DSC (closed circles) and by UV-melting profiles (open circles), respectively, as a function of sequence-length (number of CAG repeats). (C) Comparison between the calorimetric model-independent (closed circles) and the deduced from a van't Hoff analysis of the UV-melting profiles (open circles) as a function of sequence-length (number of CAG repeats). The renaturation process is considered in the single-strand-to-hairpin direction, hence negative values of ΔH°. (D) Ratio between the and the as a function of sequence-length (number of repeats).
Enzymatic probing of long CTG repeats
Nuclease sensitivity studies were performed with S1 and MB nuclease that are two single-strand specific endonucleases. The bases contained in the single-stranded regions of the structures adopted by long CTG repeats are preferentially cleaved with respect to the ones contained in the double-stranded regions. The cleavage sites for the two nucleases were determined by comparison with size markers (Materials and Methods). The enhanced MB digestion of (CTG)15 occurred at three main regions (Figure 6A): (i) two sites at the 3′ end, G13 and G14; (ii) five sequence-centred sites around the (CTG)7 (T4) marker, i.e. C7, T7, G7, C8 and T8 resulting from a TGCT-loop; and (iii) two sites around the 5′ end region identified as G1 and C2 representing a CTG-loop. S1 digestion of (CTG)15 presents only two sites around the same 5′ end region cleaved by the MB nuclease: G1 and C2 (Figure 6B). Similarly, two loops were characterized for (CAG)15: one central loop composed of two repeats (CAGCAG) and one CAG loop at the 5′ end (Supplementary Figure S5).
Figure 6.
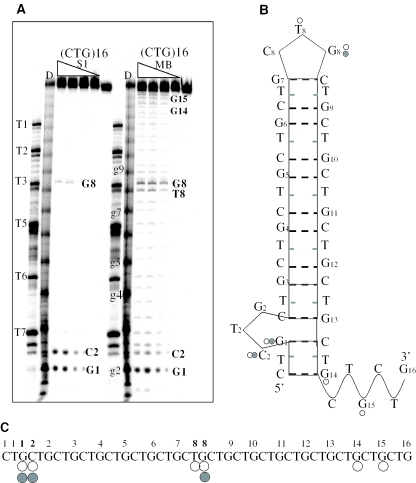
Enzymatic probing of (CTG)15 and (CTG)20. Two sequences, (CTG)15 and (CTG)20, were 5′ radiolabelled, incubated at 30°C for 5 min in a buffer composed of 10 mM Tris–HCl, pH 7.2, 10 mM MgCl2 and 30 mM KCl with (A) increasing concentrations of MB nuclease 1–3.5–7–10 U/μl. (B) Increasing concentrations of S1 nuclease (NS1) 0.2–0.4–0.8 U/μl then loaded on a denaturing 15% polyacrylamide gel. Length markers were also loaded in order to identify the residues cleaved by the nucleases. Lane F is a guanine- and adenine-specific ladder corresponding to (CAG)20 treated with formic acid (where gi indicates the G residue from the i-th CTG repeat); lane D is a guanine-specific ladder corresponding to (CTG)15 treated with DMS (where gi indicates the G residue from the i-th CTG repeat) and T1, T2, T3, T4, T5, T6 and T7 correspond to non-treated size markers, respectively, (CTG)12 (CTG)10, (CTG)8, (CTG)7, (CTG)6, (CTG)4 and (CTG)2. (C) The schematic diagram shows the (CTG)15 folding pattern which agrees with the nuclease susceptibility assays. Rather than a simple hairpin–loop model, thermodynamic analysis as well as enzymatic studies suggest a more complicated ‘bis-hairpin’ model. (D) Summary of the cleavage pattern of (CTG)15. Nucleotides cleaved by MB nuclease (open circles) or S1 nuclease (closed circles) are shown.
The enhanced digestion of (CTG)16 occurred at three main regions (Figure 7A): (i) two sites at the 3′ end: G14 and G15; (ii) two sequence-centred sites around the (CTG)8 (T3) marker: T8 and G8 resulting from a CTG-loop; and (iii) two sites around the 5′ end region identified as G1 and C2, indicating a CTG-loop. These two CTG-loops are conserved in (CTG)20 (Figure 6A and B) with the following digestion sites: (i) two sequence-centred sites around the (CTG)10 (T2) marker: T10 and G10 and (ii) two sites around the 5′ end region identified as G1 and C2.
Figure 7.
Enzymatic probing of (CTG)16 (A) (CTG)16 was 5′ radiolabelled, incubated at 30°C for 5 min in a buffer composed of 10 mM Tris–HCl, pH 7.2, 10 mM MgCl2 and 30 mM KCl with increasing concentrations of MB nuclease (1–3.5–7–10 U/μl) and increasing concentrations of S1 nuclease (0.2–0.4–0.8 U/μl). Samples were loaded on a denaturing 15% polyacrylamide gel and analysed as in Figure 6. (B) The schematic diagram shows the (CTG)16 folding pattern which agrees with the nuclease susceptibility assays. (C) Summary of the cleavage pattern of (CTG)16. Nucleotides cleaved by MB nuclease (open circles) or S1 nuclease (closed circles) are shown.
Finally, (CTG)25 presents the same kind of loops as (CTG)15 (Supplementary Figure S6): (i) four sequence-centred sites around the (CTG)12 (T1) marker: T12, G12, C13 and T13 resulting from a 4 nt loop; and (ii) two sites around the 5′ end region identified as G1 and C2, representing a CTG-loop. Thus, our data show that all these sequences present one centred loop of 3 or 4 nt and one loop at the 5′ end containing 3 nt (Figures 6C and 7B). In theory, this terminal loop could also correspond to DNA end fraying, but in this case we would have obtained four cleavage sites: C1, T1, G1 and C2 at the 5′ end region instead of the only two sites G1 and C2. Furthermore, two NMR studies on short CTG repeats sequences have demonstrated that the T–T mismatches, all over the stem of the hairpin, are very well stacked between the two adjacent CpG base pairs and are bound to each other by two hydrogen bonds (35). The DNA end fraying hypothesis is therefore unlikely, as will be confirmed in the following part.
(CTG)15 variant sequences
In order to confirm these results and to ensure that the proximal cuts of the 5′ end nucleotides do not represent DNA end fraying, the C and G bases of the single-strand regions of (CTG)15 previously defined by footprinting experiments were replaced by T bases. The and renaturation enthalpy of the modified oligonucleotide sequences were then measured by DSC and compared to the ones of the unmodified analog (Table 2). Replacement of C2 and G2 (as well as of G7 and C8) by two T bases did not influence the renaturation enthalpy. In fact, despite a little decrease in the (−2.5°C) of the modified sequences, their enthalpy remained around −80 kcal/mol as for the unmodified (CTG)15. This suggests that these four bases, i.e. C2, G2, G7 and C8, do not belong to the stem region of the hairpin but to the unpaired regions of the structure. The small difference in could be the result of little variations in the ionic conditions between the modified and the unmodified samples or errors in data acquisition/baseline determination. More importantly, this is defined as the maximum of the DSC curve (Materials and Methods) and does not necessarily correspond to the real Tm (half association/dissociation temperature): it is therefore not the best parameter to assess the stability of a structure. At this purpose, the model-independent of the renaturation process is a more suitable parameter, directly related to the number of base pairs involved in the molecule. Thus, the constancy of this value for T-replacement of C2, G2, G7 and C8, undoubtedly indicates that these four bases do not belong to the stem region of the hairpin but to unpaired regions of the structure. In contrast, we obtained a dramatic decrease, in absolute value, of the renaturation enthalpy (from −80 till −54.8, −51.8 and −51.4 kcal/mol) by replacing C3, C7 and G8 by a T base, thus showing that these three bases belong instead to the stem region of the structure.
Table 2.
Thermodynamic parameters of (CTG)15 variant sequences
aValues deduced from the DSC profiles.
DISCUSSION
The goal of this study was to analyse the assumption of a quasi ‘normal’ hairpin duplex for trinucleotide repeats, namely the CTG and CAG repeat motifs. Little is known about the exact conformation of long trinucleotide repeats. Several studies have previously demonstrated that long trinucleotide repeats adopt more compact structures than the short ones, suggesting the hypothesis of a multi-folded structure rather than a single-hairpin (16,29). Few NMR or crystallographic studies have actually analysed ‘pure’ CNG repeats, as they are usually embedded into different sequence motifs [e.g. GCGGTTTGCGG in (48) and TGGCGGC in (49); for a review see (50)]. Disease-related CNG repeats exhibit a propensity for folding at chain lengths as short as 12 residues (14). CTG-containing sequences have been studied with a variety of other techniques, including PAGE, KMnO4 modification, P1 nuclease digestion, UV-absorbance and molecular dynamic simulations [reviewed in (51)]. To date, the hairpin structure of sequences shorter than 10 repeats has been clearly established (15,28), but the nature of the compact structures observed for longer repeats is still unclear.
In this paper, CD, electrophoresis, UV and DSC data revealed sharp structural transitions, in agreement with the formation of a rather simple canonical B-DNA hairpin, with a stem length growing with the repeat number. However, independent results indicate that the energetics and/or structure of these intramolecular trinucleotide repeats were significantly altered when compared with the canonical B-DNA, especially for (relatively) long (CTG)n sequences. Qualitatively similar results were obtained for (CAG)n repeats, suggesting that this behaviour may be a general phenomenon for (CNG)n oligonucleotides.
First, the thermal stability of this structure (its Tm) was very weakly dependent on the oligonucleotide length (Figures 4B and 5B and Supplementary Figure S3). The weak sequence-length dependence of the Tm of these repeats was previously observed for (CTG)10 and (CTG)30 (16); (CTG)10 and (CTG)25 (17); (CTG)10 and (CTG)15 (31) as well, but has never been systematically studied [with a notable exception for (CUG)n RNAs; n = 5–69 (52)]. In our experience, the thermal stability of DNA hairpins increases rapidly with the stem length, in complete opposition to what is observed here. This behaviour is therefore highly irregular for a simple hairpin system. Second, the analysis of the melting curves did not lead to simple Arrhenius plots, as significant and reproducible curvatures were observed. We actually had to manually select an unlikely lower baseline in order to ‘linearize’ the Arrhenius representations (Supplementary Figure S7). At least three non-exclusive reasons may be proposed to explain this non-linear behaviour: (i) a non-two-state transition (partially melted structures are significantly populated), (ii) two (or more) intermingled transitions occur or (iii) a single two-state transition occurs with a highly negative (i.e. a temperature-dependent ΔH°). This last possibility which was, until recently (53,54), overlooked for nucleic acids transitions, should be seriously considered here. The comparison of the model-dependent and model-independent ΔH° values provides interesting clues.
Length-dependent discrepancy between van't Hoff and calorimetry enthalpy
Several groups have reported important discrepancies between the van't Hoff enthalpy () and the calorimetry enthalpy () for various nucleic acids structures (55), although not for trinucleotides. For (CTG)n and (CAG)n repeats, we observed that the melting temperatures found by DSC and van't Hoff analysis of the UV-melting curves were always in reasonable agreement; however, the model-dependent and the model-independent enthalpies were not always identical (Figures 4C and 5C and Tables 1 and S1). For relatively short (CTG)n or (CAG)n sequences (n = 4–12) the ratio was close to 1, validating the hypothesis of the two-state model used in the van't Hoff analysis. In the case of longer sequences, the ratio dramatically dropped (Figures 4D and 5D) to values ∼0.4 (for CTG repeats) or 0.5 (for CAG repeats). This length-dependent difference between the two enthalpy values suggests the hypothesis of a non-two-state transition model for longer sequences. That is, above a critical threshold number of trinucleotide repetitions (n = 10–12), a change in the structural organization of the oligonucleotides occurs, going from a simple hairpin–stem model to a structure of higher complexity, composed of different independent units. Therefore,
Below 12 repeats, no significant difference between the two ΔH° values was observed, leading to their similar increase with sequence-length. For these relatively short sequences the two-state transition model was confirmed by deconvolution of the DSC profiles (Figure 8A) into a single transition, indicating their folding into a simple hairpin structure.
Beyond 12 repeats, instead, an increasing discrepancy between the ΔH° values appeared, with the model-independent still increasing in a length-dependent manner, whereas the two-state model-dependent remained quasi-constant. In contrast with a previous report (31), no plateau was observed in for sequences longer than 15 trinucleotide repeats (Figures 4C and 5C). For long oligomers, the two-state hypothesis was no longer valid, suggesting the formation of a more complex structure with at least two independent units. This observation was confirmed by deconvolution of the DSC profiles into a three-state equilibrium with two intermediate transitions (Figure 8B).
Figure 8.
Analysis of complex differential scanning calorimetry (DSC) data. The deconvolution of the (CTG)25 and (CTG)10 DSC profiles were carried out using the deconvolution program of the Cp-Calc software (Applied Thermodynamics). In the two cases, the reconstructed curves (dashed lines) are in good agreement with the experimental ones. (A) (CTG)10 is deconvoluted with only one transition indicating a single structural domain. Deconvolution data: ΔH(total)deconv = 50.8 kcal/mol; Tm = 59.9°C; and experimental data: ΔH(total)exp = 47.2 kcal/mol. (B) (CTG)25 is deconvoluted with two transitions corresponding to the melting of two independent domains of the structure. Deconvolution data: transition 1, ΔH = 56.4 kcal/mol, Tm = 57.9°C; and transition 2, ΔH = 79.9 kcal/mol, Tm = 61.7°C. ΔH(total)deconv = 136.3 kcal/mol. Experimental data: ΔH(total)exp = 142.8 kcal/mol.
In other words, for short sequences, folding into a single-hairpin structure is relatively simple, leading to a good concordance between and . For long sequences (at least 12), rather than adopting a regular long stem, the oligonucleotide seems to maintain a relatively short and constant length of the stem region while the remaining part fold into several (at least two) independent folding units, of not necessarily the same length, which melt in an independent fashion. In that case, a multi-branched hairpin should have a relatively apparent constant van't Hoff enthalpy, as seen by UV-melting analysis, but an increased calorimetric enthalpy, as observed by DSC. This different folding trend for longer sequences may be explained in terms of a favourable enthalpy–entropy compensation. In fact, although a long single-hairpin structure should indeed have a more favourable renaturation enthalpy (owing to maximization of the number of base pairs), it has not necessarily the most favourable free energy. On the contrary, branched hairpin structures may offer additional degrees of freedom, possibly lowering the unfavourable entropic term and thus explaining their preferential formation for longer sequences.
Nuclease assays
The enzymatic assays on long sequences (Figures 6 and 7 and Supplementary Figures S5 and S6), additionally confirmed by the DSC analysis of the (CTG)15-modified sequences (Table 2), suggested that a simple hairpin was not necessarily the chosen structure, as other bases close to the 5′ end of the sequence were cleaved by single-strand specific nucleases. However this is in contrast with the previous results obtained using other enzymatic and chemical probes (43,56). According to the biophysical and biochemical data obtained in this study, we propose a possible ‘bis-hairpin’ folding pattern for (CTG)15 and (CTG)16, as shown in Figures 6C and 7B.
The different number of cut sites between the (CTG)n sequences (n = 15, 16, 20 or 25) probably reflect the different number of bases in the loops for the odd and even number of repeats (57). At least two NMR studies (15,28) have demonstrated that (CTG)n sequences (n = 1–10), with an even number of repeats, fold into a hairpin structure containing a 4 nt TGCT-loop whereas an odd number of repeats leads to a 3 nt CTG-loop hairpin. It is interesting to note that, in our study, this tendency is reversed for longer sequences: (CTG)15 and (CTG)25 present a centred 4 nt TGCT-loop, while (CTG)16 and (CTG)20 present a centred 3 nt CTG-loop. Thus, this inversion confirms the presence of a second loop composed of an odd number of CTG repeats at the 5′ end of the sequence.
CONCLUSIONS
Above a critical threshold number of trinucleotide repetitions (n = 12–15), a change in the structural organization of the oligonucleotides occurs going from a simple hairpin–stem model to a structure of higher complexity composed of different independent units such as a bis- (or multi-branched) hairpin-like structure. In contrast with a recent report (31), our calorimetric data suggest that the enthalpic stability of this structure is not compromised as the length of the hairpin overcomes 15 repeats. The possible intramolecular bis-hairpin model could be generalized as an intramolecular ‘multi-branched’ hairpin model for pathological sequences as long as 3000 repetitions. The 3 or 4 nt bulges disseminated all over the repeated sequence could be involved in the instability process during DNA replication through the formation of slipped-out DNA structures, but may also represent an important feature to discriminate trinucleotide repeats against canonical B-DNA for targeting by small repeat-specific bulge-ligands. This instability has been ascribed to the energy of the structure but in our case it could be explained in terms of number of hairpin loops that introduce an additional steric parameter to the energetic one. These results have encouraged us to carry on further investigations for a better understanding of the thermodynamics of these repetitions, and stimulating additional studies directed to the structural elucidation of long trinucleotide repeat models. We will now pursue similar studies on other DNA and RNA trinucleotides [(CGG)n, (CCG)n and (CUG)n (58,59)] to see whether these observations may be extended to other CNG repeats.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank T. Garestier, J. Pylouster, A. De Cian, L. Guittat and L. Lacroix (MNHN, Paris, France) for helpful discussions. S.A. is the recipient of a ‘Fondation Jérome Lejeune’ PhD fellowship. This work was supported by an ARC grant (no. 3365), an INSERM ‘Equipement Mi-Lourd’ grant (to J.L.M.) and a French–South African exchange grant (to J.L.M. and M.M.). Funding to pay the Open Access publication charges for this article was provided by INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Timchenko L.T., Caskey C.T. Triplet repeat disorders: discussion of molecular mechanisms. Cell. Mol. Life Sci. 1999;55:1432–1447. doi: 10.1007/s000180050383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cummings C.J., Zoghbi H.Y. Trinucleotide repeats: mechanisms and pathophysiology. Annu. Rev. Genomics Hum. Genet. 2000;1:281–328. doi: 10.1146/annurev.genom.1.1.281. [DOI] [PubMed] [Google Scholar]
- 3.Bowater R.P., Wells R.D. The intrinsically unstable life of DNA triplet repeats associated with human hereditary disorders. In: Moldave K., editor. Progress in Nucleic Acid Research. Vol. 66. San Diego, CA: Academic Press Inc.; 2001. pp. 159–202. [DOI] [PubMed] [Google Scholar]
- 4.Everett C.M., Wood N.W. Trinucleotide repeats and neurodegenerative disease. Brain. 2004;127:2385–2405. doi: 10.1093/brain/awh278. [DOI] [PubMed] [Google Scholar]
- 5.Oberle I., Rousseau F., Heitz D., Kretz C., Devys D., Hanauer A., Boue J., Bertheas M., Mandel J. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 6.Brook J.D., McCurrach M.E., Harley H.G. Molecular basis of myotonic dystrophy expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family number. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 7.Liquori C.L., Ricker K., Moseley M.L., Jacobsen J.F., Kress W., Naylor S.L., Day J.W., Ranum L.P.W. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 8.McMurray C.T. DNA secondary structure: a common and causative factor for expansion in human disease. Proc. Natl Acad. Sci. USA. 1999;96:1823–1825. doi: 10.1073/pnas.96.5.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinden R.R. Biological implications of the DNA structures associated with disease causing triplet repeats. Am. J. Hum. Genet. 1999;64:346–353. doi: 10.1086/302271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleary J.D., Pearson C.E. Replication fork dynamics and dynamic mutations: the fork-shift model of repeat instability. Trends Genet. 2005;21:272–280. doi: 10.1016/j.tig.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Hashem V.I., Sinden R.R. Chemotherapeutic induced deletion of expanded triplet repeats. Mutat Res. 2002;508:107–119. doi: 10.1016/s0027-5107(02)00190-2. [DOI] [PubMed] [Google Scholar]
- 12.Hashem V.I., Pytlos M.J., Klysik E.A., Tsuji K., Khajav M., Ashizawa T., Sinden R.R. Chemotherapeutic deletion of CTG repeats in lymphoblast cells from DM1 patients. Nucleic Acids Res. 2004;32:6334–6346. doi: 10.1093/nar/gkh976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X., Mariappan S.V.S., Catasti P., Ratliff R., Moyzis R.K., Laayoun A., Smith S.S., Bradbury E.M., Gupta G. Hairpins are formed by the single DNA strands of the fragile X triplet repeats: structure and biological implications. Proc. Natl Acad. Sci. USA. 1995;92:5199–5203. doi: 10.1073/pnas.92.11.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng M.X., Huang X.N., Smith G.K., Yang X.Y., Gao X.L. Genetically unstable CXG repeats are structurally dynamic and have a high propensity for folding. An NMR and UV spectroscopic study. J. Mol. Biol. 1996;264:323–336. doi: 10.1006/jmbi.1996.0643. [DOI] [PubMed] [Google Scholar]
- 15.Mariappan S.V.S., Garcia A.E., Gupta G. Structure and dynamics of the DNA hairpins formed by tandemly repeated CTG triplets associated with myotonic dystrophy. Nucleic Acids Res. 1996;24:775–783. doi: 10.1093/nar/24.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petruska J., Arnheim N., Goodman M.F. Stability of intrastrand hairpin structures formed by the CAG/CTG class of DNA triplet repeats associated with neurological diseases. Nucleic Acids Res. 1996;24:1992–1998. doi: 10.1093/nar/24.11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gacy A.M., McMurray C.T. Influence of hairpins on template reannealing at trinucleotide repeat duplexes: a model for slipped DNA. Biochemistry. 1998;37:9426–9434. doi: 10.1021/bi980157s. [DOI] [PubMed] [Google Scholar]
- 18.Paiva A.M., Sheardy R.D. The influence of sequence context and length on the kinetics of DNA duplex formation from complementary hairpins possessing (CNG) repeats. J. Am. Chem. Soc. 2005;127:5581–5585. doi: 10.1021/ja043783n. [DOI] [PubMed] [Google Scholar]
- 19.Pearson C.E., Sinden R.R. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry. 1996;35:5041–5053. doi: 10.1021/bi9601013. [DOI] [PubMed] [Google Scholar]
- 20.Fry M., Loeb L.A. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen F.M. Acid-facilitated supramolecular assembly of G-quadruplexes in d(CGG)4. J. Biol. Chem. 1995;270:23090–23096. doi: 10.1074/jbc.270.39.23090. [DOI] [PubMed] [Google Scholar]
- 22.Darlow J.M., Leach D.R.F. Secondary structures in d(CGG) and d(CCG) repeat tracts. J. Mol. Biol. 1998;275:3–16. doi: 10.1006/jmbi.1997.1453. [DOI] [PubMed] [Google Scholar]
- 23.Weisman-Shomer P., Naot Y., Fry M. Tetrahelical forms of the fragile X syndrome expanded sequence d(CGG)n are destabilized by two heterogeneous nuclear ribonucleoprotein-related telomeric DNA-binding proteins. J. Biol. Chem. 2000;275:2231–2238. doi: 10.1074/jbc.275.3.2231. [DOI] [PubMed] [Google Scholar]
- 24.Fojtík P., Kejnovská I., Vorlícková M. The guanine-rich fragile X chromosome repeats are reluctant to form tetraplexes. Nucleic Acids Res. 2004;32:298–306. doi: 10.1093/nar/gkh179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearson C.E., Wang Y.H., Griffith J.D., Sinden R.R. Structural analysis of slipped-strand DNA (SDNA) formed in (CTG)n·(CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res. 1998;26:816–823. doi: 10.1093/nar/26.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam M., Montgomery S.E., Kekis M., Stollar B.D., Price G.B., Pearson C.E. Slipped (CTG)–(CAG) repeats of the myotonic dystrophy locus: surface probing with anti-DNA antibodies. J. Mol. Biol. 2003;332:585–600. doi: 10.1016/s0022-2836(03)00880-5. [DOI] [PubMed] [Google Scholar]
- 27.Pearson C.E., Tam M., Wang Y.H., Montgomery S.E., Dar A.C., Cleary J.D., Nichol K. Slipped-strand DNAs formed by long (CAG)·(CTG) repeats: slipped-out repeats and slip-out junctions. Nucleic Acids Res. 2002;30:4534–4547. doi: 10.1093/nar/gkf572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chi L.M., Lam S.L. Structural roles of CTG repeats in slippage expansion during DNA replication. Nucleic Acids Res. 2005;33:1604–1617. doi: 10.1093/nar/gki307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell J.E., Newbury S.F., McClellan J.A. Compact structures of d(CNG)n oligonucleotides in solution and their possible relevance to fragile X and related human genetic diseases. Nucleic Acids Res. 1995;23:1876–1881. doi: 10.1093/nar/23.11.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Völker J., Makube N., Plum G.E., Klump H.H., Breslauer K.J. Conformational energetics of stable and metastable states formed by DNA triplet repeat oligonucleotides: implications for triplet expansion diseases. Proc. Natl Acad. Sci. USA. 2002;99:14700–14705. doi: 10.1073/pnas.222519799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paiva A.M., Sheardy R.D. Influence of sequence context and length on the structure and stability of triplet repeat DNA oligomers. Biochemistry. 2004;43:14218–14227. doi: 10.1021/bi0494368. [DOI] [PubMed] [Google Scholar]
- 32.Cantor C.R., Warshaw M.M., Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 33.Mergny J.L., Phan A.T., Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 34.Alberti P., Hoarau M., Guittat L., Takasugi M., Arimondo P.B., Lacroix L., Mills M., Teulade-Fichou M.P., Vigneron J.P., Lehn J.M., et al. Triplex vs. quadruplex specific ligands and telomerase inhibition. In: Bailly C., Demeunynck M., Wilson D., editors. Small Molecule DNA and RNA Binders: From Synthesis to Nucleic Acid Complexes. Weinheim: Wiley VCH; 2002. pp. 315–336. [Google Scholar]
- 35.Mergny J.L., Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 36.Holbrook J.A., Capp M.W., Saecker R.M., Record M.T. Enthalpy and heat capacity changes for formation of an oligomeric DNA duplex: interpretation in terms of coupled processes of formation and association of single-stranded helices. Biochemistry. 1999;38:8409–8422. doi: 10.1021/bi990043w. [DOI] [PubMed] [Google Scholar]
- 37.Chalikian T.V., Volker J., Plum G.E., Breslauer K.J. A more unified picture for the thermodynamics of nucleic acid duplex melting: a characterization by calorimetric and volumetric techniques. Proc. Natl Acad. Sci. USA. 1999;96:7853–7858. doi: 10.1073/pnas.96.14.7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouzina I., Bloomfield V.A. Heat capacity effects on the melting of DNA. 1. General aspects. Biophys. J. 1999;77:3242–3251. doi: 10.1016/S0006-3495(99)77155-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouzina I., Bloomfield V.A. Heat capacity effects on the melting of DNA. 2. Analysis of nearest- neighbor base pair effects. Biophys. J. 1999;77:3252–3255. doi: 10.1016/S0006-3495(99)77156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jelesarov I., Crane-Robinson C., Privalov P.L. The energetics of HMG box interactions with DNA: thermodynamic description of the target DNA duplexes. J. Mol. Biol. 1999;294:981–995. doi: 10.1006/jmbi.1999.3284. [DOI] [PubMed] [Google Scholar]
- 41.Shindo H., Torigoe H., Sarai A. Thermodynamic and kinetic studies of DNA triplex formation of an oligohomopyrimidine and a matched duplex by filter binding assay. Biochemistry. 1993;32:8963–8969. doi: 10.1021/bi00085a030. [DOI] [PubMed] [Google Scholar]
- 42.Chaires J.B. Possible origin of differences between van't Hoff and calorimetric enthalpy estimates. Biophys. Chem. 1997;64:15–23. doi: 10.1016/s0301-4622(96)02205-3. [DOI] [PubMed] [Google Scholar]
- 43.Mitas M., Yu A., Dill J., Kamp T.J., Chambers E.J., Haworth I.S. Hairpin properties of single-stranded DNA containing a GC-rich triplet repeat: (CTG)15. Nucleic Acids Res. 1995;23:1050–1059. doi: 10.1093/nar/23.6.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chastain P.D., II, Eichler E.E., Kang S., Nelson D.L., Levene S.D., Sinden R.R. Anomalous rapid electrophoretic mobility of DNA containing triplet repeats associated with human disease genes. Biochemistry. 1995;34:16125–16131. doi: 10.1021/bi00049a027. [DOI] [PubMed] [Google Scholar]
- 45.Chastain P.D., Sinden R.R. CTG repeats associated with human genetic disease are inherently flexible. J. Mol. Biol. 1998;275:405–411. doi: 10.1006/jmbi.1997.1502. [DOI] [PubMed] [Google Scholar]
- 46.Gacy A.M., Goellner G., Juranic N., Macura S., McMurray C.T. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 47.Riesner D., Roemer R. Differential melting techniques and typical melting curves. In: Duchesne J., editor. Physico-Chemical Properties of Nucleic Acids. Vol. 2. NY: Academic Press; 1973. pp. 277–318. [Google Scholar]
- 48.Kettani A., Kumar R.A., Patel D.J. Solution structure of a DNA quadruplex containing the fragile X syndrome triplet repeat. J. Mol. Biol. 1995;254:638–656. doi: 10.1006/jmbi.1995.0644. [DOI] [PubMed] [Google Scholar]
- 49.Patel P.K., Bhavesh N.S., Hosur R.V. Cation-dependent conformational switches in d-TGGCGGC containing two triplet repeats of Fragile X Syndrome: NMR observations. Biochem. Biophys. Res. Commun. 2000;278:833–838. doi: 10.1006/bbrc.2000.3878. [DOI] [PubMed] [Google Scholar]
- 50.Patel D., Bouaziz S., Kettani A., Wang Y. Structures of guanine-rich and cytosine-rich quadruplexes formed in vitro by telomeric, centromeric, and triplet repeat disease DNA sequence. In: Neidle S., editor. Oxford Handbook of Nucleic Acid Structure. Oxford: Oxford University Press; 1999. pp. 389–454. [Google Scholar]
- 51.Mitas M. Trinucleotide repeats associated with human disease. Nucleic Acids Res. 1997;25:2245–2253. doi: 10.1093/nar/25.12.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian B., White R.J., Xia T.B., Welle S., Turner D.H., Mathews M.B., Thornton C.A. Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA. 2000;6:79–87. doi: 10.1017/s1355838200991544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mikulecky P.J., Feig A.L. Heat capacity changes in RNA folding: application of perturbation theory to hammerhead ribozyme cold denaturation. Nucleic Acids Res. 2004;32:3967–3976. doi: 10.1093/nar/gkh723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tikhomirova A., Taulier N., Chalikian T.V. Energetics of nucleic acid stability: the effect of delta Cp. J. Am. Chem. Soc. 2004;126:16387–16394. doi: 10.1021/ja046387d. [DOI] [PubMed] [Google Scholar]
- 55.Haq I., Chowdhry B.Z., Jenkins T.C. Calorimetric techniques in the study of high-order DNA–drug interactions. In: Chaires J.B., Waring M.J., editors. Drug Nucleic Acid Interaction. Vol. 340. San Diego, CA: Academic Press Inc.; 2001. pp. 109–149. [DOI] [PubMed] [Google Scholar]
- 56.Yu A., Dill J., Wirth S.S., Huang G., Lee V.H., Haworth I.S., Mitas M. The trinucleotide repeat sequence d(GTC)15 adopts a hairpin conformation. Nucleic Acids Res. 1995;23:2706–2714. doi: 10.1093/nar/23.14.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hartenstine M.J., Goodman M.F., Petruska J. Base stacking and even/odd behavior of hairpin loops in DNA triplet repeat slippage and expansion with DNA polymerase. J. Biol. Chem. 2000;275:18382–18390. doi: 10.1074/jbc.275.24.18382. [DOI] [PubMed] [Google Scholar]
- 58.Napierala M., Michalowski D., de Mezer M., Krzyzosiak W.J. Facile FMR1 mRNA structure regulation by interruptions in CGG repeats. Nucleic Acids Res. 2005;33:451–463. doi: 10.1093/nar/gki186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sobczak K., Krzyzosiak W.J. CAG repeats containing CAA interruptions form branched hairpins structures in spinocerebellar ataxia type 2 transcripts. J. Biol. Chem. 2005;280:3898–3910. doi: 10.1074/jbc.M409984200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.