Abstract
Long INterspersed Elements (LINE-1s or L1s) are abundant non-LTR retrotransposons in mammalian genomes that are capable of insertional mutagenesis. They have been associated with target site deletions upon insertion in cell culture studies of retrotransposition. Here, we report 50 deletion events in the human and chimpanzee genomes directly linked to the insertion of L1 elements, resulting in the loss of ∼18 kb of sequence from the human genome and ∼15 kb from the chimpanzee genome. Our data suggest that during the primate radiation, L1 insertions may have deleted up to 7.5 Mb of target genomic sequences. While the results of our in vivo analysis differ from those of previous cell culture assays of L1 insertion-mediated deletions in terms of the size and rate of sequence deletion, evolutionary factors can reconcile the differences. We report a pattern of genomic deletion sizes similar to those created during the retrotransposition of Alu elements. Our study provides support for the existence of different mechanisms for small and large L1-mediated deletions, and we present a model for the correlation of L1 element size and the corresponding deletion size. In addition, we show that internal rearrangements can modify L1 structure during retrotransposition events associated with large deletions.
INTRODUCTION
Long INterspersed Elements (LINE-1s or L1s) are abundant non-LTR retrotransposons in mammalian genomes and comprise ∼17% of the human genome (1). They have reached copy numbers of ∼520 000 (1,2) and have expanded over the past 100–150 million years (3). In their full-length state, they are capable of autonomous retrotransposition through an RNA intermediate. However, ∼99.8% of extant L1s in the human genome are retrotransposition-defective (4), either due to point mutations or larger changes such as 5′ truncations, 5′ inversions or other internal rearrangements (5–8). While extant human L1-derived elements have an average size of 900 bp for all L1 copies (1), an active full-length L1 element is ∼6 kb in length, and encodes two open reading frames (ORFs) separated by a 63 bp spacer region. The first L1-encoded protein, ORF1p, is a 40 kDa RNA-binding protein, while the second, ORF2p, is a 150 kDa protein with both endonuclease (EN) and reverse transcriptase (RT) activities (9,10). The two ORFs are preceded by a 5′-untranslated region (5′-UTR), which contains an internal promoter for RNA polymerase II, and are followed by a 3′-UTR ending in a poly(A) tail. The L1-encoded proteins predominantly exhibit cis-preference, transposing the same RNA that encoded them (11,12).
The number of full-length retrotransposition-competent L1 elements that are currently estimated to be propagating in the human genome, however, is much lower than the total number of insertions, with estimates varying between 60 and 100 elements (4,13,14). The mobilization of L1 elements is based on a mechanism termed target-primed reverse transcription (TPRT), which provides useful landmarks for the identification of L1 insertion (15). During this process, a single-strand nick in the genomic DNA is made by the L1 EN at the 5′-TTTT/A-3′ consensus cleavage site (10,16–18) on the antisense strand, after which the L1 RNA transcript anneals by its poly(A) tail to the cleavage site and primes reverse transcription. After the synthesis of the complementary DNA copy and its covalent attachment to the target DNA, second strand synthesis occurs using the first strand as a template. Single-stranded regions remaining in the target DNA at either end are filled in to create target site duplications (TSDs), structural hallmarks of the TPRT process which have been used in the computational location of L1 insertions (19). However, in situations where L1 integration results in the deletion of portions of target DNA, TSDs may not be formed, and a number of studies have reported L1 insertions without TSDs of any length (17,20).
Both mammalian cell culture assays and previous genomic analyses have implicated L1s as agents in complex genomic rearrangements. Mechanisms of L1-mediated genomic instability include (i) unequal homologous recombination between L1 elements (2,21); (ii) generation of interstitial (>3 kb) deletions in the target sequence (5,22) and (iii) transduction of varying amounts of 3′ flanking sequence along with the L1 itself during retrotransposition (23). The last process is also a mechanism for L1-mediated exon shuffling (23–25). The L1 enzymatic machinery may also be utilized during pseudogene processing and Alu element mobilization (11,12).
Previous analyses of genomic deletions created upon L1 retrotransposition in human DNA have almost exclusively relied on cell culture assays and described de novo L1 retrotransposition events associated with target site deletions (5,22). Large interstitial deletions, ranging up to 71 kb, have been reported as one of the consequences of L1 retrotransposition (5). However, the artificially constructed L1 insertion cassettes utilized in these assays permit the recovery of large and full-length L1 insertions only, and the extent of genomic deletion identified in these analyses may not represent the actual extent of existing deletions associated with L1 insertions in the human genome. The recent completion of the draft chimpanzee genome sequence (PanTro1; Nov. 2003 freeze) provides the first opportunity to locate and quantify in an evolutionary framework existing human-specific and chimpanzee-specific L1 insertion-mediated deletions (L1IMDs). In this study, we identified species-specific L1IMD candidates via computational screening of the draft genomic sequences of Homo sapiens and Pan troglodytes, and confirmed them experimentally. We find that L1 insertions are directly responsible for the removal of ∼18 kb of human genomic sequence and ∼15 kb of chimpanzee genomic sequence within the past 4–6 million years and may have generated >11 000 deletion events during the radiation of the primate order, resulting in the removal of up to 7.5 Mb of DNA in the process. We also propose mechanisms to explain the correlation of L1 insertion size with the size of the deletion it causes, and suggest models for the formation of truncation/inversion structures during L1 integration processes associated with target site deletions.
MATERIALS AND METHODS
Computational analysis
To identify L1IMD candidate loci in the human genome, we first identified all L1 elements that have intact 3′ sequence in the July 2003 freeze of the human genome (hg16: UCSC genome database at http://genome.ucsc.edu/ENCODE/) by querying the genome sequence with the 50 bp of the 3′ end of the L1 consensus sequence [excluding the poly(A) tail], using the command line version of the Basic Local Alignment Search Tool (BLAST) (26). The BLAST output file was then processed by a set of in-house Perl programs to extract entries that contain matches with at least 96% sequence similarity to the query sequence over at least 40 bp, resulting in a total of 49 791 L1 entries. Using a cutoff value of 96% similarity ensured that the most recent L1 inserts (including human-specific events) were selected for further analysis. For each entry, 400 bp of sequence downstream of the start of the query (including the match to the query sequence, the poly(A) tail and the 3′ end flanking sequence) were extracted from the human genome sequence. The exact start of the 3′ end flanking sequences was determined for each entry by aligning it with the 50 bp L1 consensus sequence used as the initial query, with which a stretch of 100 adenosines was now included to simulate the poly(A) tail. The 3′ sequence immediately flanking the L1 element identified for each entry was then used as a query to search the chimpanzee genome (PanTro1; Nov. 2003 freeze). If the best match started immediately after the poly(A) tail, the locus was considered to be a human-specific L1 insertion and the start of the matching region was considered to be the insertion site in the human genome. For each identified locus, we extracted 1000 and 100 bp of sequence in the 5′ and 3′ regions of the pre-insertion site, respectively, from the chimpanzee genome. The 5′ chimpanzee sequences were then used to query the human genome. If a 1000 bp chimpanzee sequence only matched the human sequence at its 5′ end, the unmatched sequence at the 3′ end was considered as a L1IMD candidate in the human genome. In cases where there was no match in the entire 1000 bp of the query sequence, the 5′ flanking sequences from the chimpanzee genome were progressively extended until a good partial match at the 5′ end could be identified in the human sequence. These cases were considered to represent deletions that were close to or longer than 1000 bp.
Chimpanzee L1IMD candidates were identified by reversing the query and target genomes and using the same approach as described above. All candidate loci were then subjected to manual verification, resulting in a total of 30 and 33 putative L1IMDs in the human and chimpanzee genomes, respectively.
PCR amplification and DNA sequence analysis
To experimentally verify the L1IMD candidate loci, flanking oligonucleotide primers were designed using the primer design software Primer3 (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). The primers were subsequently screened against the GenBank NR and HTGS databases using BLAST queries to determine if they resided in unique DNA sequences. Detailed information for each locus including primer sequences, annealing temperature, PCR product sizes and chromosomal locations can be found in the ‘Publications’ section of our website (http://batzerlab.lsu.edu).
PCR amplification of each locus was performed in 25 μl reactions using 10–50 ng DNA, 200 nM of each oligonucleotide primer, 200 μM dNTPs in 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris–HCl (pH 8.4) and 2.5 U Taq DNA polymerase. Reactions were subjected to an initial denaturation step of 94°C for 4 min, followed by 32 cycles of 1 min of denaturation at 94°C, 1 min of annealing at optimal annealing temperature and 1 min of extension at 72°C, followed by a final extension step at 72°C for 10 min on a Biorad™ iCycler thermocycler. Resulting PCR products were separated on 2% agarose gels, stained with ethidium bromide and visualized using UV fluorescence.
Individual PCR products were purified from the gels using the Wizard® gel purification kit (Promega) and cloned into vectors using the TOPO-TA Cloning® kit (Invitrogen). For each sample, three colonies were randomly selected and sequenced on an Applied Biosystems AB3100 automated DNA sequencer using chain termination sequencing (27). All clones were sequenced in both directions using M13 forward and reverse primers to confirm the sequence, analyzed using the Seqman™ program in the DNASTAR suite and aligned using the BioEdit sequence alignment software package (http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
For each locus, this procedure was applied to one individual from each of five different primate species, including H.sapiens (HeLa cell line ATCC CCL-2), P.troglodytes (common chimpanzee; cell line AG06939B), P.paniscus (bonobo or pygmy chimpanzee; cell line AG05253B), Gorilla gorilla (western lowland gorilla; cell line AG05251) and Pongo pygmaeus (orangutan; cell line ATCC CR6301). The DNA sequences from this study are available in GenBank under accession numbers DQ017967–DQ018078.
Polymorphism analysis
To evaluate the extent of polymorphism associated with the validated L1IMD loci, each locus was further amplified in the genomes of 80 humans (20 individuals from each of four populations, see below) and 12 unrelated common chimpanzees, following the PCR protocol described above. Our human population panel was composed of DNA from African–American, European and Asian populations (isolated from peripheral blood lymphocytes) available from previous studies in our lab and South American population DNA (HD17 and HD18) purchased from the Coriell Institute for Medical Research. The common chimpanzee population panel was prepared from genomic DNA of 12 unrelated individuals of unknown geographic origin and subspecies affiliation, which was provided by the Southwest Foundation for Biomedical Research.
Phylogenetic analysis of L1IMDs
To examine the phylogenetic relationships of the human and chimpanzee L1 elements identified in this study, we constructed a median-joining network (28,29) using the software NETWORK version 4.1.1.0 (30) available at http://www.fluxus-engineering.com/sharenet.htm. The network was generated using a 94 bp stretch corresponding to positions 5930–6023 in the 3′ end consensus sequence of the L1Hs and L1PA2 reference sequences obtained from the RepeatMasker database. Elements LC9 and LH29 had to be excluded from this analysis because of truncations in the region analyzed.
Analysis of flanking sequences
For GC content analysis, we used the BLAST-Like Alignment Tool (BLAT) server (31) available at http://genome.ucsc.edu/cgi-bin/hgBlat to isolate 20 kb of flanking sequence in either direction from the reference human and chimpanzee draft sequences after adjustment at the 3′ end to prevent bias towards excessive adenosine residues (see Results). We used the EMBOSS GeeCee server (http://emboss.sourceforge.net/apps/geecee.html) to calculate GC percentages. To characterize the gene-frequency neighborhoods of the L1IMDs, we pinpointed exact chromosomal location of the L1 insertions with BLAT, and then used the NCBI MapViewer interface (http://www.ncbi.nlm.nih.gov/mapview/) to map all known genes within 4, 2 and 0.5 Mb windows surrounding the 5′ and 3′ ends of the L1IMDs.
RESULTS
A genome-wide analysis of human- and chimpanzee-specific L1IMDs
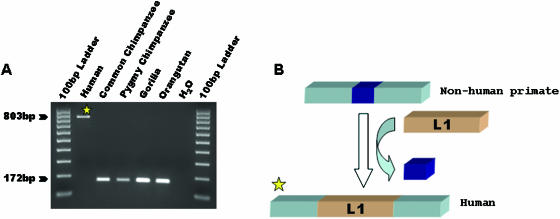
To locate L1IMD loci in the human and common chimpanzee lineages, we first compared data from the draft human and common chimpanzee genomic sequences. We computationally detected 30 human-specific and 33 chimpanzee-specific L1 insertion candidates associated with extra (non-homologous) sequences at the orthologous loci in the other genome. PCR display and manual inspection of the DNA sequences resulted in the exclusion of four human loci and six chimpanzee loci as false positives for L1IMD. These cases were due to poly(N) stretches in the chimpanzee genome assembly (corresponding to unsequenced regions) or species-specific Alu insertions at the 5′ end of the loci, leading to partial mismatches at the orthologous locus in the other species, one of the prerequisites in our computational approach to identify candidate L1IMD loci. This resulted in the validation of 26 and 27 L1IMDs identified from the human and chimpanzee genomes, respectively. PCR analysis of all but one (LH4) L1IMD loci in five primate species showed that all the L1IMDs were specific to the species from which they were identified (Figure 1). Locus LH4 could not be amplified due to the presence of other repeat elements in the flanking sequence. However, on the basis of (i) the 99.5% similarity of the L1 element inserted at this locus to the consensus sequence of the human-specific L1Hs subfamily and (ii) the presence of extra (non-homologous) genomic sequence at this locus in the common chimpanzee genome, the L1 insertion and associated deletion at locus LH4 were included in our dataset of human-specific genomic deletions directly associated with L1 insertion.
Figure 1.
L1IMD in the human genome. (A) Gel chromatograph of PCR products from a phylogenetic analysis of the human-specific L1IMD. The DNA template used in each lane is shown at top. The product sizes for filled and empty alleles are indicated at the left. (B) Schematic diagram depicting the insertion of the L1 element (orange boxes) and the deletion of genomic DNA (blue boxes). Flanking unique DNA sequences are shown as light blue boxes.
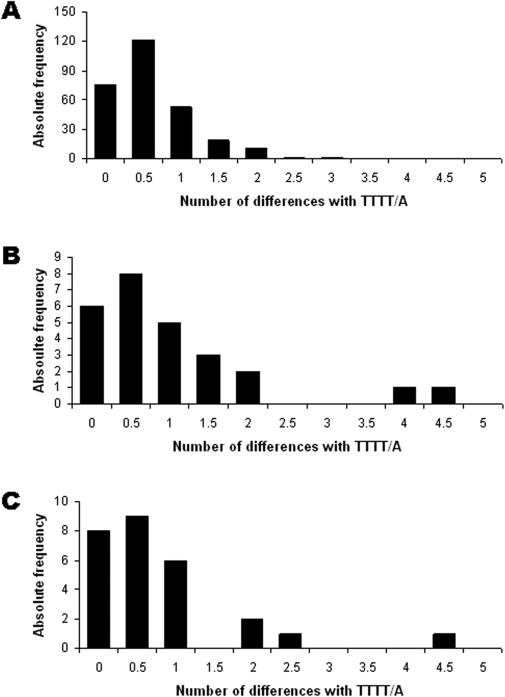
Because the L1 elements associated with L1IMD were not flanked by TSDs, the only possible hallmark of TPRT in our L1IMD events was the presence of L1 EN cleavage sites. To confirm that the deletions observed in the human and chimpanzee genomes were generated during the process of L1 insertion rather than prior to (and therefore independently of) the L1 insertion, we looked for L1 EN cleavage motifs in our L1IMD loci and divided the loci into categories based on the number of differences with the 5′-TTTT/A-3′ consensus L1 EN cleavage site (17,18,32). For each locus, we compared the sequence corresponding to the insertion site predicted to the consensus EN cleavage motif to see if it was L1 EN-generated or not. To conservatively exclude ‘false’ cleavage motifs arising from post-insertion mutations mimicking the L1 EN consensus cleavage sequence, we down-weighted the number of transition differences with the consensus EN cleavage motif by a factor 0.5 because transitions in the cleavage site that conserve the homopurine or homopyrimidine runs are generally better tolerated by the EN than transversions (33). Additionally, we further down-weighted transitions by a second factor 0.5, because of their more frequent occurrence than transversions in GC-poor regions (34). In both humans and chimpanzees, the frequency spectra of the integration site preferences showed unimodal distributions with modes at 0.5 differences from the consensus sequence 5′-TTTT/A-3′ (Figure 2). The L1 EN site preference of our L1IMDs is thus very similar to that of L1-Ta subfamily elements (n = 282) identified in a previous study (17). However, three of the 53 loci (LH11, LH12 and LC6) identified computationally as L1IMD candidates had cleavage sites substantially differing from the consensus by four or more substitutions while the maximum number of substitutions observed in the L1-Ta subfamily is three (Figure 2), hence casting doubt on the use of EN during insertion of these elements. We believe that these deletions are the products of EN-independent insertions similar to those reported in previous cell culture assays (17). To be conservative, these three elements were removed from the analyses, resulting in a final dataset of 24 and 26 L1IMD loci in the human and chimpanzee genomes, respectively, with deletions produced unambiguously by an L1 EN-dependent mechanism.
Figure 2.
EN cleavage site preferences for the L1IMDs. The number of differences from the consensus L1 endonuclease cleavage site (TTTT/A) are shown after down-weighting transitions. The data are analyzed for (A) The L1-Ta subfamily elements identified in Morrish et al. (17); (B) Human lineage-specific L1 insertions; (C) Chimpanzee lineage-specific L1 insertions.
Characteristics of the L1 insertions associated with L1IMDs
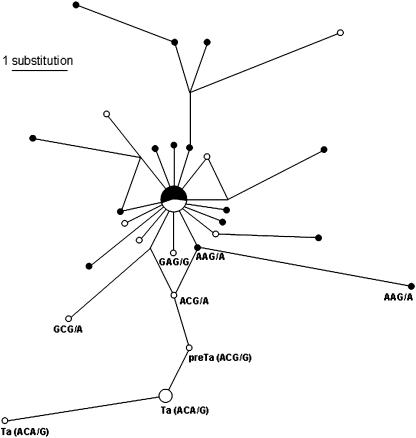
The L1 insertions in our study ranged in size from 61 to 5174 bp. Of the 24 human L1 insertions, 8 belonged to the L1Hs subfamily according to RepeatMasker, 14 to L1PA2 and 2 could not be confidently assigned to any subfamily. As for the 26 chimpanzee L1 insertions, 23 belonged to the L1PA2 subfamily, one to L1PA5 while two could not be confidently assigned to any subfamily. Median-joining network analysis (Figure 3) of the L1 elements in our study, using substitutions at the four key subfamily-diagnostic sequence positions (i.e. 5930–5932 and 6015 in the 3′-UTR of the full-length L1 consensus sequence) show that the chronological order in evolutionary time (from youngest to oldest) of the L1 elements in our study is Ta (ACA/G)–PreTa (ACG/G)–ACG/A–GCG/A or AAG/A–GCG/G–L1PA2 (GAG/A). This evolutionary order is consistent with previous analyses of L1 insertions utilizing other phylogenetic approaches such as neighbor-joining, maximum-likelihood and maximum parsimony analyses (35).
Figure 3.
Median-joining network of the L1 elements associated with L1IMD. Empty circles denote human-specific L1 elements. Filled circles denote chimpanzee-specific L1 elements. The size of circles indicates the number of L1 loci with that sequence type. The lines denote substitution steps, with a one-step distance indicated in the top-left corner. The subfamily-specific diagnostic sequence positions (corresponding to positions 5930–5932 and 6015 in the 3′-UTR of the full-length L1 consensus sequence) are specified below each relevant node.
All the elements were 5′ truncated to different degrees (36), with most having their 5′ start position located in the 3′-UTR of the consensus full-length L1.3 reference sequence (37) (Table 1). The size distribution of the L1 insertions is similar to that obtained in a previous human cell culture assay of L1-mediated genomic instability (22). As to chromosomal distribution, the majority of the L1IMDs were located on chromosomes 1–12, which probably relates to both the larger size of these chromosomes and their higher density of truncated (3′ intact) L1 insertions (19).
Table 1.
Structural summary of L1IMD
| Feature | Human | Chimpanzee |
|---|---|---|
| Full-length L1 insertions | 0 | 0 |
| 5′ truncated L1 insertions | 24 | 26 |
| Internal rearrangements | 4 | 2 |
| Non-inverted | 4 | 0 |
| 5′ truncation/inversions | 0 | 2 |
| With TSDs of any length | 0 | 0 |
| Total L1 size (bp) | 31 617 | 25 031 |
| Mean of L1 size (bp) | 1322 | 963 |
| Total deletion size (bp) | 17 671 | 14 923 |
| Mean of deletion size (bp) | 736 | 574 |
| Median of deletion size (bp) | 21 | 73 |
Four human-specific L1 insertions (at loci LH17, LH19, LH26 and LH31) showed the presence of partially duplicated or internally rearranged L1 segments, suggesting either an atypical structure for the particular L1 insertion or two independent L1 insertions into the same locus during a relatively short time. Given the size of the human genome (∼3300 Mb), two L1 insertions occurring at exactly the same location four times in 24 human loci is very improbable, considering that there have been no instances of L1 element insertion homoplasy ever reported (38–40). Loci LH17 and LH31 each consist of two L1PA2 segments in the same orientation with 300 and 286 bp gaps between the two segments, respectively, relative to the L1PA2 consensus sequence. These loci probably represent single L1 insertion events associated with internal deletions. The other two loci, LH19 and LH26, each consist of two identical L1PA2 segments in tandem, with 53 and 189 bp stretches respectively being repeated in the same orientation without any intervening region. Two chimpanzee loci (LC26 and LC27) also presumably resulted from 5′ truncation/inversion events, with overlapping junctions between the inverted segments (19).
The poly(A) tails of the L1 inserts ranged in length from 2 to 64 bases, with similar averages of 19 bases in humans and 21 bases in chimpanzees. Our value for the average poly(A) tail lengths for human L1 insertions is thus much lower than those from two previous cell culture assays of de novo L1 retrotransposition in HeLa cells, that reported averages of ∼60 residues (5) and 88 ± 27 residues (22). Furthermore, the 23 bp average length of the poly(A) tail among members of the youngest L1Hs subfamily was slightly higher than the 16 bp average for the older L1PA2 subfamily elements. Our data thus suggest the occurrence of post-insertional shortening of poly(A) tails over time, possibly due to replication slippage (36,41). While the poly(A) tails in the de novo insertions identified in the aforementioned studies are exclusive runs of adenosine residues, the tails of the L1s identified in our study show considerable patterning and incidence of other nucleotide residues, with TA(n) being the most common pattern (six cases in the chimpanzee L1s and four cases in human L1s), which corroborates the findings of Szak et al. (19). We found no significant correlation between the size of the poly(A) tail and the size of the L1 insertion in our dataset (r = 0.12, P = 0.84).
Characteristics of the L1IMDs
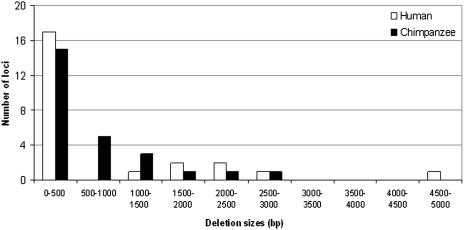
L1IMD events resulted in the deletion of 17 671 nt from the human genome and 14 921 nt from the chimpanzee genome (Table 1). The size distribution of the deletions (Figure 4) showed a strong bias towards the smaller sizes, with 50% of the chimpanzee L1IMDs and 58% of the human L1IMDs showing sizes of <200 bp. However, both human and chimpanzee events were also characterized by 20–30% of L1IMDs longer than 1 kb. These observations were further reflected by the medians of the L1IMD sizes being an order of magnitude smaller than the average L1IMD size in both human and chimpanzee (Table 1). The L1IMD loci in our study in both human and chimpanzee lineages showed significant (P < 0.05 in both species) positive correlations between the size of the L1IMD and the size of its associated L1 insertion.
Figure 4.
Size distribution of the L1IMDs. The size distribution of all the L1IMD events identified in the human and chimpanzee lineages is displayed in 500 bp intervals or bins.
L1IMD polymorphism
To estimate the level of polymorphism associated with human-specific L1IMD loci, we amplified them in 80 individuals from four geographically diverse populations. In all, 5 out of 23 loci (∼22%) were polymorphic (Table 2), 3 of which contained L1Hs elements and 2 contained L1PA2 elements. Within our common chimpanzee panel of 12 individuals, 4 out of 26 loci (∼15%) were polymorphic (Table 2), 3 of which contained L1PA2 elements and 1 contained a L1PA5 element. Overall, this indicates that human L1IMDs are associated with slightly higher polymorphism rates than their chimpanzee counterparts. These results contrast with those obtained for Alu retrotransposition-mediated deletions (ARDs) (42) and Alu insertions (43) in the context of human/chimpanzee comparisons, in which the polymorphism rates were found to be about twice as high in chimpanzee as in human. These data could be indicative of a slowdown of L1 retrotransposition within the chimpanzee lineage as compared to the human lineage.
Table 2.
L1IMD frequency and polymorphism levels within the human and chimpanzee lineages
| Human | Chimpanzee | Human to chimpanzee ratio | |
|---|---|---|---|
| Total observed L1IMDs | 24 | 26 | 0.92 |
| PCR amplified | 23 | 26 | – |
| Fixed present | 18 | 22 | – |
| Polymorphic loci | 5 | 4 | – |
| Polymorphic fraction | 0.22 | 0.15 | 1.41 |
| Adjusted polymorphic loci | 10 | 8 | |
| Adjusted number of L1IMDs | 29 | 30 |
Genomic environment of L1IMDs
Contrary to non-autonomous Alu elements, L1s seem to have a preference for GC-poor regions of the genome (36,44), which may be a consequence of either the L1 EN site preference (16) or of faster removal of L1s from GC-rich regions (45). To analyze whether L1 insertions causing deletions in the target sequence behaved differently from typical insertions, we analyzed GC content of 40 kb of the flanking sequences (20 kb each from the 5′ and 3′ ends) of the L1IMDs. Because poly(A) tails are shortened over time by the combined effects of mutation and replication slippage (36) causing the presence of ‘fossil’ poly(A) tails in the 3′ flanking sequence, we avoided bias towards excessive adenosine residues by counting 20 kb at the 3′ end after excluding 100 bp from the end of the poly-adenylation signal (AATAAA) of the L1 inserts. The mean GC content for the flanking regions of the human-specific and chimpanzee-specific L1IMDs was 38 and 39%, respectively. Compared to ∼42% average GC content of the draft human and chimpanzee genomes (1,46), L1IMD loci thus seem to be concentrated in AT-rich areas of the genome. Remarkably, ARDs in the human and chimpanzee genomes also show a preference for AT-rich locations (42). The reduced GC content (∼36%) around the eight youngest human L1 elements belonging to the L1Hs subfamily in our dataset (LH4, LH15, LH17, LH19, LH20, LH22, LH23, LH24) is consistent with previous findings (44).
To further characterize the genomic context in which L1IMDs occur, we calculated known and predicted gene densities in 4, 2 and 0.5 Mb windows lying immediately 5′ and 3′ to the L1IMDs (see Supplementary data for gene counts). Our results indicate that L1IMDs are concentrated in regions of low gene density (i.e. 1 gene per ∼200 kb, which contrasts with the human genomic average of 1 gene per ∼100 kb) (47). To test whether the size of the L1 insertions at L1IMD loci showed any relation to its surrounding gene density, we performed correlation tests for each window size (4, 2 and 0.5 Mb) in both chimpanzee and human. While we found no significant correlation (−0.16 < r < 0.34, P > 0.05 in all cases), the r-value itself was negative in 5 out of 6 tests, opening the possibility that analysis of a larger dataset of L1 insertions may show a trend towards shorter L1 insertions in gene-rich areas of the genome. Because the chimpanzee LC23 locus was located in an unusually gene-dense region in the short arm of chromosome 9 (i.e. 1 gene per ∼30 kb), we performed our correlation tests involving chimpanzee loci including and excluding this locus. However, the results were similar.
To characterize L1 insertions causing deletions within genes, we analyzed the 14 L1IMD loci (10 in human and 4 in chimpanzee) that were located within the introns of known or predicted genes. Eight of these were in collinear orientation with the gene transcript, while six were in antisense orientation. The average length of the L1 insertions within introns was considerably lower than the average L1 insertion length observed at non-intron L1IMD loci in both human and chimpanzee (849 versus 1601 bp and 474 versus 1053 bp, respectively). These 47 and 55% reductions, respectively, might indicate that smaller L1 insertions are better tolerated than longer ones within the introns of genes.
DISCUSSION
The role of Alu and L1 retrotransposons in the creation of genomic instability is no longer a matter of dispute (5,14,22,42). While extensive cell culture analyses have documented in detail the types and prevalence of genomic rearrangements by L1 insertion in vitro, the possibility remains that in vivo, evolutionary factors such as selection, variation in the number of actively retrotransposing elements and differences in effective population size (43,45) may substantially impact the spectrum of these rearrangements. To test the latter, we made use of the genome sequence of our closest living relative, the common chimpanzee (Pan troglodytes), and performed a human/chimpanzee comparison of L1IMD events.
Evolutionary levels of L1IMD
The previous cell culture analyses of Symer et al. (22) and Gilbert et al. (5), have both reported the presence of large (>3 kb) deletions associated with L1 retrotransposition, with one candidate in Gilbert et al. (5) even deleting at least 24 kb and possibly as much as 71 kb of target sequence. However, such massive deletions are very unlikely to persist in the population because of the likelihood that such events would delete regions of the genome required for survival and thus would subsequently be removed by selection. Consistent with this view, we find that the vast majority of L1IMDs with some degree of evolutionary success are shorter than a few hundred bases in both the human and chimpanzee lineages. In fact, the total amount of lineage-specific deleted sequences through L1IMD in the latest draft of the human genome is estimated to be only ∼17.7 kb, corresponding to an average deletion rate of ∼3.5 kb per haploid genome per million years within the ∼5 million years since the divergence of humans and chimpanzees (48,49). The rate of deletion in the chimpanzee genome is also similar at ∼3 kb per haploid genome per million years.
To estimate the number of human-specific L1 insertions, we reasoned that all human-specific L1 elements belong to only three subfamilies (L1-Ta, L1-preTa and L1PA2) (8,50,51). Given that both empirical (44) and theoretical (43) evidence suggests that the analysis of a single genome results in the recovery of only ∼50% of all polymorphic elements in a subfamily, we estimated each L1 subfamily copy number as the sum of the number of fixed elements and twice the number of polymorphic elements detected in the human genome reference sequence. This resulted in a total of ∼5800 L1 elements for these three subfamilies. However, not all of these L1 elements are specific to humans (52). Using the method of identification of human-specific L1 insertions from Buzdin et al. (52), we conclude that ∼1300 L1 elements have inserted in the human genome since the human/chimpanzee divergence. Given that L1 elements in the human genome have an average size of ∼1 kb (1), we calculate that the insertion of L1 elements within the past 5 million years resulted in the addition of ∼1.3 Mb of sequence to the human genome. This is two orders of magnitude higher than the ∼18 kb length of sequence deleted in the same period by L1IMDs. On a larger time scale, assuming that ∼2.2% of L1 insertions are associated with L1IMD in primates (29/1300 in humans) and the median deletion size of 21 bp from the L1IMD events in our study, the ∼520 000 L1 elements that inserted in primate genomes were responsible for the deletion of a minimum of ∼240 kb of DNA sequences. However, if we perform the same calculation using the average L1IMD size of 655 bp, then almost 7.5 Mb of primate genomic DNA would have been deleted during the retrotransposition of L1 elements. It is also interesting to note that ∼520 Mb (520 000 L1 elements with an average size of 1 kb) of sequence has been added to the genome by the insertion of L1s in the same time period. This is reflective of the ongoing process of renewal of genomic sequences through the retrotransposition process.
Chronological framework of L1IMD events
We were able to place our L1IMD events in a chronological framework on the basis of (i) the results of the median-joining network analysis (Figure 3); (ii) the observation that about two-thirds of the human-specific L1IMDs are caused by L1PA2 insertions versus about one-third caused by members of the younger L1Hs subfamily and (iii) only 20% of the chimpanzee-specific L1IMD events were specific to the common chimpanzee and 80% are shared with the pygmy chimpanzee. Taken together, these results suggest that L1IMD events in the human genome may have occurred to a large extent soon after the human/chimpanzee divergence when the L1PA2 subfamily was active, although they may be continuing to accumulate, as suggested by the non-trivial contribution of L1Hs members. In the chimpanzee lineage as well, the majority of L1IMDs are older than 1–2 million years, which corresponds to the divergence time of common and pygmy chimpanzees (48,49). However, these observations may, at least partly, be influenced by the overrepresentation of older insertions within genomic sequences (i.e. younger events are more likely to be polymorphic than older events and could remain undetected when a small number of individuals are sequenced). Nevertheless, the fact that 23 out of 26 L1IMDs in the common chimpanzee involve L1PA2 elements suggests that the L1PA2 subfamily may still be actively undergoing retrotransposition in the chimpanzee lineage.
Interestingly, among the chimpanzee-specific L1IMDs, we found an ancient L1PA5 element (LC8) that was polymorphic. The L1PA5 subfamily is ∼25 million years old (51). We excluded the possibility of polymorphism being maintained by balancing selection acting on this locus because of the low gene density in its vicinity. It is worthy to note that Bennett et al. (53) also recently identified four polymorphic old AluS elements and one L1PA3 polymorphism. Therefore, this suggests that at least some copies of older L1 retrotransposon subfamilies can retain the ability of retrotransposition for extended periods of time similar to Alu elements (29). Alternatively, it is possible that these polymorphisms have been maintained over a very long period of time by chance. Although this is expected to happen very rarely, it may not be surprising to find a few such cases in view of the hundreds of thousands of L1 and Alu elements (51,54,55) that have inserted during primate evolution. However, we favor the former explanation in the case of the polymorphic L1PA5 element we detected, because DNA sequencing of the locus showed that the L1PA5 insert was specific to the chimpanzee lineage and absent from all other primate genomes we examined.
Different mechanisms may exist for different deletion sizes
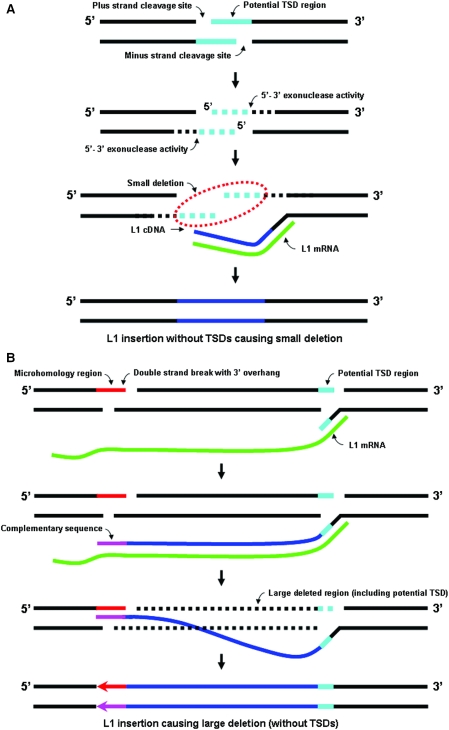
The sizes of the L1IMDs we identified are in general agreement with the size range of similar deletions (13 deletion events ranging from 2 to 14 kb) identified in a recent study of L1 retrotransposition in cell culture (20). However, our sample size for L1IMDs is substantially larger. Very large deletions like those seen in cell culture analyses (5,22) did not appear in our study, presumably because they are more likely to have been removed from the populations rapidly due to their deleterious nature (especially if they were located in gene-rich regions). Interestingly, in both the human and chimpanzee datasets, we noticed a tendency for the deletions to be either very short (i.e. <100 bp) or, to a lesser extent, relatively large (>1 kb), which possibly indicates the concomitant action of two different mechanisms of L1IMD acting on different scales. This dichotomy in deletion sizes was also observed by Gilbert et al. (5), and our data would seem to fit their general models for small and large L1IMD events, to which we propose further extensions to better explain some of the L1 structures that are unique to our study. In general, small deletions may be caused by the creation of 5′ overhangs by top strand cleavage being inexactly opposed to bottom strand cleavage in an upstream direction, with subsequent 5′–3′ exonuclease activity on both the exposed 5′ ends (Figure 5A). In contrast, larger deletions may be explained if the nascent L1 cDNA invades a double-strand break with a 3′-overhang located upstream to the initial integration site (Figure 5B), with gap repair removing the intervening single-stranded segment and causing a large deletion (5,20). Additionally, we suggest that large deletions could result if palindromic stretches downstream of the original site of integration, mechanically or enzymatically held in single-strand conformation during the physical integration of the L1 DNA, formed hairpin loops which were subsequently removed by repair enzymes. Remarkably, a similar pattern of deletion size differences (small or large) also characterizes the deletions caused in the target sequence by the retrotransposition of Alu elements (42). Taken together, the data from genomic deletions caused by L1 and Alu retrotransposon insertions are consistent with the view that two different mechanisms underlie the deletions of small and large stretches of target sequence, especially as both ARDs (42) and the L1IMDs in our study are whole-genome analyses that should represent the comprehensive picture of such deletions.
Figure 5.
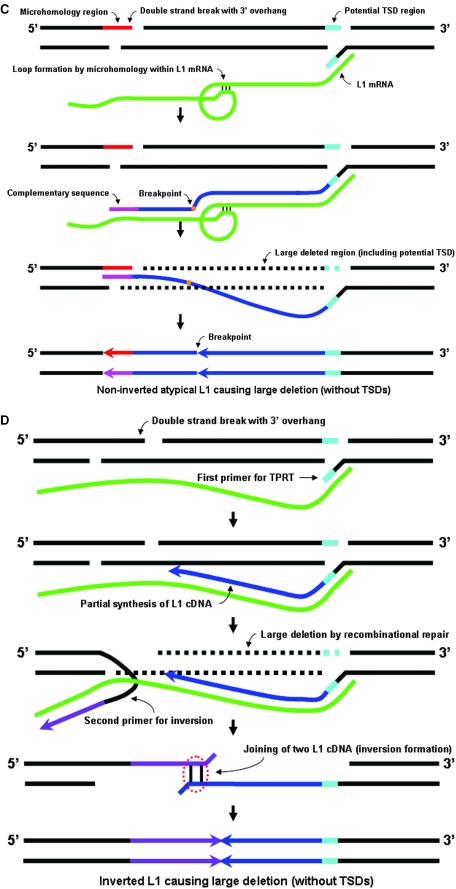
Models for the creation of L1IMDs and formation of deletion associated inverted L1 elements. (A) Formation of small deletions. 5′ overhangs created by inexact cleavage of the top strand by the L1 EN are subject to 5′–3′ exonuclease activity that removes small single-stranded stretches from both the plus and minus strands (dotted light blue lines), which would otherwise have been the templates for the formation of TSDs. Subsequent ligation of the L1 cDNA to the upstream minus-strand sequence and plus-strand sequence synthesis by cellular enzymes results in the creation of small deletions and an L1 insertion without TSDs. (B) Formation of large deletions. For any preexisting double-strand break that has a 3′ overhang (red) for base pairing of the L1 cDNA (blue), a longer cDNA transcript is more likely to contain a stretch of sequence that has adequate complementary bases for annealing (pink) than a shorter one. Subsequent recombinational repair would remove a large segment of the target sequence, extending downstream to the original integration site (dotted black line) and resulting in a L1 insertion without TSDs. (C) Formation of a non-inverted atypical L1 insertion resulting in a large deletion. The L1 mRNA (green) forms a loop, with microhomology stretches within its sequence annealing to each other. The resulting L1 cDNA (blue) has an internal breakpoint (orange) where a stretch of the consensus sequence (complementary to the loop) is missing. Arrows show the orientation of the two parts of the L1 insertion. (D) Formation of a 5′ truncation/inversion resulting in a large deletion. Annealing of the L1 mRNA (green) to a complementary sequence in the 3′ overhang of a preexisting double-strand break leads to the transcription of a second stretch (purple) apart from the original cDNA (blue). Subsequently, both dissociate from the mRNA and form an ‘inversion junction’ (circled in red). Recombinational repair removes the stretch of DNA between the double-strand break and the original site of integration. Plus-strand synthesis results in a 5′ truncated L1 with the inverted portion being reverse complementary to the consensus sequence. Arrows show the orientation of the L1 segments in the inversion.
A model for correlation between insert size and deletion size
In both our human and chimpanzee data sets, we noted a significant positive correlation between the size of the L1 insertion and the size of the deletion caused thereupon. In the extension of the model of Gilbert et al. (5) described above for the creation of large deletions, we propose a probability-based mechanism to further explain the observed correlation (Figure 5B). Our model assumes that given the prior presence of a 3′ overhang in the double-strand break (which is a necessary prerequisite for the occurrence of the deletion by this mechanism) a longer segment of newly transcribed minus strand L1 cDNA is more likely to contain the adequate number of complementary bases (and thus be able to bind with sufficient strength) than a shorter segment. A longer stretch of complementarity than expected by chance between the end of the L1 cDNA and the region surrounding the 5′ end of the L1 insertion in the ancestral (pre-insertion) sequence would provide support for this model. To quantify this parameter, we located (i) the 5′ start position of the L1 insertions with respect to the L1.3 consensus sequence and (ii) the site corresponding to the 5′ start position of the human-specific L1 insertions in the chimpanzee genomic sequence and vice versa. Next, we isolated 15 bp stretches of sequence in the 5′ direction from both these locations in the L1.3 consensus sequence and the genomic sequences, respectively, and aligned them. In all the 12 L1IMD loci that had large deletions corresponding to large L1 insertions (both sizes >500 bp), we found between 27% and 53% complementary bases, which would indicate that potential binding sites were present in all the cases (see Supplementary data for alignments). Additionally, in 7 out of the 15 loci, the first two (LH28, LH30, LC4, LC31) to three (LH17, LH27, LC29) bases in the 3′ end of the alignments were complementary. This further indicates that these bases could have been utilized for binding between the L1 transcript and the target sequence. Recent computational analyses of the 5′ junctions of young L1 insertions in the human genome (56) suggest that microhomology-mediated end-joining is the probable mechanism for 5′ end attachment during the retrotransposition of 5′ truncated L1 elements. Thus, our results support this hypothesis and indicate that longer L1 cDNA strands, because of the higher probability of possessing such microhomology with the pre-integration site, are better suited to the creation of longer genomic deletions by bridging double-strand breaks. The presence of two double-strand breaks (one at the original integration site and one upstream of it) would also lessen the chance of mechanical obstruction to the annealing of the L1 cDNA across the potential deleted region. We note that as proposed in Gilbert et al. (in press), the site of integration is very likely to be a ‘host/parasite battleground’, with the L1 cDNA trying to finish reverse transcription and the host enzymatic machinery opposing it (20). Given the odds against the simultaneous occurrence of L1 insertion reaching comparatively near full-length and the presence of a double-strand break with a 3′ overhang conducive to binding, the lower number of large deletions corresponding to large insertions (6/26 in chimpanzee and 6/24 in human) lends support to our model.
Rearrangements within the L1 elements associated with L1IMD
Six of the L1IMD loci were also characterized by rearrangements within the sequence of the L1 insertion, resulting in atypical L1 structures. Of these, two were both 5′ truncated and partially inverted (LC26 and LC27) while the other four (LH17, LH19, LH26 and LH31) were 5′ truncated non-inverted L1 elements that showed internal rearrangements. Previous cell culture studies have also shown that L1 rearrangements can occur during the process of retrotransposition (5,20). In our study, the presence of the homologous sequence from the respective closest ancestors allowed us to confirm that these loci did not have prior insertions of endogenous L1 elements at the pre-integration sites. The probability of two independent L1 insertions into the same locus after the human–chimpanzee divergence is extremely small, given the large size of the human and chimpanzee genomes, and the estimated number of L1 insertions specific to these lineages (e.g. ∼1300 in humans), which leads us to suggest that mechanistic processes led to the generation of these particular structures during the retrotransposition events. Of the non-inverted atypical L1 elements, LH19 and LH26 are strong candidates for gene duplication, with portions of the L1.3 consensus sequence repeated in parallel orientation without any intervening region (53 and 189 bp, respectively). LH17 and LH31 were 5′ truncated L1 insertions that showed two stretches of the consensus L1.3 sequence with a gap of ∼300 bp in between them. We propose a novel mechanism for this structure, by which stretches of microhomology within the L1.3 consensus sequence might have led to the L1 mRNA looping back on itself (Figure 5C), resulting in the formation of an L1 insertion with the characteristic structure observed and an associated deletion of target site DNA. The presence of at least one such 8 bp homologous stretch was visually confirmed by us in both the cases.
With respect to the 5′ truncation/inversions in our study (LC26 and LC27), a mechanism termed ‘twin priming’ has been suggested for the creation of such structures during L1 retrotransposition (7). However, the existing model does not incorporate the possibility of creation of large deletions during this process. To provide a possible explanation for the large deletions caused at these loci (2973 and 1175 bp, respectively), we suggest a ‘modified twin priming’ model, whereby a stretch of complementarity between the extended L1 mRNA and a 3′ overhang formed at a preexisting double-strand break would lead to a second site of priming on the mRNA (Figure 5D). Subsequently, dissociation of the two newly synthesized cDNA segments from the mRNA and the formation of an ‘inversion junction’, followed by double-strand synthesis, would lead to the removal of the intervening DNA (between the original site of TPRT and the double-strand break) with the formation of a rearranged L1 element with the truncation/inversion structure observed.
CONCLUSION
In conclusion, our study demonstrates that L1IMDs are not restricted to transformed cells but are also a feature of in vivo insertions as well, and that this process has been active in causing deletions in both the human and chimpanzee lineages. Our in vivo evolutionary analysis and prior in vitro cell culture studies of deletions caused by L1 retrotransposition provide pictures that differ at first sight, but can be reconciled by evolutionary factors. While 16–25% of L1 insertions identified in the cell culture studies cause deletions at the target site (5,20,22), only ∼2.2% of existing human-specific L1 insertions seem to be directly linked to genomic deletions [compared to 0.2–0.4% for Alu elements (42)]. As the currently available chimpanzee assembly covers ∼95% of the genome sequence while the human genome sequence is considered to be ‘finished’ (UCSC genome database), our human–chimpanzee comparison probably recovered most species-specific L1IMD events. A slight underestimation due to different levels of completion of the human and chimpanzee genome sequences could not account for the ∼10-fold difference between in vivo and in vitro L1IMD rates. The difference in the rate of L1IMD estimated from cell culture-based analyses and genome-based analyses may more likely reflect the differences in the number of these events that are tolerated in the genome after natural selection has occurred. Thus, our study validates the use of cell culture retrotransposition assays as surrogate models to deduce the underlying mechanisms for these complex genomic rearrangements.
The extent of genomic deletion is reduced compared to the amount of sequence inserted by the L1 retrotransposition process. In addition evidence from our study indicates that many large L1IMDs such as those identified in cell culture assays do not persist in the primate lineage over time. We propose new mechanisms for the creation of some of the specific L1 structures reported in our analysis. Most of the existing human-specific deletions appear to have taken place soon after the divergence of the human and chimpanzee lineages. The atypical L1 elements created during the deletion process could also be sources for new L1 subfamilies in both the human and chimpanzee lineages (5,57).
Acknowledgments
We thank Drs N. Gilbert and J. Moran for sharing their cell culture retrotransposition data prior to publication. We also thank Drs S. Herke, R. Garber and J. Xing for critical reading and suggestions during the preparation of this manuscript. This research was supported by a development fund from Roswell Park Cancer Institute (P.L.), National Institutes of Health RO1 GM 59290 (M.A.B.), CA101515 (P.L.) and CA16056 (RPCI), National Science Foundation BCS-0218338 (M.A.B.) and EPS-0346411 (M.A.B.), and the State of Louisiana Board of Regents Support Fund (M.A.B.). Funding to pay the Open Access publication charges for this article was provided by Louisiana State University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Ostertag E.M., Kazazian H.H., Jr Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 3.Smit A.F., Toth G., Riggs A.D., Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J. Mol. Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- 4.Sassaman D.M., Dombroski B.A., Moran J.V., Kimberland M.L., Naas T.P., DeBerardinis R.J., Gabriel A., Swergold G.D., Kazazian H.H., Jr Many human L1 elements are capable of retrotransposition. Nature Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert N., Lutz-Prigge S., Moran J.V. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 6.Kazazian H.H., Jr, Moran J.V. The impact of L1 retrotransposons on the human genome. Nature Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 7.Ostertag E.M., Kazazian H.H., Jr Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 2001;11:2059–2065. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myers J.S., Vincent B.J., Udall H., Watkins W.S., Morrish T.A., Kilroy G.E., Swergold G.D., Henke J., Henke L., Moran J.V., et al. A comprehensive analysis of recently integrated human Ta L1 elements. Am. J. Hum. Genet. 2002;71:312–326. doi: 10.1086/341718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathias S.L., Scott A.F., Kazazian H.H., Jr, Boeke J.D., Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 10.Feng Q., Moran J.V., Kazazian H.H., Jr, Boeke J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 11.Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewannieux M., Esnault C., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nature Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 13.Brouha B., Schustak J., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazazian H.H., Jr, Goodier J.L. LINE drive. retrotransposition and genome instability. Cell. 2002;110:277–280. doi: 10.1016/s0092-8674(02)00868-1. [DOI] [PubMed] [Google Scholar]
- 15.Luan D.D., Korman M.H., Jakubczak J.L., Eickbush T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 16.Cost G.J., Boeke J.D. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37:18081–18093. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- 17.Morrish T.A., Gilbert N., Myers J.S., Vincent B.J., Stamato T.D., Taccioli G.E., Batzer M.A., Moran J.V. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nature Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 18.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl Acad. Sci. USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szak S.T., Pickeral O.K., Makalowski W., Boguski M.S., Landsman D., Boeke J.D. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-10-research0052. research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert N., Lutz S., Morrish T.A., Moran J.V. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 2005 doi: 10.1128/MCB.25.17.7780-7795.2005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burwinkel B., Kilimann M.W. Unequal homologous recombination between LINE-1 elements as a mutational mechanism in human genetic disease. J. Mol. Biol. 1998;277:513–517. doi: 10.1006/jmbi.1998.1641. [DOI] [PubMed] [Google Scholar]
- 22.Symer D.E., Connelly C., Szak S.T., Caputo E.M., Cost G.J., Parmigiani G., Boeke J.D. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 23.Pickeral O.K., Makalowski W., Boguski M.S., Boeke J.D. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 2000;10:411–415. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran J.V., DeBerardinis R.J., Kazazian H.H., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 25.Goodier J.L., Ostertag E.M., Kazazian H.H., Jr Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum. Mol. Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cordaux R., Hedges D.J., Batzer M.A. Retrotransposition of Alu elements: how many sources? Trends Genet. 2004;20:464–467. doi: 10.1016/j.tig.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Han K., Xing J., Wang H., Hedges D.J., Garber R.K., Cordaux R., Batzer M.A. Under the genomic radar: the Stealth model of Alu amplification. Genome Res. 2005;15:655–664. doi: 10.1101/gr.3492605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bandelt H.J., Forster P., Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 31.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boeke J.D., Devine S.E. Yeast retrotransposons: finding a nice quiet neighborhood. Cell. 1998;93:1087–1089. doi: 10.1016/s0092-8674(00)81450-6. [DOI] [PubMed] [Google Scholar]
- 33.Cost G.J., Golding A., Schlissel M.S., Boeke J.D. Target DNA chromatinization modulates nicking by L1 endonuclease. Nucleic Acids Res. 2001;29:573–577. doi: 10.1093/nar/29.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nachman M.W., Crowell S.L. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ovchinnikov I., Rubin A., Swergold G.D. Tracing the LINEs of human evolution. Proc. Natl Acad. Sci. USA. 2002;99:10522–10527. doi: 10.1073/pnas.152346799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ovchinnikov I., Troxel A.B., Swergold G.D. Genomic characterization of recent human LINE-1 insertions: evidence supporting random insertion. Genome Res. 2001;11:2050–2058. doi: 10.1101/gr.194701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dombroski B.A., Scott A.F., Kazazian H.H., Jr Two additional potential retrotransposons isolated from a human L1 subfamily that contains an active retrotransposable element. Proc. Natl Acad. Sci. USA. 1993;90:6513–6517. doi: 10.1073/pnas.90.14.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho H.J., Ray D.A., Salem A.H., Myers J.S., Batzer M.A. Straightening out the LINEs: LINE-1 orthologous loci. Genomics. 2005;85:201–207. doi: 10.1016/j.ygeno.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 39.Salem A.H., Kilroy G.E., Watkins W.S., Jorde L.B., Batzer M.A. Recently integrated Alu elements and human genomic diversity. Mol. Biol. Evol. 2003;20:1349–1361. doi: 10.1093/molbev/msg150. [DOI] [PubMed] [Google Scholar]
- 40.Salem A.H., Ray D.A., Batzer M.A. Identity by descent and DNA sequence variation of human SINE and LINE elements. Cytogenet. Genome Res. 2005;108:63–72. doi: 10.1159/000080803. [DOI] [PubMed] [Google Scholar]
- 41.Roy-Engel A.M., Salem A.H., Oyeniran O.O., Deininger L., Hedges D.J., Kilroy G.E., Batzer M.A., Deininger P.L. Active Alu element ‘A-tails’: size does matter. Genome Res. 2002;12:1333–1344. doi: 10.1101/gr.384802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Callinan P.A., Wang J., Herke S.W., Garber R.K., Liang P., Batzer M.A. Alu retrotransposition-mediated deletion. J. Mol. Biol. 2005;348:791–800. doi: 10.1016/j.jmb.2005.02.043. [DOI] [PubMed] [Google Scholar]
- 43.Hedges D.J., Callinan P.A., Cordaux R., Xing J., Barnes E., Batzer M.A. Differential alu mobilization and polymorphism among the human and chimpanzee lineages. Genome Res. 2004;14:1068–1075. doi: 10.1101/gr.2530404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boissinot S., Entezam A., Young L., Munson P.J., Furano A.V. The insertional history of an active family of L1 retrotransposons in humans. Genome Res. 2004;14:1221–1231. doi: 10.1101/gr.2326704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boissinot S., Entezam A., Furano A.V. Selection against deleterious LINE-1-containing loci in the human lineage. Mol. Biol. Evol. 2001;18:926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe H., Fujiyama A., Hattori M., Taylor T.D., Toyoda A., Kuroki Y., Noguchi H., BenKahla A., Lehrach H., Sudbrak R., et al. DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature. 2004;429:382–388. doi: 10.1038/nature02564. [DOI] [PubMed] [Google Scholar]
- 47.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 48.Goodman M., Porter C.A., Czelusniak J., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P. Toward a phylogenetic classification of Primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- 49.Chen F.C., Li W.H. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salem A.H., Myers J.S., Otieno A.C., Watkins W.S., Jorde L.B., Batzer M.A. LINE-1 preTa elements in the human genome. J. Mol. Biol. 2003;326:1127–1146. doi: 10.1016/s0022-2836(03)00032-9. [DOI] [PubMed] [Google Scholar]
- 51.Furano A.V., Duvernell D.D., Boissinot S. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 2004;20:9–14. doi: 10.1016/j.tig.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 52.Buzdin A., Ustyugova S., Gogvadze E., Lebedev Y., Hunsmann G., Sverdlov E. Genome-wide targeted search for human specific and polymorphic L1 integrations. Hum. Genet. 2003;112:527–533. doi: 10.1007/s00439-002-0904-2. [DOI] [PubMed] [Google Scholar]
- 53.Bennett E.A., Coleman L.E., Tsui C., Pittard W.S., Devine S.E. Natural genetic variation caused by transposable elements in humans. Genetics. 2004;168:933–951. doi: 10.1534/genetics.104.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Britten R.J. Evidence that most human Alu sequences were inserted in a process that ceased about 30 million years ago. Proc. Natl Acad. Sci. USA. 1994;91:6148–6150. doi: 10.1073/pnas.91.13.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Batzer M.A., Deininger P.L. Alu repeats and human genomic diversity. Nature Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 56.Zingler N., Willhoeft U., Brose H.-P., Schoder V., Jahns T., Hanschmann K.-M.O., Morrish T.A., Löwer J., Schumann G.G. Analysis of 5′ junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5′ end attachment requiring microhomology-mediated end-joining. Genome Res. 2005;15:780–789. doi: 10.1101/gr.3421505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saxton J.A., Martin S.L. Recombination between subtypes creates a mosaic lineage of LINE-1 that is expressed and actively retrotransposing in the mouse genome. J. Mol. Biol. 1998;280:611–622. doi: 10.1006/jmbi.1998.1899. [DOI] [PubMed] [Google Scholar]