Abstract
Indolicidin, a l3-residue antimicrobial peptide-amide, which is unusually rich in tryptophan and proline, is isolated from the cytoplasmic granules of bovine neutrophils. In this study, the structures of indolicidin in 50% D3-trifluoroethanol and in the absence and presence of SDS and D38-dodecylphosphocholine were determined using NMR spectroscopy. Multiple conformations were found and were shown to be due to different combinations of contact between the two WPW motifs. Although indolicidin is bactericidal and able to permeabilize bacterial membranes, it does not lead to cell wall lysis, showing that there is more than one mechanism of antimicrobial action. The structure of indolicidin in aqueous solution was a globular and amphipathic conformation, differing from the wedge shape adopted in lipid micelles, and these two structures were predicted to have different functions. Indolicidin, which is known to inhibit DNA synthesis and induce filamentation of bacteria, was shown to bind DNA in gel retardation and fluorescence quenching experiments. Further investigations using surface plasmon resonance confirmed the DNA-binding ability and showed the sequence preference of indolicidin. Based on our biophysical studies and previous results, we present a diagram illustrating the DNA-binding mechanism of the antimicrobial action of indolicidin and explaining the roles of the peptide when interacting with lipid bilayers at different concentrations.
INTRODUCTION
A rapid increase in the emergence of microbes resistant to conventionally used antibiotics has occurred in recent years (1). This has not only made treatment of infectious diseases more problematic, but has also resulted in the reappearance of many diseases that were thought to be under control. To treat infections caused by multidrug-resistant micro-organisms, searching for new antibiotics has become inevitable and urgent.
Antimicrobial cationic peptides, isolated from a wide range of animal (2), plant (3) and bacterial (4,5) species, are ubiquitous in nature and are thought to be an important component in innate host defenses against infectious agents (4,5). There are four structural classes of cationic antimicrobial peptides: the amphipathic α-helical peptides, such as the cecropins and melittins; the disulfide-bonded β-sheet peptides, including the defensins; the loop-structured peptides, such as bactenecin (6); and the extended peptides, which often have a predominant amino acid (e.g. indolicidin).
Indolicidin (ILPWKWPWWPWRR-NH2), isolated from cytoplasmic granules of bovine neutrophils, has a unique composition consisting of 39% tryptophan and 23% proline (7), and the native peptide is amidated at the C-terminus. Indolicidin is the smallest of the known naturally occurring linear antimicrobial peptides, contains the highest percentage of tryptophan of any known protein and consists of only six different amino acids. Owing to the presence of tryptophan residues interspersed with proline residues throughout the sequence, it probably assumes a structure distinct from the well-described α-helical and β-structured peptides. Indolicidin has been shown to be a fairly potent antimicrobial peptide active against a variety of microorganisms (7), fungi (8) and protozoa (9). Direct viral inactivation of HIV-1 (10) and strong cytotoxicity for rat and human T lymphocytes (11) by indolicidin have also been described.
Most studies on indolicidin have focused on its membrane permeability and have shown that it has a tendency to partition into lipid bilayers of various compositions (8,12,13). Although it is presumed to act by disrupting membranes (8), its mechanism of action remains to be established. Many cationic antimicrobial peptides have been extensively studied in terms of their ability to form pores in membranes, and the killing mechanism for the majority of these peptides has been shown to be the attack on the outer and cytoplasmic membranes, leading to target cell lysis. Interestingly, indolicidin has also been shown to permeabilize the outer and cytoplasmic membranes of Escherichia coli, but is distinct from other cationic peptides in that the membrane permeabilization does not lead to lysis (14). Furthermore, the minimal inhibitory concentration (MIC) of indolicidin leading to a gradual loss of viability on treatment of susceptible cells at 0.4 μM is lower than the minimal hemolytic concentration causing membrane leakage at 15.6 μM (15–17). Further investigations showed that indolicidin mainly reduces synthesis of DNA, rather than RNA and protein, and that inhibition of DNA synthesis causes E.coli filamentation and contributes to the antimicrobial activity of indolicidin (15). Thus, it is conceivable that indolicidin uses its membrane binding properties so as to enter the cytoplasm and exert its antimicrobial activity by attacking targets other than the membrane.
In the present study, with the aim of elucidating the antimicrobial mechanism of indolicidin, we examined its solution structure using circular dichroism (CD) and nuclear magnetic resonance (NMR). The DNA-binding ability of the peptide was also studied by gel retardation, fluorescence measurement, and surface plasmon resonance (SPR) to study in relationship to inhibition of DNA synthesis (15). Here, we report the first demonstrations for the relationship of indolicidin and DNA. Furthermore, we provide the new insight of the antimicrobial action of indolicidin.
METHODS
Sample preparation
Indolicidin (ILPWKWPWWPWRR-NH2) was synthesized in automated mode by Fmoc [N-(9-fluorenyl)methoxycarbonyl] chemistry on a solid-phase synthesizer (model 431 A; Applied Biosystems, Foster City, CA). The synthetic peptide was cleaved from the resin with trifluoroacetic acid and purified by gel filtration, followed by reverse-phase high-pressure liquid chromatography and lyophilization. Electrospray mass spectrometry and amino acid analysis were used to confirm the peptide sequence. D3-trifluoroethanol (TFE), D25-SDS and D38-dodecylphosphocholine (DPC) were purchased from Cambridge Isotope (Woburn, MA). Unless otherwise specified, all reagents and solvents were of reagent grade and used without further purification.
CD experiments
CD data were recorded on an AVIV CD 202 spectrometer (AVIV, Lakewood, NJ) calibrated with (+)-10-camphorsulfonic acid at 25°C using a 2 mm path-length cuvette. The CD spectra of 100 μM indolicidin were recorded at molar ratios of 1:100 (peptide/SDS) in 20 mM phosphate buffer containing 0, 10, 30 or 50% TFE at different pH values (pH 3.5–9.0). The CD data are the average of three scans with a 1 nm bandwidth. The spectra were recorded from 180 to 260 nm at a scanning rate of 38 nm/min with a wavelength step of 0.5 nm and a time constant of 100 ms. After background subtraction and smoothing, all CD data were converted from CD signal (millidegrees) into mean residue ellipticity (deg cm2 dmol−1).
NMR experiments
NMR spectra were recorded on a Bruker AMX-500 or an Avance-600 spectrometer. Samples of indolicidin in a membrane-mimicking environment (∼0.35 ml of 2 mM peptide; peptide/detergent ratio 1:100) were prepared by adding solutions of perdeuterated SDS-d25 or DPC-d38 in 20 mM sodium phosphate buffer, pH 3.5, containing 10% D2O, to lyophilized indolicidin. Samples in TFE were dissolved in TFE-d3/H2O (1:1 v/v) at pH 3.0. pH values were measured using a pH meter (DO microelectronic pH-Vision, model PHB-9901) equipped with a 4 mm electrode; all reported pH values were direct readings from the pH meter without correction for the isotope effect. To monitor the exchange rates of labile protons, the concentrated sample in H2O was lyophilized once and re-dissolved in D2O (99.99% D), and NMR spectra were acquired immediately and thereafter at appropriate time intervals. All chemical shifts were externally referenced to the methyl resonance of 2,2-dimethyl-2-silapentane-5-sulfonate (0 p.p.m.). DQF-COSY (18), TOCSY (19) and NOESY (20) spectra were collected with 512 t1 increments with 2K complex data points. All spectra were recorded in time-proportioned phase-sensitive mode (21). Low temperature studies employed a temperature-controlled stream of cooled air generated using a Bruker BCU refrigeration unit and a B-VT 2000 control unit. Water suppression was achieved by 1.4 s presaturation at the water frequency or by the gradient method (22). All spectra were collected with 6024.1 or 7788.16 Hz spectral widths for the AMX-500 or AVANCE-600 spectrometer, respectively.
The data were transferred to an SGI O2 workstation (200 MHz R5000SC, Silicon Graphics, CA) for processing and further analysis using Bruker XWINNMR and AURELIA software packages (Karlsruhe, Germany). All datasets acquired were zero-filled to equal points in both dimensions prior to further processing. A 60°-shifted skewed sine bell window function was used for all NOESY and TOCSY spectra, while a 20° or 30°-shifted skewed sine bell function was used for all COSY spectra. To help resolve spectral overlap, data were collected at different temperatures.
Structure calculations
Distance restraints of indolicidin were derived primarily from the 200 ms NOESY spectrum recorded at 310K in TFE/H2O (1:1 v/v), pH 3.0, or in membrane-mimicking detergents (DPC and SDS), pH 3.5; this was compared with the 100 ms NOESY spectrum to assess possible contributions from spin diffusion. Peak intensities were classified as large, medium, small and very small, corresponding to upper bound interproton distance restraints of 2.5, 3.5, 4.5 and 6.0 Å, respectively. An additional correction of 1.0 Å was added for methylene and methyl groups. Backbone dihedral restraints were inferred from 3JNHα coupling constants, with restrained to −120 ± 40° for 3JNHα > 8 Hz and to −60 ± 30° for 3JNHα < 6 Hz. Three-dimensional structures were generated using a simulated annealing and energy minimization protocol in the program X-PLOR 98 (23). Hydrogen bond restraints were included only in the final stage of refinement. Average structures were calculated using the final set of refined structures and were further energy minimized to ensure correct local geometry. The INSIGHTII (Molecular Simulation Inc., San Diego, CA), MOLMOL (24) and GRASP (25) software programs were used to visualize sets of structures and to calculate and draw the electrostatic surface potential of the final 3D models. The convergence of the calculated structures was evaluated in terms of the structural parameters, i.e. the root-mean-square deviation (RMSD) from the experimental distance and dihedral constraints, the values of the energy statistics (Fnoe, Ftor and Frepel), and the RMSD from the idealized geometry. The distributions of the backbone dihedral angles of the final converged structures were evaluated by the representation of the Ramachandran dihedral pattern, indicating the deviations from the sterically allowed (φ, ψ) angle limits, using PROCHECK-NMR (26).
DNA/RNA gel retardation assay
Plasmid PET-16b was purified using a QIAprep Spin Miniprep Kit (Qiagen, GMbH). Yeast RNA was resuspended in RNase-free water. The plasmid DNA (200 ng) or yeast RNA (10 μg) was mixed with increasing amounts of peptides in 15 μl of 10 mM Tris, 1 mM EDTA buffer, pH 8.0, and the mixtures incubated at room temperature for 2 min, then subjected to electrophoresis on a 0.5 or 1% agarose gel in the TBE buffer provided with the kit.
Tryptophan fluorescence measurements
Intrinsic fluorescence changes of indolicidin on addition of plasmid DNA were recorded using an F-4500 fluorescence spectrophotometer (Hitachi, Japan). The emission spectra were recorded from 300 to 400 nm with excitation at 295 nm. The fluorescence spectra of indolicidin (0.10 mg/ml in 50 mM Tris–HCl, pH 8.0, 10 mM MgCl2 and 150 mM NaCl) were determined at 25°C alone or in the presence of different amounts of plasmid DNA pET21a.
Surface plasmon resonance
The following four DNA hairpins and four single-strand DNAs with PAGE-purified and 5′-biotin-labeled (MDBio Inc, Taiwan) were used (hairpin loop underlined): d(biotin-CATATATATCCCCATATATATG) (ds[AT]); d(biotin-CGCGCGCGTTTTCGCGCGCG) (ds[GC]); d(biotin-CAGAGAGAGTTTTCTCTCTCTG) (ds[AG]); d(biotin-CGTGTGTGTCCCCACACACACG) (ds[GT]); d(biotin-CATAATATTC) (ss[AT]); d(biotin-CGCGGCGGCT) (ss[GC]); d(biotin-CAGAGAGAGT) (ss[AG]); and d(biotin-CGTGTGTGTC) (ss[GT]).
SPR measurements were performed on a BIAcore X instrument (BIAcore AB, Uppsala, Sweden) using the BIAlogue kinetics evaluation program (BIAEVALUATION V.3.1, Pharmacia Biosensor). Samples of hairpin DNA oligomers or single-strand DNA (25 nM) in HBS-EP buffer (0.01 M HEPES, pH 7.4, 0.15 M NaCl, 3 mM EDTA and 0.005% surfactant P20) were applied to flow cells in streptavidin-derivatized sensor chips (BIAcore SA chips) in a BIAcore X optical biosensor system at a flow rate of 2 μl min−1 to achieve long contact times with the surface and to control the amount of DNA bound to the surface. The sensor chips were conditioned with three consecutive 1 min injections of 1 M NaCl in 50 mM NaOH followed by extensive washing with HBS-EP buffer. Similar amounts of each oligomer were immobilized on the surface by non-covalent capture. The DNA ligand-modified chip surface was regenerated by injecting 20 ml of 1 M NaCl in 50 mM NaOH. The sensor chip surface without a DNA ligand coating was used as the control and was injected with indolicidin simultaneously for each binding experiment. Steady-state binding analysis was performed at 25°C by sequential injection of different concentrations of indolicidin over the immobilized DNA surfaces, each for a 10 min period at a flow rate of 20 μl min−1. Solutions of known concentrations of indolicidin were prepared in filtered and degassed HBS-EP buffer by serial dilutions from a stock solution in HBS-EP buffer and were injected from 7 mm plastic vials with pierceable plastic crimp caps (BIAcore Inc.). The results were plotted on a sensorgram and expressed as resonance units against time.
RESULTS
Indolicidin has an unordered conformation in aqueous and bulk organic solutions and a somewhat more ordered, but not alpha-helical, conformation in SDS micelles and DPC vesicles
In order to investigate the secondary structure of indolicidin, CD experiments were carried out under several conditions.
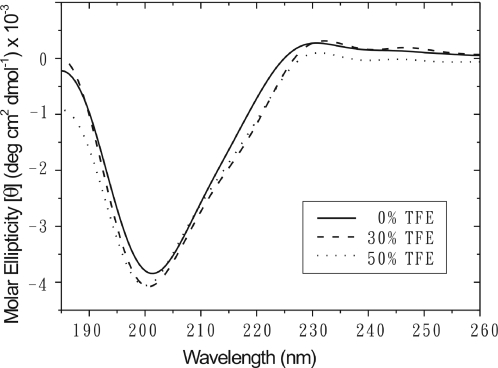
The CD spectra of indolicidin were recorded in 20 mM phosphate buffer alone and in the presence of different concentrations of TFE (0–50% v/v) at pH 7. Intriguingly, as shown in Figure 1, the CD spectrum of indolicidin in aqueous buffer alone showed a single negative band with a minimum at ∼202 nm, which is normally assigned to unordered peptides, but has also been observed for poly-l-proline II (27) and for β-turns (28). On increasing the percentage of TFE, the negative band shifted from 202 to 200 nm. Thus, the similar CD spectra for indolicidin indicate that there is no dramatic conformational change between 0 and 50% TFE at pH 7.0. In addition, the spectra in 50% TFE at different pH values were almost the same (data not shown).
Figure 1.
CD spectra of indolicidin in 0, 30 and 50% TFE.
The CD spectrum at pH 7.0 of indolicidin in SDS micelles was similar to that in DPC vesicles (data not shown). Compared with the spectrum in 50% TFE at pH 7.0, an additional negative band was seen at 225–227 nm, and the minimum at 202 nm shifted to 205 nm due to light scattering caused by large unilamellar vesicles (29). This additional band has been attributed to the tryptophan side chains of indolicidin (12), which interact with the lipid bilayer, and indicates the formation of a β-turn with the B-type topology (30).
Comparison of CD spectra in water, in 50% TFE, and in SDS and DPC micelles suggest that indolicidin adopts diverse conformations at these different conditions, but it is not easy to make conclusion for the small peptide with irregular secondary structure. The further NMR determinations of indolicidin in several environments would provide more structural information to elucidate the arduous question.
NMR spectral evidence for multiple conformations of indolicidin in aqueous solution and membrane-mimicking environments
To investigate the conformation of indolicidin, NMR experiments were carried out in water, 50% TFE (31) and in membrane-mimicking detergents (SDS and DPC) (32). The NMR signals of indolicidin in water were severely overlapped and the sensitivity of NOESY experiment is quite poor. Therefore, we did not attempt to determine the structure of indolicidin in water. The downfield region (∼9.5–11 p.p.m.) of the 1D NMR spectrum of indolicidin represents the indole HN of tryptophan; however, the number of peaks in this region was greater than the number of tryptophan residues (data not shown), indicating multiple conformations of indolicidin in all determined conditions.
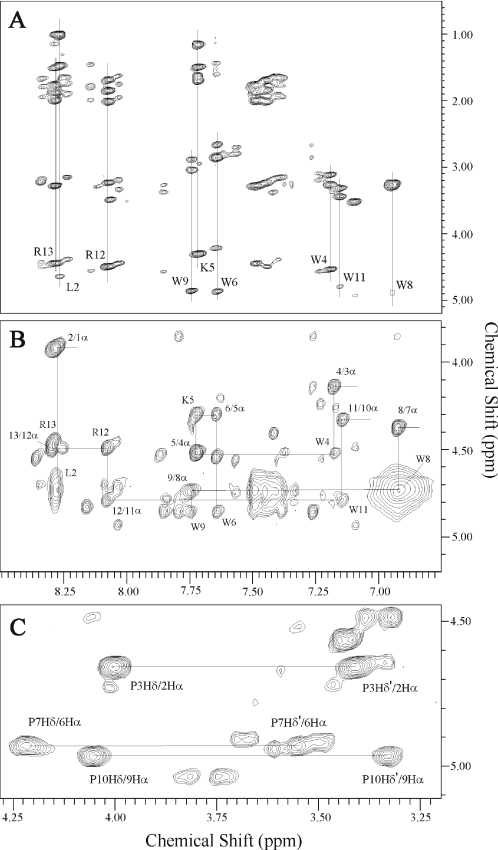
In the fingerprint region of the NMR spectrum of indolicidin in 50% TFE (Figure 2A), it was apparent that, besides a major set of proton resonance, there were additional minor signals, most of which were assigned to at least the amino acid type. At the peptide termini, a doubling or tripling of the proton resonances was observed, whereas most of the central proline and tryptophan residues showed no or one additional set of signals. Even though the spectra of indolicidin in 50% TFE, DPC and SDS were complicated, resonance assignment of the major peaks was accomplished using standard procedures (33). The TOCSY and DQF-COSY spectra were used to assign the spin systems and NOESY spectra were used to make sequential connections between spin systems. In addition, NMR experiments at 285, 295 and 310K at different pH values allowed the complete assignment of the overlapping peaks (the proton chemical shifts are shown in the Supplemental Material). Figure 2B shows the NOESY spectrum at pH 3.0 in 50% TFE, illustrating sequential amino acid assignments of the major species using cross-peaks between a backbone amide proton and the α-proton of the preceding reside. Interruptions in the ‘sequential walk’ occur at proline residues due to the absence of a backbone amide proton. The sequential resonance assignments of indolicidin in DPC and SDS at pH 3.5 have also been completed (data not shown). In 50% TFE, the proline residues were in the trans-conformation, as evidenced by strong NOE contacts between the δ-protons of proline and the α-protons of the preceding tryptophan residue (Figure 2C). Cross-peaks between the α-protons of proline and the preceding residue of the minor species, indicating the cis-proline isomer, were not observed.
Figure 2.
Portions of the 2D proton NMR spectra of indolicidin in 50% TFE at pH 3.0 and 310K, illustrating resonance assignments. (A) TOCSY spectrum showing the backbone amide side-chain region. Individual amino acid spin systems are indicated by the vertical lines. (B) NOESY spectrum showing sequential amino acid residue assignments using cross-peaks between a backbone amide proton and the α-protein of the preceding residue. (C) NOESY spectrum showing the strong NOE cross-peaks between the δ-protons of proline and the α-protons of the preceding residue (connected by a horizontal line) that indicate the all-trans conformation of proline resides.
Indolicidin in 50% TFE shares similar Trp-Pro-Trp motif structures with those in SDS and DPC, but different modes of combination of these two motifs
Previous structural analysis of indolicidin based on CD data has been somewhat controversial. While an unordered structure was detected in aqueous solution, either a poly-l-proline type II helix (34) or a β-turn conformation had been proposed for the lipid-bound molecule (12,30). NMR structures of indolicidin in TFE and in lipid model systems would provide a more definite answer to this question.
While our study was in progress and the assignments of the NMR spectra at different conditions were finished, an independent report appeared on the structure of indolicidin bound to DPC and SDS micelles at pH 4.5 (35). The results of this study are in good agreement with our own observations on the behavior of indolicidin in membrane-mimicking environments at pH 3.5 and are discussed below.
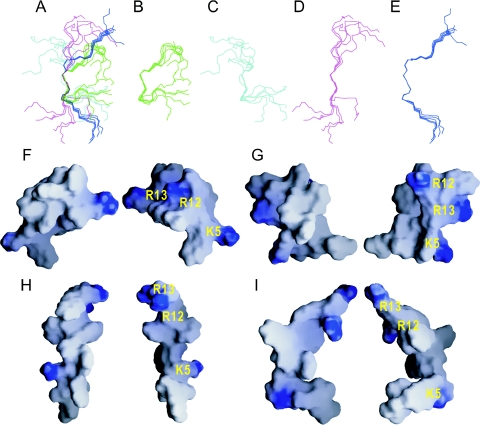
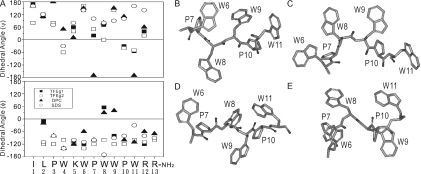
The structures of indolicidin in 50% TFE were computed using 76 NOE-based distance restraints, including 34 intra-residue NOEs and 42 inter-residue NOEs in which there is no long-range NOE greater than (i,i + 4). Interestingly, the calculated structures could be divided into two groups, the backbone in both being rigid between residues 5 and 11 and the RMSDs being 0.59 ± 0.23 Å and 1.03 ± 0.37 Å, respectively (Figure 3B and C), whereas the four N-terminal residues were poorly defined. The previous report for residues 5–11 of indolicidin in DPC and SDS (35) is also presented in Figure 3D and E, respectively. The assembly structures of these four groups (TFE group 1, TFE group 2, DPC and SDS) are best fitted as shown in Figure 3A. The statistics of the calculated structures for indolicidin in the presence of 50% TFE or SDS or DPC micelles are listed in Table 1. The number of inter-residues restraints for the structure calculation in the different environments is almost the same except indolicidin in DPC micelles. In all four subgroups, the backbones of the four N-terminal and two C-terminal residues were flexible and extended, and the well-characterized central region (5–11) adopted a rigid, but slightly different, conformation dominated by two WPW motifs (Figure 3A–E). Interestingly, in the structures of TFE groups 1 and 2, the central region contained two turns (Trp4-Lys5-Trp6-Pro7 and Trp9-Pro10-Trp11-Arg12), whereas, in those in DPC and SDS, it contained two half-turns (Trp4-Lys5-Trp6 and Pro7-Trp8-Trp9) (Figure 4A). The presence of more turns in TFE results in a more compact structure than in the lipid environment. Among the turns, hydrogen bonds between the amide proton of Arg12 and carbonyl oxygen of Trp9 in group 1 as well as the amide protons of Trp8 and Trp6 and/or the carbonyl oxygens of Arg12 and Trp9 in group 2 are observed in some of the NMR structures. Compared among the NMR structures in 50% TFE and DPC as well as SDS, indolicidin undergoes significant conformational rearrangement upon micelles interaction. It is reasonable that the central Trp/Pro-rich region should assume a linear structure for hydrophobic interaction with lipid, whereas an amphipathic and globular structure is suited to an aqueous environment or the docking molecules. These multiple conformers of the central region were formed by different side-chain contacts of the tryptophans in the two WPW motifs (Figure 4B and C). The distances between Trp and Pro are closed. Thus, most of the contacts between Trp and Pro should be van der Waals. No Trp/Trp stacking interaction is obviously seen, but one possible pseudo-stacking interaction between Trp11 and Pro10 is observed in group 1. The structure of indolicidin bound to SDS micelles showed close contact of the side chains of W8 and W9 and contact of W8 and P10, which extends the backbone to form a β-structure (Figure 4D). The bend at residues 7–9 seen in indolicidin in DPC was absent in the SDS-bound molecule, probably because of the long distance between the side chains of W8 and W9, and the side-chain contacts of W9 and W11 with P10 seen in DPC (Figure 4E) were replaced with W8–P10 contacts in SDS (Figure 4D). Furthermore, the NOEs between P7 and W8 were stronger in DPC than in SDS. In aqueous solution, the side chains of the tryptophans in the WPW motifs exhibited at least two contact modes. Figure 4B shows the stick model for the indolicidin structure of TFE group 1, with contacts between W6 and P7 and between W9, P10 and W11. The close proximity of the side chains of W6 and W9 showed that the two WPW motifs interacted. The central WPW structures for TFE group 2 exhibit contact of P7 and W8 with W6 and of W9 and W11 with P10 (Figure 4C). Interestingly, these two WPW motifs seem to be the symmetric enantiomers of each other. The different combinations of contacts between the tryptophan side chains result in multiple conformations of the two WPW, as evidenced by the similarity of the dihedral angles, but large differences in the P7 ψ and W8 φ angles (Figure 4A).
Figure 3.
Backbone superposition and surface potential representation of the calculated NMR structures under different conditions. (A) TFE group 1, (B) TFE group 2, (C) SDS, (D) DPC. (E) Total assembly of the four subgroups shown with 5–11 overlaid. Two opposite surfaces of (F) TFE group 1, (G) TFE group 2, (H) SDS; (I) DPC are shown with the positively charged residues labeled in yellow.
Table 1.
Statistics for the calculated structures for indolicidin in the presence of 50% TFE or SDS or DPC micelles
| 50% TFE | SDSa | DPCa | ||
|---|---|---|---|---|
| Group 1 | Group 2 | |||
| No. of NOE-based inter-residue distance restraints | 42 | 42 | 47 | 61 |
| Mean global RMSD (Å) | ||||
| Backbone (N, Cα, C′) residues (5–11) | 0.59 ± 0.23 | 1.03 ± 0.37 | 0.68 ± 0.23 | 0.10 ± 0.05 |
| Heavy atom residues (5–11) | 1.62 ± 0.45 | 2.36 ± 0.97 | 1.91 ± 0.63 | 0.51 ± 0.19 |
| Ramachandran data (%) | ||||
| Residues in most favored region | 53.5 | 45.0 | 42.5 | 72.5 |
| Residues in allowed region | 35.0 | 52.5 | 57.5 | 12.5 |
| Residues in generously allowed region | 7.5 | 2.5 | 0.0 | 15.0 |
| Residues in disallowed regions | 0.0 | 0.0 | 0.0 | 0.0 |
aData taken from Rozek et al. (35).
Figure 4.
Average dihedral angles and stick models of the WPW motifs of the four subgroups. (A) Average dihedral angles φ and ψ of TFE group 1, TFE group 2, SDS and DPC. (B–E) Stick models of the two WPW motifs in TFE group 1 (B), TFE group 2 (C), SDS (D) and DPC (E) with the residues labeled.
The electrostatic potential maps of the contact surface of the calculated structures of the four indolicidin subgroups were computed using the GRASP program (25). Figure 3F and G shows the surface structures of TFE group 1 and TFE group 2, respectively. These amphipathic and globular structures would be suitable for an aqueous environment and for contact with macromolecules waiting to be docked. In contrast, a wedge shape structure, with most of the tryptophan indole rings packed against the backbone of the peptide, was seen in SDS and DPC. The charge distribution showed a hydrophobic core, flanked by positively charged regions located near both peptide termini. Furthermore, the wedge shape of indolicidin bound to SDS (Figure 3H) or DPC (Figure 3I) appears to be ideal for intercalation between the lipid molecules of a bilayer.
It would be very interesting that such large discrepancies between the structures of indolicidin in TFE and in DPC or SDS micelles are observed, since the TFE-stabilized peptide structure is similar to the micelles-induced conformation in general membrane-associated peptide, such as antimicrobial (36), disease-related (37) or fusion peptide (38). However, TFE is known as an intra-molecular hydrogen bond enhancer and protects exposed hydrophobic residues from aggregation (31), which is applied more like as a structural stabilizer. Several cases of N- and C-terminal peptides of histone were studied and indicated that the peptides become fully structured either in TFE solution or upon interaction with the DNA (39,40). Furthermore, the conformation of the helical peptide from random coil, which is induced by TFE and determined by CD and NMR, is considerably similar to the DNA-induced peptide structure (41). The different conformations of indolicidin in TFE compared with that of micelles indicate that it might be the other molecule-induced form and the result encourages us to investigate the other possible antimicrobial action.
Indolicidin binds DNA in gel retardation experiments
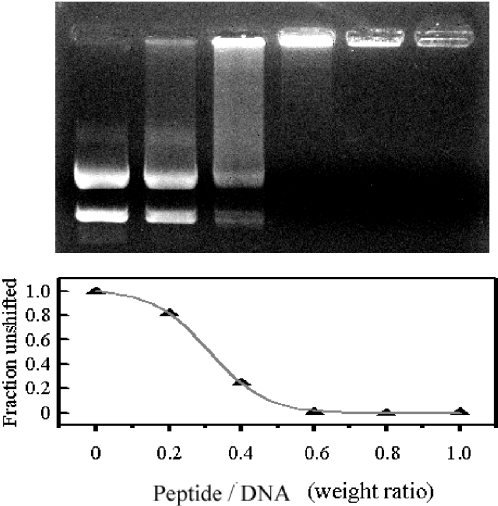
Since the NMR structures of indolicidin in aqueous and lipid environments showed multiple conformations, the structural–functional relationship of indolicidin against microbes is clearly more complicated than that of other cationic antimicrobial peptides. The multiple conformations of indolicidin in different environments suggest that each conformation may play a different functional role. Since indolicidin has been reported to inhibit DNA synthesis (15), its DNA binding ability was evaluated in a gel retardation assay. Peptide (0–200 ng) was mixed with a fixed amount (200 ng) of plasmid DNA and the complexes electrophoresed on a 0.5% agarose gel (Figure 5). At a peptide/DNA weight ratio of 0.2, a fraction of the plasmid DNA was still able to migrate into the gel in the same way as non-complexed DNA, whereas, at a weight ratio of 0.4, almost all of the DNA remained at the origin. At higher weight ratios, complete retardation of the DNA was observed, showing that the DNA was aggregated by indolicidin. Similar patterns of migration were observed with K5 (the fusion peptide analog of influenza virus with the primary sequence: GLFKAIAKFIKGGWKGLIKG), except that free DNA was still seen at a weight ratio of 1.25–2.5 (C. H. Hsu et al., unpublished data). RNA-binding ability of indolicidin was also evaluated by gel retardation (Supplementary Material 2). Yeast RNA was aggregated by indolicidin above the peptide/RNA weight ratio of 6.0, which indicates the poor affinity of indolicidin bound to RNA. These results show that indolicidin has intrinsic DNA-binding but not RNA-binding ability.
Figure 5.
Gel retardation analysis of the binding of indolicidin to DNA. The top panel shows different mixtures of DNA (200 ng) and indolicidin (0, 40, 80, 120, 160 and 200 ng) run on a gel; the bottom panel shows the unshifted fraction of DNA versus the same peptide/plasmid weight ratio.
DNA binding causes conformational changes within indolicidin
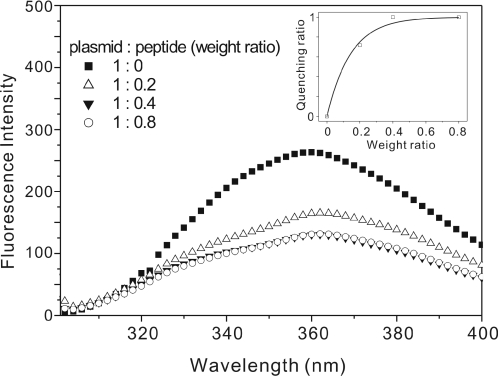
We propose that the DNA binding of indolicidin is regulated by conformational changes within the peptide. The intrinsic tryptophan fluorescence spectrum was used to examine the conformational changes and the interactions between DNA and indolicidin, since the intensity and the emission maximum depend on the surroundings of the indole ring of the tryptophan residue and change when different conformations are observed under different environments. The fluorescence maximum shifts to longer wavelength and the intensity of the fluorescence decreases as the polarity of the solvent surrounding the tryptophan residue increases (42). Since indolicidin contains three tryptophan residues and DNA binding would be expected to increase the polarity of the environment around these residues, we examined whether addition of DNA resulted in quenching of tryptophan fluorescence. As shown in Figure 6, in the absence of DNA, indolicidin exhibited a fluorescence emission maximum at 360 nm, and addition of DNA caused the fluorescence intensity to decrease and the emission maximum to shift from 360 to 362 nm, showing that conformational changes occurred in the environment of tryptophan residues on DNA binding. The inset in Figure 6 is a plot of peptide/DNA weight ratio versus DNA quenching ratio. The results of the quenching experiment are in agreement with our previous gel retardation assay, which showed that retardation was first seen at a peptide/DNA weight ratio near 0.3.
Figure 6.
Quenching of the intrinsic tryptophan fluorescence of indolicidin on addition of DNA. The inset shows the quenching fraction as a function of the peptide/plasmid weight ratio.
DNA-binding sequence preference of indolicidin
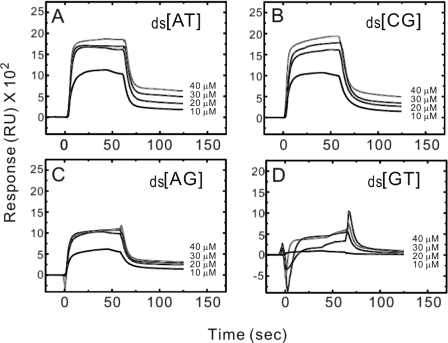
The above gel retardation and fluorescence spectra data indicate that indolicidin interacts with DNA. To further evaluate the sequence selectivity of indolicidin, DNA hairpin oligomers with four alternating ds[AT], ds[GC], ds[AG] or ds[GT] base pair duplex sequences were separately immobilized on BIAcore SA sensorchips and SPR experiments were performed at 25°C. The experiments were based on the SPR optical phenomenon, which would result in light extinction if indolicidin bound to a DNA duplex; this process, detected as a change in a particular angle, is recorded in a sensorgram. A typical set of sensorgrams (response units versus time) at different peptide concentrations for indolicidin binding to the ds[AT], ds[GC], ds[AG] and ds[GT] oligomers is shown in Figure 7. In all the cases, the DNA surface was regenerated by buffer flow without additional regeneration agents. The individual binding and dissociation curves were all aligned at t = 0 (the start of each injection) and the background noise removed to give the final dataset, which consists of a set of curves showing association and dissociation at several different peptide concentrations. Indolicidin bound rapidly to the ds[AT], ds[GC] and ds[AG] sequences and reached a stable steady-state plateau (Figure 7A–C), but only bound poorly to ds[GT] (Figure 7D). Moreover, the dissociation rates were quite different depending on the sequence, since the SPR sensorgrams for binding of indolicidin to ds[AT] and ds[GC] revealed that it took a long time to wash off the peptide from the DNA. The set of curves was used to mathematically fit association and dissociation rates and the dissociation constant calculated using the equation KD = kd/ka; the results are shown in Table 2. Visual inspection of the binding sensorgram in Figure 7D was sufficient to conclude that indolicidin exhibited poor binding to GT compared with the other three DNA sequences. The equilibrium binding constants clearly indicated that the affinity of ds[GT] for indolicidin was much lower than those of ds[AT], ds[GC] and ds[AG]. The kinetic parameters also reflect this behavior.
Figure 7.
SPR sensorgrams for the binding of indolicidin to (A) ds[AT], (B) ds[GC], (C) ds[AG] and (D) ds[GT] hairpin oligomers in HBS-EP buffer at 25°C. The concentration of the unbound ligand in the flow solution varies from 10 μM in the lowest curve to 40 μM in the top curve. RU denotes response units.
Table 2.
Kinetic and affinity constants for the interaction of indolicidin with various double-strand and single-strand DNA sequences
| DNA sequences | Ka (M−1 s−1) | Kd (s−1) | KD (μM) |
|---|---|---|---|
| ds[AT] | 4.77 × 103 | 0.190 | 39.8 |
| ds[CG] | 4.67 × 103 | 0.197 | 42.2 |
| ds[AG] | 3.56 × 103 | 0.156 | 43.8 |
| ds[GT] | 7.61 × 102 | 0.146 | 191.9 |
| ss[AT] | 1.06 × 102 | 0.221 | 2090 |
| ss[CG] | 1.27 × 102 | 0.195 | 1530 |
| ss[AG] | 7.25 × 10 | 0.177 | 2440 |
| ss[GT] | 1.42 × 103 | 0.163 | 115 |
It would be meaningful on the binding comparison of indolicidin bound to duplex and single-strand DNAs with similar sequences. Single-strand DNA oligomers with four alternating [AT], [GC], [AG] or [GT] sequences were separately immobilized on BIAcore SA sensorchips and SPR experiments were then performed at 25°C.
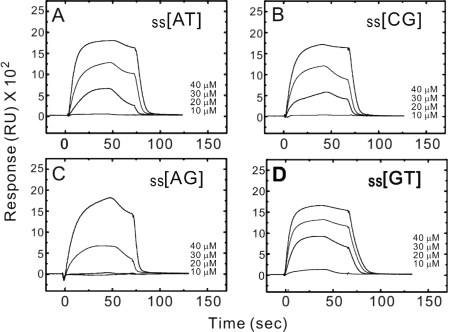
Figure 8 shows the sensorgrams at different peptide concentrations for indolicidin binding to the ss[AT], ss[GC], ss[AG] and ss[GT], respectively. The apparent response curves with higher peptide concentrations and rapid association/dissociation indicate that indolicidin seems to bind single-strand DNA weakly. The mathematic fittings and calculations of the curves also present the low affinity between single-strand DNA and indolicidin, as shown in Table 2. Strikingly, the KD value of ss[GT] is lower than ss[AT], ss[GT], ss[AG] and even ds[GT] that showed the poorest duplex DNA-binding property among four duplex DNAs tested. Together, the SPR experiments demonstrated indolicidin prefers to bind duplex DNA than single-strand DNA generally.
Figure 8.
SPR sensorgrams for the binding of indolicidin to (A) ss[AT], (B) ss[GC], (C) ss[AG] and (D) ss[GT] hairpin oligomers in HBS-EP buffer at 25°C. The concentration of the unbound ligand in the flow solution varies from 10 μM in the lowest curve to 40 μM in the top curve. RU denotes response units.
DISCUSSION
Many different types of organisms use antimicrobial peptides, typically 20–40 amino acids in length, for defense against infection. Most are capable of rapidly killing a wide range of microbial cells. It is not yet clear how these peptides kill bacterial cells, but it is widely believed that some cationic antimicrobial peptides kill by disrupting bacterial membranes, allowing the free exchange of intra- and extra-cellular ions. Indeed, many antimicrobial peptides isolated from a variety of sources have been shown to have a membrane-permeabilizing mode of action.
Although short, indolicidin resembles the Pro/Arg-rich peptides, as it is highly cationic and rich in proline. It induces a voltage-dependent current across planar lipid bilayers (14,16) and has been shown to permeabilize both the outer and inner membrane of E.coli, but is distinct from other amphiphilic peptides in that the membrane permeabilization does not lead to lysis (14,15). Indolicidin fails to fully dissipate the cytoplasmic membrane potential even at concentrations four times higher than the MIC, and the concentration-dependency is very flat; in addition, treatment of susceptible cells with indolicidin at the MIC results in loss of viability without cell lysis; in addition, contrary to expectations, an increase in the optical density of the cell is seen (16). These results point to a mechanism of action that is different from well-defined channel formation. Indeed, indolicidin inhibits DNA synthesis in E.coli, leading to filamentation (15), which would require the peptide to be present in the cytoplasm. The mechanism may be dependent on the presence of a ‘docking molecule’ or receptor-like molecule present in susceptible bacteria and which is required for full antimicrobial activity.
The structure–function relationship of indolicidin has been examined using several biophysical methods. Solution structures of indolicidin in different environments have been determined by fluorescence, CD and NMR spectroscopy, as presented in this paper and elsewhere (12,14,30,35). The fluorescence spectra of indolicidin in aqueous solution or bound to different lipid and detergent particles revealed a blue shift in the emission maximum with a concomitant increase in intensity, indicating that the tryptophan side chains of indolicidin have moved into a more hydrophobic environment. The CD spectrum of indolicidin changes upon partitioning into bilayers (12,14), but the interpretation in terms of secondary structure formation is problematic because of the unusual composition of this peptide. The NMR spectra of indolicidin showed multiple conformations of the peptide in both aqueous solution and lipid environments. Bound to SDS micelles or DPC vesicles, indolicidin adopted a wedge shape comprising a hydrophobic core flanked by two positively charged regions, which appears ideal for intercalation between the lipid molecules of a bilayer. However, spin-label experiments (35) showed that an interfacial membrane location was preferred by indolicidin, and it was speculated that a bilayer-spanning orientation of indolicidin may be transitional and a way of redistributing the antibiotic onto the inner leaflet of the cytoplasmic membrane, from where it can diffuse to other sites in the cell. The ability to cross the membrane would explain the bactericidal activity of indolicidin and the observed transient perturbation of planar membranes (16).
In addition, in aqueous solution, indolicidin adopted a globular and amphipathic conformation, which should be suitable for interaction with macromolecules in the cytoplasm of the cell. Since indolicidin is a relatively symmetric macromolecule, particularly in the region Lys5 to Arg12, which includes the two WPW motifs, different combinations of contacts between the two motifs can result in multiple conformations, as shown by the NMR spectra. Thus, previous studies using retro-indolicidin (43) and our structural approach indicate that plasticity in structure and interactions is critical for the action of indolicidin. These results provide a new insight into the more efficient mechanism of antimicrobial action of indolicidin. To test our hypothesis that indolicidin bound to DNA, gel retardation and fluorescence experiments were carried out and confirmed that a complex was formed when the weight ratio of indolicidin to DNA was ∼0.3. This is the first time that the mechanism of the DNA-binding action of indolicidin has been investigated. Furthermore, different duplex and single-strand DNA sequences were immobilized on a biosensor surface and the association/dissociation of indolicidin monitored. The sensorgrams clearly showed that indolicidin bound strongly to ds[AT], ds[CG] and ds[AG], but only weakly to ds[GT]. Interestingly, the SPR experiments demonstrated the higher affinity of duplex DNA than single-strand DNA with an exception of [GT] sequence.
Together, the SPR studies and structural studies also support the idea that the cluster of positively charged amino acids is the structural character responsible for the ability of indolicidin to bind to nucleic acid and thus kill bacteria. The fact that a two-state reaction model is the best possible fit to the experimental data suggests the mechanism of action of indolicidin involves an initial step of electrostatic binding to the phosphate groups of the DNA duplex, followed by insertion into the DNA groove. Intuitively, indolicidin should prefer to bind strongly to phosphate groups via its positively charged amino acids and the tryptophans should stack between the nucleotide bases or deoxyriboses in each strand of the DNA duplex. Further investigations of DNA binding by indolicidin and the structure determination of the indolicidin/DNA complex are in progress in our laboratory.
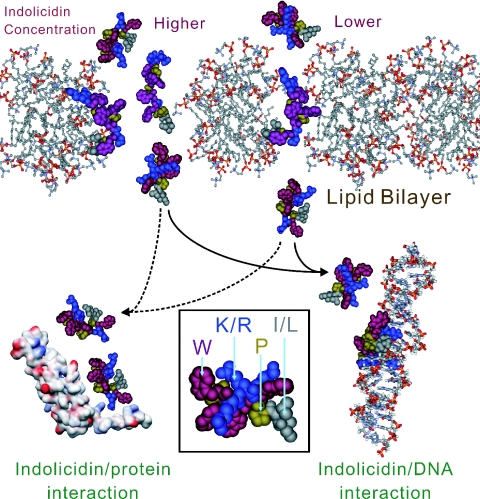
In conclusion, we present the first evidence for multiple conformations of indolicidin and for its binding to DNA, which may explain its known ability to inhibit DNA synthesis and to perturb membranes. Recently, indolicidin was shown to bind to calmodulin (44). As shown in Figure 9, higher concentrations of indolicidin may perturb and even lyse the cell membrane, but when the MIC is lower than the lytic concentration, indolicidin mainly kills bacteria by other processes. It can be reasonably assumed that indolicidin enters the cytoplasm to exert its action and that the changes in cytoplasmic permeability may reflect the transport of the peptide across the membrane. After the transient perturbation and transfer to the cytoplasm, indolicidin then performs other functions in the cytoplasm. The multiple conformers of indolicidin can act as inhibitors of DNA synthesis by binding to DNA or a protein involved in the process. Other biosynthesis pathway or signal transduction pathways may be binding targets for indolicidin, which appears to exert its antimicrobial action by a mechanism different from that used by the well-studied lytic peptides.
Figure 9.
Cartoon representation showing the DNA-binding mechanism of the antimicrobial action of indolicidin and the roles of the peptide interacting with the lipid bilayer at different concentrations. At a concentration higher than the MIC indolicidin perturbs and lyses the cell membrane like normal cationic antimicrobial peptides. However, when the peptide concentration is lower (‘lower concentration’), indolicidin mainly kills bacteria by other processes, such as binding to DNA (full lines) and by docking with proteins (dotted lines). The inset show the CPK presentation of indolicidin, colored for different amino acid types. It can be reasonably assumed that indolicidin enters the cytoplasm to exert its action and that the changes in cytoplasmic permeability reflect the transport of the peptide across the membrane. After the transient perturbation and transfer to the cytoplasm, indolicidin then performs other functions. The multiple conformers of indolicidin can act as DNA synthesis inhibitors by binding to DNA or proteins involved in the process. Indolicidin may also have targets in other biosynthesis pathways or cell cycle signal transduction.
Antimicrobial peptides are attractive candidates for clinical development because of their selectivity and speed of action and because bacteria may not easily develop resistance to them. The multiple conformations of indolicidin have many highly desirable structural characteristics, which are responsible for the broad spectrum of activity, including against most pathogenic bacteria, fungi and even viruses, an ability to kill known antibiotic-resistant clinical isolates, synergy with conventional antibiotics and an endotoxin neutralization activity. Knowledge of the multiple functions and structures of indolicidin may not only result in an understanding of the mechanism of action of this antimicrobial peptide family, but also provide a new lead in the design of potent antimicrobial peptides with therapeutic application.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
C.-H.H. is awarded and funded by the National Health Research Institutes (NHRI) postdoctoral fellowship award (PD-9202) and Academia Sinica distinguished postdoctoral scholar. The authors thank the Academia Sinica and the National Science Council in Taiwan for financial support. The NMR spectra were obtained at the High-Field Biomacromolecular NMR Core Facility for Proteomic Research, National Research Program for Genomic Medicine (NRPGM), Taiwan. Funding to pay the Open Access publication charges for this article was provided by Academia Sinica.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cohen M.L. Epidemiology of drug resistance: implications for a post-antimicrobial era. Science. 1992;257:1050–1055. doi: 10.1126/science.257.5073.1050. [DOI] [PubMed] [Google Scholar]
- 2.Lehrer R.I., Lichtenstein A.K., Ganz T. Defensins: antimicrobial and cytotoxic peptides of mammalian cells. Annu. Rev. Immunol. 1993;11:105–128. doi: 10.1146/annurev.iy.11.040193.000541. [DOI] [PubMed] [Google Scholar]
- 3.Broekaert W.F., Terras F.R., Cammue B.P., Osborn R.W. Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiol. 1995;108:1353–1358. doi: 10.1104/pp.108.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boman H.G. Peptide antibiotics and their role in innate immunity. Annu. Rev. Immunol. 1995;13:61–92. doi: 10.1146/annurev.iy.13.040195.000425. [DOI] [PubMed] [Google Scholar]
- 5.Hancock R.E. Peptide antibiotics. Lancet. 1997;349:418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 6.Radermacher S.W., Schoop V.M., Schluesener H.J. Bactenecin, a leukocytic antimicrobial peptide, is cytotoxic to neuronal and glial cells. J. Neurosci. Res. 1993;36:657–662. doi: 10.1002/jnr.490360606. [DOI] [PubMed] [Google Scholar]
- 7.Selsted M.E., Novotny M.J., Morris W.L., Tang Y.Q., Smith W., Cullor J.S. Indolicidin, a novel bactericidal tridecapeptide amide from neutrophils. J. Biol. Chem. 1992;267:4292–4295. [PubMed] [Google Scholar]
- 8.Ahmad I., Perkins W.R., Lupan D.M., Selsted M.E., Janoff A.S. Liposomal entrapment of the neutrophil-derived peptide indolicidin endows it with in vivo antifungal activity. Biochim. Biophys. Acta. 1995;1237:109–114. doi: 10.1016/0005-2736(95)00087-j. [DOI] [PubMed] [Google Scholar]
- 9.Aley S.B., Zimmerman M., Hetsko M., Selsted M.E., Gillin F.D. Killing of Giardia lamblia by cryptdins and cationic neutrophil peptides. Infect. Immun. 1994;62:5397–5403. doi: 10.1128/iai.62.12.5397-5403.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson W.E., Jr, McDougall B., Tran D., Selsted M.E. Anti-HIV-1 activity of indolicidin, an antimicrobial peptide from neutrophils. J. Leukoc. Biol. 1998;63:94–100. doi: 10.1002/jlb.63.1.94. [DOI] [PubMed] [Google Scholar]
- 11.Schluesener H.J., Radermacher S., Melms A., Jung S. Leukocytic antimicrobial peptides kill autoimmune T cells. J. Neuroimmunol. 1993;47:199–202. doi: 10.1016/0165-5728(93)90030-3. [DOI] [PubMed] [Google Scholar]
- 12.Ladokhin A.S., Selsted M.E., White S.H. Bilayer interactions of indolicidin, a small antimicrobial peptide rich in tryptophan, proline, and basic amino acids. Biophys. J. 1997;72:794–805. doi: 10.1016/s0006-3495(97)78713-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schibli D.J., Epand R.F., Vogel H.J., Epand R.M. Tryptophan-rich antimicrobial peptides: comparative properties and membrane interactions. Biochem. Cell Biol. 2002;80:667–677. doi: 10.1139/o02-147. [DOI] [PubMed] [Google Scholar]
- 14.Falla T.J., Karunaratne D.N., Hancock R.E.W. Mode of action of the antimicrobial peptide indolicidin. J. Biol. Chem. 1996;271:19298–19303. doi: 10.1074/jbc.271.32.19298. [DOI] [PubMed] [Google Scholar]
- 15.Subbalakshmi C., Sitaram N. Mechanism of antimicrobial action of indolicidin. FEMS Microbiol. Lett. 1998;160:91–96. doi: 10.1111/j.1574-6968.1998.tb12896.x. [DOI] [PubMed] [Google Scholar]
- 16.Wu M., Maier E., Benz R., Hancock R.E. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry. 1999;38:7235–7242. doi: 10.1021/bi9826299. [DOI] [PubMed] [Google Scholar]
- 17.Staubitz P., Peschel A., Nieuwenhuizen W.F., Otto M., Gotz F., Jung G., Jack R.W. Structure–function relationships in the tryptophan-rich, antimicrobial peptide indolicidin. J. Pept. Sci. 2001;7:552–564. doi: 10.1002/psc.351. [DOI] [PubMed] [Google Scholar]
- 18.Rance M., Sorensen O.W., Bodenhausen G., Wagner G., Ernst R.R., Wuthrich K. Improved spectral resolution in cosy 1H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun. 1983;117:479–485. doi: 10.1016/0006-291x(83)91225-1. [DOI] [PubMed] [Google Scholar]
- 19.Bax A., Davis D.G. MLEV-17 based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985;65:355–360. [Google Scholar]
- 20.Kumar A., Ernst R.R., Wuthrich K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton–proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 1980;95:1–6. doi: 10.1016/0006-291x(80)90695-6. [DOI] [PubMed] [Google Scholar]
- 21.Marion D., Wuthrich K. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H–1H spin–spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 1983;113:967–974. doi: 10.1016/0006-291x(83)91093-8. [DOI] [PubMed] [Google Scholar]
- 22.Piotto M., Saudek V., Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 23.Brunger A.T. X-PLOR. New Haven, CT: Yale University Press; 1998. [Google Scholar]
- 24.Koradi R., Billeter M., Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. 29–32. [DOI] [PubMed] [Google Scholar]
- 25.Nicholls A., Sharp K.A., Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 26.Laskowski R.A., Rullmannn J.A., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 27.Bovey F.A., Hood F.P. The circular dichroism spectrum of poly-l-acetoxyproline. Biopolymers. 1967;5:915–919. doi: 10.1002/bip.1967.360051005. [DOI] [PubMed] [Google Scholar]
- 28.Perczel A., Hollosi M., Foxman B.M., Fasman G.D. Conformational analysis of pseudocyclic hexapeptides based on quantitative circular dichroism (CD), NOE, and X-ray data. The pure CD spectra of type I and type II beta-turns. J. Am. Chem. Soc. 1991;113:9772–9784. [Google Scholar]
- 29.Wallace B.A., Mao D. Circular dichroism analyses of membrane proteins: an examination of differential light scattering and absorption flattening effects in large membrane vesicles and membrane sheets. Anal. Biochem. 1984;142:317–328. doi: 10.1016/0003-2697(84)90471-8. [DOI] [PubMed] [Google Scholar]
- 30.Ladokhin A.S., Selsted M.E., White S.H. CD spectra of indolicidin antimicrobial peptides suggest turns, not polyproline helix. Biochemistry. 1999;38:12313–12319. doi: 10.1021/bi9907936. [DOI] [PubMed] [Google Scholar]
- 31.Buck M. Trifluoroethanol and colleagues: cosolvents come of age. Recent studies with peptides and proteins. Q. Rev. Biophys. 1998;31:297–355. doi: 10.1017/s003358359800345x. [DOI] [PubMed] [Google Scholar]
- 32.Henry G.D., Sykes B.D. Methods to study membrane protein structure in solution. Methods Enzymol. 1994;239:515–535. doi: 10.1016/s0076-6879(94)39020-7. [DOI] [PubMed] [Google Scholar]
- 33.Wuthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 34.Falla T.J., Hancock R.E. Improved activity of a synthetic indolicidin analog. Antimicrob. Agents Chemother. 1997;41:771–775. doi: 10.1128/aac.41.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rozek A., Friedrich C.L., Hancock R.E. Structure of the bovine antimicrobial peptide indolicidin bound to dodecylphosphocholine and sodium dodecyl sulfate micelles. Biochemistry. 2000;39:15765–15774. [PubMed] [Google Scholar]
- 36.Lequin O., Bruston F., Convert O., Chassaing G., Nicolas P. Helical structure of dermaseptin B2 in a membrane-mimetic environment. Biochemistry. 2003;42:10311–10323. doi: 10.1021/bi034401d. [DOI] [PubMed] [Google Scholar]
- 37.Benaki D., Zikos C., Evangelou A., Livaniou E., Vlassi M., Mikros E., Pelecanou M. Solution structure of humanin, a peptide against Alzheimer's disease-related neurotoxicity. Biochem. Biophys. Res. Commun. 2005;329:152–160. doi: 10.1016/j.bbrc.2005.01.100. [DOI] [PubMed] [Google Scholar]
- 38.Hsu C.H., Wu S.H., Chang D.K., Chen C. Structural characterizations of fusion peptide analogs of influenza virus hemagglutinin. Implication of the necessity of a helix–hinge–helix motif in fusion activity. J. Biol. Chem. 2002;277:22725–22733. doi: 10.1074/jbc.M200089200. [DOI] [PubMed] [Google Scholar]
- 39.Clark D.J., Hill C.S., Martin S.R., Thomas J.O. Alpha-helix in the carboxy-terminal domains of histones H1 and H5. EMBO J. 1988;7:69–75. doi: 10.1002/j.1460-2075.1988.tb02784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vila R., Ponte I., Collado M., Arrondo J.L., Suau P. Induction of secondary structure in a COOH-terminal peptide of histone H1 by interaction with the DNA: an infrared spectroscopy study. J. Biol. Chem. 2001;276:30898–30903. doi: 10.1074/jbc.M104189200. [DOI] [PubMed] [Google Scholar]
- 41.Vila R., Ponte I., Collado M., Arrondo J.L., Jimenez M.A., Rico M., Suau P. DNA-induced alpha-helical structure in the NH2-terminal domain of histone H1. J. Biol. Chem. 2001;276:46429–46435. doi: 10.1074/jbc.M106952200. [DOI] [PubMed] [Google Scholar]
- 42.Ladokhin A.S. Fluorescence spectroscopy in peptide and protein analysis. In: Meyers R.A., editor. Encyclopedia of Analytical Chemistry. Chichester: John Wiley & Sons Ltd.; 2000. pp. 5762–5779. [Google Scholar]
- 43.Nagpal S., Kaur K.J., Jain D., Salunke D.M. Plasticity in structure and interactions is critical for the action of indolicidin, an antibacterial peptide of innate immune origin. Protein Sci. 2002;11:2158–2167. doi: 10.1110/ps.0211602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sitaram N., Subbalakshmi C., Nagaraj R. Indolicidin, a 13-residue basic antimicrobial peptide rich in tryptophan and proline, interacts with Ca(2+)-calmodulin. Biochem. Biophys. Res. Commun. 2003;309:879–884. doi: 10.1016/j.bbrc.2003.08.095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.