Abstract
The genomes of most Nicotiana species contain three different subfamilies of the Tnt1 retrotransposon, which differ completely in their U3 sequence, whereas the rest of the sequence is relatively constant. The results presented here show that all three Tnt1 subfamilies are expressed in tobacco (Nicotiana tabacum) and that the U3 sequence variability correlates with differences in the pattern of expression of the Tnt1 elements. Each of the three Tnt1 subfamilies is induced by stress, but their promoters have a different response to different stress-associated signaling molecules. The Tnt1A subfamily is particularly strongly induced by elicitors and methyl jasmonate, whereas expression of the Tnt1C subfamily is more sensitive to salicylic acid and auxins. The direct relationship between U3 sequence variability and differences in the stress-associated expression of the Tnt1 elements present in a single host species gives support to our model that postulates that retrotransposons have adapted to their host genomes through the evolution of highly regulated promoters that mimic those of the stress-induced plant genes. Moreover, here we show that the analysis of the transcriptional control of a retrotransposon population such as Tnt1 provides new insights into the study of the complex and still poorly understood network of defense- and stress-induced plant signal transduction pathways.
Retrotransposons are mobile genetic elements ubiquitously present in eukaryote genomes. In some cases, such as the BARE1 element from barley, a single retrotransposon family can reach a copy number as high as 50,000 copies per haploid genome (Suoniemi et al., 1996). This high invasivity is facilitated by their replicative mechanism of transposition: the transcription of the element generates an RNA copy that is reverse transcribed into cDNA prior to re-insertion into the genome (Boeke and Corces, 1989). As retrotransposons do not excise, the copy number of retrotransposons increases exponentially with transposition. On the other hand, retrotransposition is potentially a highly mutagenic event, which makes the invasivity of these elements a hindrance for their survival in evolution. As retrotransposons are noninfective agents that cannot leave the host they inhabit, their survival depends on a fine-tuning of activity—high enough to maintain the ability to transpose, but below a threshold that would compromise the viability of the host genome. How this equilibrium is reached is an interesting and still open question that will probably have different answers for different retroelement-host genome couples. In the case of the yeast Ty elements, which are probably the most well-characterized retrotransposons, this equilibrium is achieved by a strict specificity of insertion to regions devoid of genes (Boeke and Devine, 1998), combined with a frequent intra-element long terminal repeat (LTR) recombination that eliminates newly inserted elements (Jordan and McDonald, 1999). This high turnover of Ty elements leads to the maintenance within the yeast genome of a small population of active Ty elements with a high level of sequence homogeneity (Jordan and McDonald, 1999). The case of plant genomes seems to be very different, as they contain a high copy number of retrotransposon sequences that can account for more than 50% of their DNA content (Kumar and Bennetzen, 1999). In addition, these sequences display a high degree of variability (for example, see Casacuberta et al., 1995; Marillonnet and Wessler, 1998) and, in most cases, they represent elements that have lost the ability to transpose. This is probably a consequence of the fact that although intra-element LTR recombination has been shown to occur for plant retrotransposons (Vicient et al., 1999), it does not seem to be a general or important mechanism for reducing retrotransposon copy number in most plant genomes (Bennetzen and Kellogg, 1997). Moreover, plant retrotransposons do not display target site specificity and can insert within or close to genes, creating mutations. Therefore, it seems that plant retrotransposon populations are controlled by mechanisms unlike those of the yeast Ty elements.
All the active plant retrotransposons characterized so far are only expressed under very precise stress situations, being silent during most of the plant life cycle (Wessler, 1996; Grandbastien, 1998). This suggests that transcriptional regulation is a major mechanism of control for retrotransposition in plants (Wessler, 1996; Grandbastien, 1998). This is the case for the tobacco (Nicotiana tabacum) retrotransposons Tnt1A and Tto1, whose promoters have been analyzed in detail (Grandbastien et al., 1997; Takeda et al., 1999). How plant retrotransposons have evolved such stress-activated promoters is an open question. We have previously shown that Tnt1 is expressed in protoplasts and roots as a heterogeneous population of RNA molecules that resembles retroviral quasispecies (Casacuberta et al., 1995). This lead us to suggest that as for retroviral quasispecies, this high sequence variability could endow Tnt1 with a high sequence plasticity that could facilitate the evolution of its promoter regions to improve its coexistence with the host (Casacuberta et al., 1997). To validate this hypothesis, we have looked for a possible correlation between sequence variability and differences in the expression patterns of Tnt1 elements. The results presented here show that the three previously defined Tnt1 subfamilies (Casacuberta et al., 1997; Vernhettes et al., 1998) are expressed in tobacco with different expression patterns. These specific patterns of expression are probably a consequence of the sequence variability of their U3 regions, which in each case contain a different stress-inducible promoter.
RESULTS
The Three Tnt1 Subfamilies Are Expressed in Tobacco Cell Cultures
Tnt1 is present in hundreds of copies in the genome of tobacco and related Nicotiana spp. (Casacuberta et al., 1997). The U3 region of Tnt1 is highly variable, and three different subfamilies of Tnt1 have been defined according to their U3 sequence (Casacuberta et al., 1997; Vernhettes et al., 1998). All three different subfamilies, Tnt1A, Tnt1B, and Tnt1C, are present, but they differ in relative abundance in different Nicotiana genomes, which suggests that all three subfamilies remain active (Vernhettes et al., 1998). However, until now only the Tnt1A subfamily has been shown to be expressed. Tnt1A is expressed in protoplasts and roots and, although we have analyzed more than 100 partial Tnt1 sequences by reverse transcriptase (RT)-PCR, we have failed to detect expression of Tnt1B and Tnt1C elements in these tissues (Casacuberta et al., 1995).
Tnt1 is transiently expressed during protoplast isolation, and its RNA level decreases rapidly when protoplasts are cultured (Grandbastien et al., 1997). However, it also has been reported that Tnt1 copy number increases slightly in cultured cells, suggesting that Tnt1 could be expressed in those cells (Hirochika, 1993). As different gene programs are activated during the different cell culture stages, we decided to analyze the expression of Tnt1 in cultured tobacco cells to look for a possible expression of Tnt1B and Tnt1C elements by RT-PCR analysis.
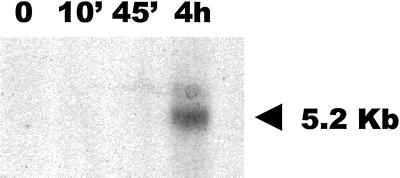
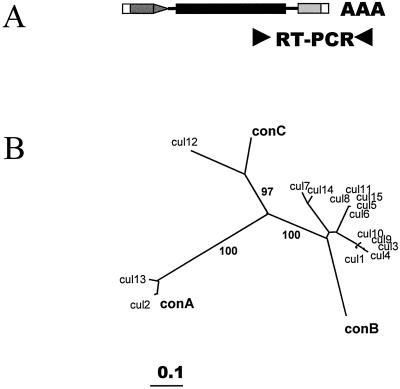
The results presented in Figure 1 show that whereas Tnt1 is not expressed in 1-week-old cultures (Fig. 1, 0′), there is a transient induction of Tnt1 expression 4 h after the addition of fresh media to the culture. Preliminary results suggest that this induction could be associated to the presence of auxin hormones in the fresh media (data not shown). To determine the Tnt1 subfamilies expressed under these conditions, 15 partial Tnt1 RNA sequences comprising the U3 region (see Fig. 2A) were obtained by RT-PCR amplification with RNA from tobacco cell cultures and were cloned and sequenced. A phylogenetic analysis of the sequences, including consensus sequences of the Tnt1A, Tnt1B, and Tnt1C subfamilies (Casacuberta et al., 1997) by the neighbor-joining method, is shown in Figure 2B. This analysis shows that only two of the sequences obtained are closely similar to the Tnt1A consensus, whereas most of the sequences are highly similar to the Tnt1B subfamily consensus. One of the sequences was found to be more similar to the consensus for Tnt1C than to the consensus for the two other Tnt1 subfamilies. These results show that the three Tnt1 subfamilies are expressed in freshly subcultured tobacco cells and that Tnt1B RNA is predominant in the conditions. To our knowledge, this is the first report of different subfamilies of a retrotransposon being expressed in a particular host species.
Figure 1.
Induction of Tnt1 expression in tobacco cultured cells. Northern analysis of RNAs obtained from tobacco cells cultured in NK1 medium after 1-week culture (0) or after 10 min (10′), 45 min (45′), or 4 h of subculture in fresh media, hybridized with a probe corresponding to a conserved endonuclease region of Tnt1. The 5.2-kb band corresponding to the Tnt1 genomic RNA is indicated by an arrow.
Figure 2.
Analysis of the Tnt1 RNA molecules expressed in cultured cells. A, Scheme showing the region of Tnt1 amplified by RT-PCR. Coding sequences are shown in black, the U5 region is shown in dark gray, and the U3 region is shown in light gray. ▸, The approximate position of oligonucleotides used for the RT-PCR amplification. B, Neighbor-joining tree obtained with an alignment of the 15 Tnt1 partial sequences obtained from cultured cells RNA (cul#) together with the consensus sequences for the Tnt1A (conA), Tnt1B (conB), and Tnt1C (conC) subfamilies. Bootstrap values over 60 for the main branches are shown.
Induction of Different Tnt1 RNA Populations with Different Stress-Associated Signaling Molecules
To get an insight into the signal transduction pathways that lead to the induction of the different Tnt1 subfamilies, we infiltrated tobacco leaf discs with different elicitors and stress-associated signaling molecules. Previous results showed that cryptogein, a protein from Phytophtora cryptogea that elicits tobacco defense responses (Ricci et al., 1989), was able to induce the Tnt1A promoter, as was salicylic acid, the signal transduction intermediate of the plant responses to wounding and pathogen infection (Grandbastienet al., 1997; Vernhettes et al., 1997). Nevertheless, it has recently been proposed that another defense-related signaling molecule, methyl jasmonate, could be also part of the signaling cascade triggered by cryptogein (Rusterucci et al., 1999). On the other hand, preliminary results suggest that addition of fresh auxins to the culture medium could be responsible for the transient induction of Tnt1 we have observed in tobacco subcultured cells. Therefore, we investigated the effect on Tnt1 expression of infiltrating leaf discs with cryptogein, salicylic acid, methyl jasmonate, and 2,4-dichlorophenoxyacetic acid (2,4-D) using discs infiltrated with water as a control.
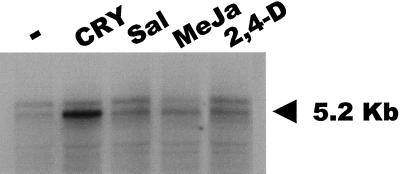
Figure 3 presents a northern-blot analysis of the RNAs obtained after each treatment. It can be seen that, as previously reported (Vernhettes et al., 1997), the 5.2-kb RNA of Tnt1 is expressed to low levels in leaf discs infiltrated with water, due to the wounding stress associated with the infiltration process. However, infiltration with chemicals results in a visible increase in induction of Tnt1. A quantitative analysis of the hybridization using a phosphoimager (Bio-Rad, Hercules, CA) shows that infiltration with cryptogein leads to a 10-fold induction of the steady-state level of Tnt1 RNA, whereas the increase in induction with salicylic acid, methyl jasmonate, and 2,4-D is 3- to 4-fold. A 6.5-kb leaf mRNA is also detected, but it has been shown that this does not correspond to the genomic transcript of the retrotransposon (Pouteau et al., 1991).
Figure 3.
Induction of Tnt1 expression by infiltration of tobacco leaf discs with stress-associated signaling molecules. Northern analysis of RNAs obtained from tobacco leaf discs infiltrated with water medium (–) or 1 μg mL−1 cryptogein (CRY), 2 mm salicylic acid (Sal), 10 mm methyl jasmonate (MeJa), and 1 mm 2,4-D, hybridized with a probe corresponding to a conserved endonuclease region of Tnt1. The 5.2-kb band corresponding to the Tnt1 genomic RNA is indicated by an arrow.
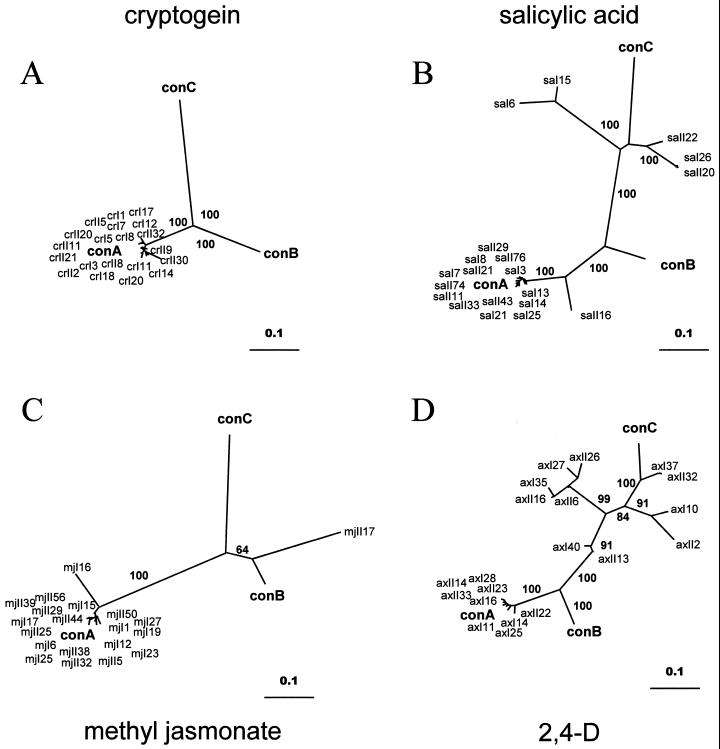
To analyze which Tnt1 subfamilies are expressed after each treatment, we amplified the 3′ end of the Tnt1 RNA, which comprises the U3 region, by RT-PCR. Two sets of 10 sequences obtained from two independent RT-PCR reactions were cloned and sequenced from RNA obtained from each treatment. These sequences were compared with the consensus of the three Tnt1 subfamilies using a phylogenetic approach. The neighbor-joining trees obtained with the sequences from each treatment are shown in Figure 4. These results show that although cryptogein infiltration induces the expression of the Tnt1A subfamily only (Fig. 4A), the rest of the treatments induce a more complex population of Tnt1 RNAs. This is particularly clear for the salicylic acid (Fig. 4B) and 2,4-D (Fig. 4D) treatments in which five and 11 out of 20 sequences, respectively, closely resemble the consensus sequence for the Tnt1C subfamily. A sequence highly similar to the Tnt1B consensus was detected in the methyl jasmonate-treated material (Fig. 4C).
Figure 4.
Analysis of the Tnt1 RNA molecules expressed in infiltrated leaf discs. Neighbor-joining trees obtained with an alignment of the 20 Tnt1 partial sequences obtained by two independent RT-PCR amplifications from infiltrated leaf disc RNA together with the consensus sequences for the Tnt1A (conA), Tnt1B (conB), and Tnt1C (conC) subfamilies. A, Sequences obtained from cryptogein-infiltrated leaf discs (crI# and crII# for the sequences obtained from the first or the second RT-PCR experiment, respectively); B, sequences obtained from salicylic acid-infiltrated leaf discs (saI# and saII#); C, sequences obtained from methyl jasmonate-infiltrated leaf discs (mjI# and mjII#); D, sequences obtained from 2,4-D-infiltrated leaf discs (axI# and axII#). Bootstrap values over 60 for the main branches are shown.
All the sequences obtained from the water-infiltrated control leaf discs were closely similar to the consensus for the Tnt1A subfamily (data not shown).
The U3 Regions of the Three Tnt1 Subfamilies Contain Promoter Elements Differentially Induced by Different Stress-Associated Signaling Molecules
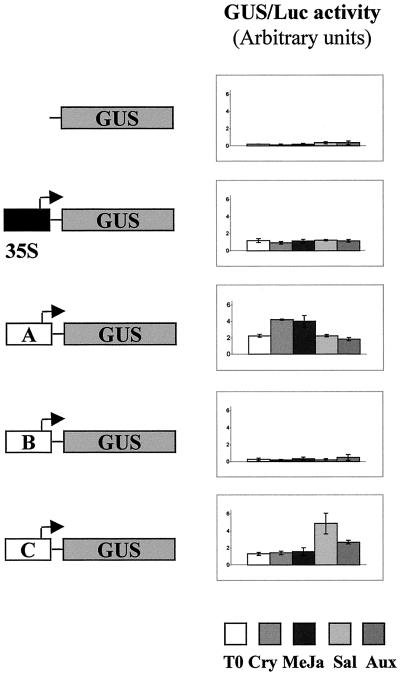
Our results clearly show that the stress-associated signaling molecules, salicylic acid, methyl jasmonate, and 2,4-D, as well as the fungal elicitor cryptogein, differentially induce the expression of the three Tnt1 retrotransposon subfamilies. To analyze whether the U3 regions of each Tnt1 subfamily contain different stress-inducible promoters, we bombarded leaf discs with constructs of U3 regions representative of each subfamily upstream of a β-glucuronidase (GUS) reporter gene, and then analyzed GUS expression after incubation with cryptogein, salicylic acid, methyl jasmonate, or 2,4-D. As secondary effects could be generated by single nucleotide differences within the conserved TATA box or transcription start region, we cloned Tnt1B and Tnt1C U3 sequences upstream of the TATA box in front of the TATA-box region of the sequence chosen as representative for the Tnt1A subfamily (see “Materials and Methods”). Leaf discs bombarded with a GUS reporter gene devoid of promoter and a GUS reporter gene driven by the 35S constitutive promoter were used, respectively, as negative and positive controls.
Results presented in Figure 5 show that the U3 region of the Tnt1B subfamily is not able to efficiently promote GUS expression in the conditions tested. On the contrary, the Tnt1A and Tnt1C U3 regions are able to drive a high level of GUS expression in tobacco leaves. It is interesting that whereas the U3 region of Tnt1A seems to induce a particularly high level of expression after incubating the bombarded leaves with cryptogein or methyl jasmonate, the U3 region of the Tnt1C subfamily is particularly induced in leaf discs incubated with salicylic acid and 2,4-D. On the other hand, no GUS activity is detected in leaf disc bombarded with the negative control, whereas, as expected, a strong noninducible GUS activity is detected in leaf discs bombarded with the 35S-GUS construct. These results are in agreement with the RT-PCR results obtained and are consistent with a major role of the U3 region in conferring a specific pattern of expression on each Tnt1 subfamily.
Figure 5.
Transient expression analysis of Tnt1 promoters. Relative GUS/Luc activity obtained after incubating the leaf discs, bombarded with the different GUS constructs, with different chemicals.
We have previously shown that the Tnt1A promoter is activated by cryptogein (Vernhettes et al., 1997), and Figures 3 and 4 show that it is also highly activated by methyl jasmonate. To analyze if jasmonate induces Tnt1A through the same promoter elements as cryptogein, we compared the Tnt1 RNA sequences obtained after induction by these different chemicals. A neighbor-joining tree of the 61 Tnt1A sequences obtained failed to form groups supported by a bootstrap value of more than 10%, indicating that it is not possible to differentiate populations within them (not shown). The populations of Tnt1A sequences expressed after the different treatments here analyzed are thus indistinguishable. In addition to this, the BII elements, which were shown to be important for cryptogein induction (Vernhettes et al., 1997), are conserved in all the sequences amplified, 67% of them having four tandem repetitions of this element and only one sequence out of 61 having less than three elements (data not shown). These results suggest that methyl jasmonate and cryptogein induce Tnt1A through the same cis-acting elements.
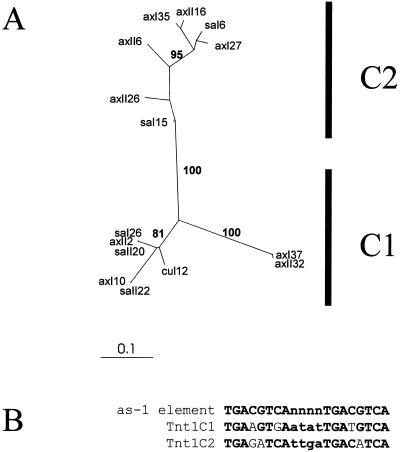
On the other hand, our results show that the promoter of Tnt1C is particularly sensitive to 2,4-D and salicylic acid. Although the populations of Tnt1C RNA induced by 2,4-D and salicylic acid are much more variable than those of Tnt1A RNA, they cannot be differentiated from one another by phylogenetic analysis. Figure 6A presents a neighbor-joining tree of all the Tnt1C sequences obtained, and it can be seen that different groups supported by high bootstrap values can be defined. Nevertheless, sequences belonging to each of these groups were found after salicylic acid and 2,4-D treatments, and we have not been able to define any induction-specific group.
Figure 6.
Analysis of the Tnt1C RNA molecules expressed in infiltrated leaf discs. A, Neighbor-joining trees obtained with an alignment of all the Tnt1C partial sequences obtained. Sequences obtained from salicylic acid-infiltrated leaf discs are shown as in Figure 4. Bootstrap values over 60 for the main branches are shown. B, Comparison of the consensus as-1 motif with those found in Tnt1C1 and Tnt1C2 sequences. Conserved nucleotides are shown in bold.
It has been shown that salicylic acid and auxins can induce plant genes through the interaction of inducible transcription factors with the as-1-related cis-elements (Chen and Singh, 1999; Niggeweg et al., 2000). The Tnt1C sequence analyzed here contains a nearly palindromic sequence within the U3 region that closely resembles the as-1 element (TGACGTCAnnnnTGACGTCA), and this could be important for the induction of this Tnt1 subfamily by auxins and SA. It is interesting that although the two Tnt1C groups of sequences differ within the U3 region, both contain an almost identical as-1-like element located 20 to 30 nucleotides upstream of the corresponding TATA boxes (see Fig. 6B), suggesting that this element is important for the expression of these retrotransposon elements.
DISCUSSION
Different Subfamilies of the Retrotransposon Tnt1 Have Maintained Their Transcriptional Activity in Tobacco
Although three different subfamilies of the tobacco retrotransposon Tnt1 have been found to be present at a high copy number in the genome of different Nicotiana species, until now evidence for expression activity has only been obtained for the Tnt1A subfamily (Grandbastien et al., 1997). The results presented in this work show that all three Tnt1 subfamilies are expressed in tobacco and that they have different patterns of expression. We have previously shown that the fungal elicitor cryptogein induces the expression of the Tnt1A promoter (Pouteau et al., 1994; Vernhettes et al., 1997). We show here that this treatment induces an important accumulation of RNA that corresponds to Tnt1 elements that belong to the A subfamily. This result is similar to what we have previously reported from the analysis of Tnt1 RNA present in root tips and protoplasts in which only Tnt1A elements are expressed (Casacuberta et al., 1995). In contrast, the infiltration of leaf discs with methyl jasmonate, and particularly with salicylic acid or 2,4-D, induces a more complex population of Tnt1 elements that belong to all three subfamilies. To our knowledge this is the first report of different subfamilies of a retrotransposon being expressed in a particular host genome, which makes Tnt1 a particularly interesting model for analyzing the evolution of retrotransposons within plant genomes.
The Three Different Tnt1 Subfamilies Are Differentially Induced by Stress-Associated Signaling Molecules through Specific Promoter Elements
The results presented here not only show that the three Tnt1 subfamilies are expressed in tobacco, but they also show that these subfamilies are differentially regulated. The RT-PCR analysis shows that infiltration treatments with different signaling molecules are able to induce particular subsets of Tnt1 elements. We found expression of Tnt1A RNA in leaves infiltrated with all the chemicals analyzed. Tnt1A RNA was also found in the water-infiltrated leaf discs (not shown), probably because of the wounding stress associated with the infiltration procedure (Vernhettes et al., 1997). Also, it is likely that the bombardment procedure induces a wound response on its own and, thus, we cannot rule out the possibility that the weak induction of Tnt1A detected with 2,4-D in these experiments is the result of a wounding-associated stress response. In line with this hypothesis, it has been previously reported that the promoter of Tnt1A is not inducible by 2,4-D in transgenic tobacco calli (Pauls et al., 1994). On the contrary, the clear induction of the Tnt1A promoter observed after cryptogein and methyl jasmonate incubation in the transient expression analysis reveals an elevated sensitivity of the Tnt1A promoter to both chemicals, and is in agreement with the RT-PCR results obtained.
The presence of several sequences belonging to the Tnt1C subfamily among those obtained from salicylic acid- and 2,4-D-treated leaf discs suggests that Tnt1C expression is probably induced in stress situations regulated by these signal molecules. The transient expression analysis presented here shows that the first 150 nucleotides of the U3 region of Tnt1C are sufficient to produce high levels of expression from a minimal promoter in bombarded leaves incubated with salicylic acid and 2,4-D and suggests that this region contains the promoter elements of Tnt1C that respond to these signaling molecules.
We have not been able to detect any promoter activity of the U3 region of the Tnt1B sequence analyzed. This is not completely unexpected considering that most of the Tnt1B RNA sequences obtained were amplified from subcultured cells and almost no Tnt1B RNA sequence was obtained from infiltrated leaf discs in which the transient expression experiments were performed. These results could suggest that the Tnt1B promoter is not expressed in leaf tissues. We are, at present, transforming tobacco plants with constructs similar to those used in this report to analyze in more detail the expression pattern of the different Tnt1 subfamilies.
The strong correlation of the RT-PCR results and the transient expression experiments suggest that the promoter regions that control the transcription of the Tnt1 elements in response to wounding, cryptogein, or methyl jasmonate are all contained within the U3 region of the different Tnt1 elements.
In conclusion, the results presented here show that the different Tnt1 subfamilies are expressed in tobacco with different patterns, driven by different inducible promoter elements located within their U3 regions. It is interesting to note that the expression of all three Tnt1 subfamilies is strongly regulated, with the different Tnt1 elements only being expressed under stress situations or after inoculation with stress-associated signaling molecules. This emphasizes the importance of transcriptional regulation in the control of plant retrotransposon activity and the tight relationship that exists between retrotransposition and stress situations in plants.
Retrotransposons as Models to Analyze Stress-Associated Expression in Plants
Defense-related responses of plant cells depend on a complex network of signal transduction pathways. Salicylic acid is probably the most well-characterized signaling molecule in plant defense reactions (Klessig et al., 2000), but signal transduction pathways involving methyl jasmonate and ethylene have also been characterized (Reymond and Farmer, 1998). On the other hand, auxin has been shown to activate some defense-related genes through the as-1 element, which also confers inducibility by salicylic acid (Strompen et al., 1988; Xiang et al., 1996). In some cases, these different pathways seem to be independent (Penninckx et al., 1996), but genes concomitantly activated by two of these pathways have also been reported (Strompen et al., 1988; Xiang et al., 1996; Asai et al., 2000). A recent microarray-based analysis has shown that a high number of defense-related genes are coordinately induced by different signal transduction pathways, although genes controlled by only one of these signals, and examples of signal antagonism, were also found (Schenk et al., 2000). This illustrates the complexity of the network of regulatory interactions that controls plant defense reactions, and the risk of drawing general conclusions from the analysis of a single gene promoter. The presence of retrotransposon promoter sequences within the RNA molecule, as well as the high sequence variability found for these elements in plants, allows for the combination of classical promoter analysis with population-based approaches to study transcriptional regulation. We have applied this strategy to analyze the promoter of Tnt1A (Casacuberta and Grandbastien, 1993; Casacuberta et al., 1995; Vernhettes et al., 1997), and here we used a similar approach to improve our analysis of this promoter, as well as to begin to analyze the promoters of the Tnt1B and Tnt1C subfamilies.
Our results show that the Tnt1A RNA populations induced by methyl jasmonate and cryptogein are indistinguishable, suggesting that both signaling molecules induce Tnt1A through the same cis-acting elements. Our results thus reinforce the recent hypothesis that jasmonic acid could be part of the signaling cascade triggered by cryptogein (Rusterucci et al., 1999).
On the other hand, the Tnt1C populations induced by salicylic acid and 2,4-D are also indistinguishable, suggesting that also in this case, both chemicals induce Tnt1C through the same cis-acting elements. It has been already shown that salicylic acid and 2,4-D can induce plant gene expression through as-1-related elements (Chen and Singh, 1999; Niggeweg et al., 2000). It is interesting that although sequence variability among the Tnt1C sequences amplified is high and two different subfamilies can be defined, all of them maintain a consensus as-1-related element within the U3 region. We are currently analyzing the possible implication of this element in the induction of the Tnt1C promoter by auxins and salicylic acid, and we will examine the promoters of the two Tnt1C groups here defined for differences in induction by pathogen-associated stresses.
Sequence Plasticity and Evolution of Stress-Associated Promoters: A Strategy for Retrotransposon Maintenance in Plant Genomes
We show in this paper that the sequence variability found within the U3 regions of Tnt1 elements correlates with differences in their pattern of expression. The three Tnt1 subfamilies contain different promoter elements that induce their activity in different stress situations. Such expression variability should have consequences for the evolution of these elements, as repeated exposure of the host to a particular stress situation should favor amplification of a particular subfamily and a series of different induction events will thus lead to the amplification of different populations of elements in different host genomes. We have previously shown that the three Tnt1 subfamilies are differently represented in different Nicotiana genomes (Vernhettes et al., 1998).
It is interesting to note that the U3 region displays a high degree of sequence variability even within a single Tnt1 subfamily. For example, two different groups of Tnt1C sequences can be defined based on their U3 sequence, so even though both groups of elements are induced by the same chemicals, slight differences in the pattern of expression might exist between them. The simultaneous expression of heterogeneous populations of Tnt1 elements with such differences in expression should allow the selection of different subsets of elements in different genomes, allowing them to evolve and to adapt to their hosts. The existence of Tnt1-related retrotransposon, Retrolyc1, in tomato and related wild species showing extensive sequence similarities to Tnt1 except for its U3 region, which also contains the promoter sequences, seems to confirm that adaptation to the host genome correlates with U3 sequence divergence (Costa et al., 1999; Araujo et al., 2001).
Thus, we propose that the high sequence variability of Tnt1 and other plant retrotransposons, which is a consequence of the infidelity of the retrotransposition process (Gabriel et al., 1996) and the high copy number of these elements in plant genomes (Casacuberta et al., 1995), could have allowed their promoter sequences to evolve the optimum pattern of expression to be maintained in each host genome. This strategy obviously would be very different from that of Ty elements in yeast and could be a consequence of the important differences of the organization of these different host genomes. The high proportion of intragenic regions and repetitive DNA in plant genomes compared with the compact genome of Saccharomyces cerevisiae, as well as the high frequency of polyploidy in plant genome evolution, could have minimized the negative impact of retrotransposons being inserted randomly. In addition, this could have allowed these elements to reach the high copy number that is essential to develop such a quasispecies-like strategy of adaptation.
MATERIALS AND METHODS
Cell Culture
Tobacco (Nicotiana tabacum cv Wisconsin 38) cells cultured in NK1 medium (Xiang et al., 1996) were subcultured once a week by the addition of one-half-volume of fresh media to one-half-volume of cells. After subculturing, cells were harvested at different times, pelleted by centrifugation, and frozen in liquid nitrogen.
Infiltration of Leaf Discs
Tobacco (cv Xanthi, wild-type line XHFD8, Bourgin and Missonier, 1973) leaf discs were infiltrated with water, 1 μg mL−1 of cryptogein, 2 mm salicylic acid, and 10 μm methyl jasmonate or 1 mm 2,4-D as previously described (Vernhettes et al., 1997)
RNA Extractions, Northern-Blot Hybridizations, RT-PCR Amplifications, Cloning, and Sequencing
RNA from cultured cells and infiltrated leaves was obtained by standard methods (Casacuberta et al., 1995). RNA blotting and hybridization with a conserved Tnt1 endonuclease probe was performed as previously described (Mhiri et al., 1997). RT-PCR amplifications with the Avi and dT primers were performed as previously described (Casacuberta et al., 1995), and the PCR fragments were cloned in a pGEM-T vector (Stratagene, La Jolla, CA) and were sequenced on both strands.
Phylogenetic Analysis
Sequences were aligned using the CLUSTAL W multiple-alignment program (version 1.5; Thompson et al., 1994) with some minor refinements. DNADIST in Felsestein's PHYLIP package (Felsenstein, 1989) was used to generate a distance matrix based on the Jukes-Cantor algorithm (Jukes and Cantor, 1969). This was used to generate neighbor-joining trees (Saitou and Nei, 1987). Bootstrap analyses were performed using the Seqboot and Consense programs from Felsestein's PHYLIP package (Felsenstein, 1989).
Promoter-GUS Constructs
A previously described tab7 clone (Vernhettes et al., 1998) was chosen as representative of the U3 region of the Tnt1A subfamily. To construct the A-GUS clone, the tab7 clone was digested by SalI, filled in with Klenow, and re-digested with BamHI. The fragment containing the U3 of tab7 was cloned in a previously described GP plasmid (Casacuberta et al., 1993) containing a GUS reporter gene devoid of promoter and was digested with HindIII (blunt-ended with Klenow) and BamHI. To obtain the B-GUS and C-GUS plasmids, the region upstream of the TATA box of the clones chosen as representatives of the Tnt1B subfamily (cul4) and Tnt1C (cul12) was amplified by PCR by standard procedures, using the universal primer located within the polylinker region of the pGEM plasmid and an oligonucleotide complementary to 25 to 30 nucleotides of each clone 10 nucleotides upstream of their respective TATA box, followed by an HindIII site (5′-GCC AAA GCT CTA CCA ACC TTG ACC-3′ for Tnt1B, 5′-GCC AAA GCT TGC ACA TAT TGA CTT ATG CAA TGA CAT C-3′ for Tnt1C). The amplified fragments were digested with SalI and HindIII and this fragment was used to substitute the SalI-HindIII fragment of the tab7 clone. The corresponding fragments were cloned within the GP plasmid to generate the B-GUS and the C-GUS constructs as for the A-GUS construct.
Microprojectile Bombardment and Enzyme Assays
Tobacco leaf discs were transformed by particle bombardment, with 4 μg of the different Tnt1-GUS constructs and 1 μg of a pCAMV35SLUC used as an internal standard (Marzabal et al., 1998). Enzyme assays were performed as described (Marzabal et al., 1998).
ACKNOWLEDGMENTS
We thank Pilar Fontanet for excellent technical assistance in the greenhouse, and Maria-Lluïsa Espinás and Salomé Prat (Institut de Biologia Molecular de Barcelona-Consejo Superior de Investigaciones Científicas, Barcelona) for helpful discussions and critical reading of the manuscript.
LITERATURE CITED
- Araujo PJ, Casacuberta JM, Costa AP, Hashimoto RY, Grandbastien MA, Van Sluys MA (2001) Retrolyc1 subfamilies defined by different U3 LTR regulatory regions in the Lycopersicon genus. Mol Gen Genet (in press) [DOI] [PubMed]
- Asai T, Stone JM, Heard JE, Kovtun Y, Yorgey P, Sheen J, Ausubel F. Fumonisin B1-induced cell death in Arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell. 2000;12:1823–1835. doi: 10.1105/tpc.12.10.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Kellogg EA. Do plants have a one-way ticket to genomic obesity? Plant Cell. 1997;9:1509–1514. doi: 10.1105/tpc.9.9.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Corces V. Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol. 1989;45:403–434. doi: 10.1146/annurev.mi.43.100189.002155. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Devine SE. Yeast retrotransposons: finding a nice quiet neighborhood. Cell. 1998;93:1087–1089. doi: 10.1016/s0092-8674(00)81450-6. [DOI] [PubMed] [Google Scholar]
- Bourgin JP, Missonier C. Vegetative propagation and cold preservation of haploid plants of Nicotiana tabacum and Nicotiana paniculata. Haploid Inf Serv. 1973;8:7. [Google Scholar]
- Casacuberta JM, Grandbastien MA. Characterization of LTR sequences involved in the protoplast-specific expression of the tobacco Tnt1 retrotransposon. Nucleic Acids Res. 1993;21:2087–2093. doi: 10.1093/nar/21.9.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta JM, Vernhettes S, Grandbastien M-A. Sequence variability within the tobacco retrotransposon Tnt1 population. EMBO J. 1995;14:2670–2678. doi: 10.1002/j.1460-2075.1995.tb07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta JM, Vernhettes S, Grandbastien M-A. Quasispecies in retrotransposons: a role for sequence variability in Tnt1 evolution. Genetica. 1997;100:109–117. [PubMed] [Google Scholar]
- Chen W, Singh KB. The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J. 1999;19:667–677. doi: 10.1046/j.1365-313x.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- Costa AP, Scortecci KC, Hashimoto RY, Araujo PG, Grandbastien MA, Van Sluys MA. Retrolycl-1, a member of the tntl retrotransposon super-family in the Lycopersicon peruvianum genome. Genetica. 1999;107:65–72. [PubMed] [Google Scholar]
- Gabriel A, Willems M, Mules EH, Boeke JD. Replication infidelity during a single cycle of Ty1 retrotransposition. Proc Natl Acad Sci USA. 1996;93:7767–7771. doi: 10.1073/pnas.93.15.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbastien M-A. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 1998;3:181–187. [Google Scholar]
- Grandbastien M-A, Lucas H, Morel J-B, Mhiri C, Vernhettes S, Casacuberta JM. The expression of the tobacco Tnt1 retrotransposon is linked to plant defense responses. Genetica. 1997;100:241–252. [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP: phylogeny inference package (version 3.56) Cladistics. 1989;5:164–166. [Google Scholar]
- Hirochika H. Activation of tobacco retrotransposons during tissue culture. EMBO J. 1993;12:2521–2528. doi: 10.1002/j.1460-2075.1993.tb05907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan IK, McDonald JF. Tempo and mode of Ty element evolution in Saccharomyces cerevisiae. Genetics. 1999;151:1341–1351. doi: 10.1093/genetics/151.4.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian Protein Metabolism. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- Klessig DF, Durner J, Noad R, Navarre DA, Wendehenne D, Kumar D, Zhou JM, Shah J, Zhang S, Kachroo P. Nitric oxide and salicylic acid signaling in plant defense. Proc Natl Acad Sci USA. 2000;97:8849–8855. doi: 10.1073/pnas.97.16.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Bennetzen JL. Plant retrotransposons. Annu Rev Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- Marillonnet S, Wessler SR. Extreme structural heterogeneity among the members of a maize retrotransposon family. Genetics. 1998;150:1245–1256. doi: 10.1093/genetics/150.3.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzabal P, Busk PK, Ludevid MD, Torrent M. The bifactorial endosperm box of gamma-zein gene: characterization and function of the Pb3 and GZM cis-acting elements. Plant J. 1998;16:41–52. doi: 10.1046/j.1365-313x.1998.00272.x. [DOI] [PubMed] [Google Scholar]
- Mhiri C, Morel JB, Vernhettes S, Casacuberta JM, Lucas H, Grandbastien MA. The promoter of the tobacco Tnt1 retrotransposon is induced by wounding and by abiotic stress. Plant Mol Biol. 1997;33:257–266. doi: 10.1023/a:1005727132202. [DOI] [PubMed] [Google Scholar]
- Niggeweg R, Thurow C, Kegler C, Gatz C. Tobacco transcription factor TGA2.2 is the main component of as-1-binding factor ASF-1 and is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J Biol Chem. 2000;275:19897–19905. doi: 10.1074/jbc.M909267199. [DOI] [PubMed] [Google Scholar]
- Pauls PK, Kunert K, Huttner E, Grandbastien MA. Expression of the tobacco Tnt1 retrotransposon promoter in heterologous species. Plant Mol Biol. 1994;26:393–402. doi: 10.1007/BF00039548. [DOI] [PubMed] [Google Scholar]
- Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Metraux JP, Manners JM, Broekaert WF. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouteau S, Grandbastien M-A, Boccara M. Microbial elicitors of plant defense responses activate transcription of a retrotransposon. Plant J. 1994;5:535–542. [Google Scholar]
- Pouteau S, Huttner E, Grandbastien M-A, Caboche M. Specific expression of the tobacco Tnt1 retrotransposon in protoplasts. EMBO J. 1991;10:1911–1918. doi: 10.1002/j.1460-2075.1991.tb07717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Farmer EE. Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol. 1998;1:404–411. doi: 10.1016/s1369-5266(98)80264-1. [DOI] [PubMed] [Google Scholar]
- Ricci P, Bonnet P, Huet JC, Sallantin M, Beauvais-Cante F, Bruneteau M, Billard V, Michel G, Pernollet JC. Structure and activity of proteins from pathogenic fungi Phytophtora eliciting necrosis and acquired resistance in tobacco. Eur J Biochem. 1989;183:555–563. doi: 10.1111/j.1432-1033.1989.tb21084.x. [DOI] [PubMed] [Google Scholar]
- Rusterucci C, Montillet JL, Agnel JP, Battesti C, Alonso B, Knoll A, Bessoule JJ, Etienne P, Suty L, Blein JP. Involvement of lipoxygenase-dependent production of fatty acid hydroperoxides in the development of the hypersensitive cell death induced by cryptogein on tobacco leaves. J Biol Chem. 1999;274:36446–36455. doi: 10.1074/jbc.274.51.36446. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei N. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strompen G, Gruner R, Pfitzner UM. An as-1-like motif controls the level of expression of the gene for the pathogenesis-related protein 1a from tobacco. Plant Mol Biol. 1988;37:871–883. doi: 10.1023/a:1006003916284. [DOI] [PubMed] [Google Scholar]
- Suoniemi A, Narvanto A, Schulman AH. The BARE-1 retrotransposon is transcribed in barley from an LTR promoter active in transient assays. Plant Mol Biol. 1996;31:295–306. doi: 10.1007/BF00021791. [DOI] [PubMed] [Google Scholar]
- Takeda S, Sugimoto K, Otsuki H, Hirochika H. A 13-bp cis-regulatory element in the LTR promoter of the tobacco retrotransposon Tto1 is involved in responsiveness to tissue culture, wounding, methyl jasmonate and fungal elicitors. Plant J. 1999;18:383–393. doi: 10.1046/j.1365-313x.1999.00460.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernhettes S, Grandbastien M-A, Casacuberta JM. In vivo characterization of transcriptional regulatory sequences involved in the defense-associated expression of the tobacco retrotransposon Tnt1. Plant Mol Biol. 1997;36:673–679. doi: 10.1023/a:1005826605598. [DOI] [PubMed] [Google Scholar]
- Vernhettes S, Grandbastien M-A, Casacuberta JM. The evolutionary analysis of the Tnt1 retrotransposon in Nicotiana species reveals the high variability of its regulatory sequences. Mol Biol Evol. 1998;15:827–836. doi: 10.1093/oxfordjournals.molbev.a025988. [DOI] [PubMed] [Google Scholar]
- Vicient CM, Suoniemi A, Anamthawat-Jonsson K, Tanskanen J, Beharav A, Nevo E, Schulman AH. Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell. 1999;11:1769–1784. doi: 10.1105/tpc.11.9.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler SR. Plant retrotransposons: turned on by stress. Curr Biol. 1996;6:959–961. doi: 10.1016/s0960-9822(02)00638-3. [DOI] [PubMed] [Google Scholar]
- Xiang C, Miao ZH, Lam E. Coordinated activation of as-1-type elements and a tobacco glutathione S-transferase gene by auxins, salicylic acid, methyl jasmonate and hydrogen peroxide. Plant Mol Biol. 1996;32:415–426. doi: 10.1007/BF00019093. [DOI] [PubMed] [Google Scholar]