Abstract
In raspberry (Rubus idaeus), development of fruit color and flavor are critically dependent on products of the phenylpropanoid pathway. To determine how these metabolic functions are integrated with the fruit ripening program, we are examining the properties and expression of key genes in the pathway. Here, we report that l- phenylalanine ammonia-lyase (PAL) is encoded in raspberry by a family of two genes (RiPAL1 and RiPAL2). RiPAL1 shares 88% amino acid sequence similarity to RiPAL2, but phylogenetic analysis places RiPAL1 and RiPAL2 in different clusters within the plant PAL gene family. The spatial and temporal expression patterns of the two genes were investigated in various vegetative and floral tissues using the reverse transcriptase competitor polymerase chain reaction assay. Although expression of both genes was detected in all tissues examined, RiPAL1 was associated with early fruit ripening events, whereas expression of RiPAL2 correlated more with later stages of flower and fruit development. Determination of the absolute levels of the two transcripts in various tissues showed that RiPAL1 transcripts were 3- to 10-fold more abundant than those of RiPAL2 in leaves, shoots, roots, young fruits, and ripe fruits. The two RiPAL genes therefore appear to be controlled by different regulatory mechanisms.
Ripening fruits undergo a complex developmental process that radically remodels both the morphology and metabolism of their tissues. In most species, ripening involves expansion of the ovule-derived tissue, a loss of chlorophyll, and a progressive decrease in tissue firmness. However, the traits that make fruits uniquely attractive to harvesting species are the accumulation of specific metabolites, notably pigments and aroma volatiles (Brady, 1987). The quality of ripe raspberry (Rubus idaeus) fruits derives, in part, from accumulation of specific anthocyanin pigments (cyanidin glycosides) and characteristic flavor components (predominantly p-hydroxyphenylbutan-2-one; Perkins-Veazie and Nonnecke, 1992; Borejsza-Wysocki and Hrazdina, 1994). Both of these metabolites are biosynthetically derived from l-Phe through the phenylpropanoid pathway (Barrit and Torre, 1975; Borejsza-Wysocki and Hrazdina, 1994).
Elevated levels of phenylpropanoid derivatives are also associated with ripening in other fruits. The aroma of ripe strawberry (Fragaria × ananassa) fruits is attributed to the accumulation of volatile cinnamate derivatives (Latza et al., 1996), and the formation of flavor in developing fruits of Vanilla planifolia is due to the accumulation of phenolics such as 4-hydroxybenzaldehyde, 4-hydroxybenzoic acid, vanillin, and vanillic acid (Brodelius, 1994). Green fruits of Litchi chinensis contain the anthocyanin, malvidin 3-acetylglucoside, whereas ripe fruits contain cyanidin-3-rutinoside (Rivera-Lopez et al., 1999). In Capsicum frutescens, maturation of the fruit is accompanied by the disappearance of three cinnamyl glycosides and two flavonoids, whereas “lignin-like” substances and the glycosides of vanillic acid and p-hydroxybenzaldehyde accumulate (Sukrasno and Yeoman, 1993). However, although such changes in phenylpropanoid metabolite profiles have been documented, there is little information concerning the underlying regulatory mechanisms involved.
Synthesis of phenylpropanoids in fruits could be regulated at multiple steps, including the entry of sugars into the shikimic acid pathway, the entry of Phe into the general phenylpropanoid pathway, or the entry of activated coenzyme A (CoA) esters into various subbranches of the phenylpropanoid pathway. l-Phe ammonia-lyase (PAL; EC 4.3.1.4) is the first enzyme of the phenylpropanoid pathway. PAL catalyzes the nonoxidative deamination of l-Phe to yield cinnamic acid, a reaction that is generally considered to represent a key point at which carbon flux into this pathway is controlled.
PAL has been characterized from a limited number of fruits, including citrus (Dubery and Schabort, 1986), grape (Vitis vinifera) berries (Roubelakis-Angelakis and Kliewer, 1985), and strawberry (Given et al., 1988a). Changes in PAL activity have been described for ripening fruits of grape (Hrazdina et al., 1984), apple (Malus pumila; Faragher and Brohier, 1984), and strawberry (Given et al., 1988b; Cheng and Breen, 1991), whereas PAL gene expression has been profiled in ripening fruits of melon (Cucumis melo; Diallinas and Kanellis, 1994) and sweet cherry (Prunus arium; Wiersma and Wu, 1998). In strawberry, it is noteworthy that two peaks of PAL activity have been reported, one in green fruits and a second in nearly ripe fruits (Cheng and Breen, 1991), but whether this represents different PAL gene products is not known.
PAL appears to exist universally in higher plants as a family of genes, and the presence of PAL isoforms is a common observation. The significance of this diversity is unclear, but evidence for a degree of metabolic channeling within phenylpropanoid metabolism suggests that partitioning of photosynthate into particular branches of phenylpropanoid metabolism may involve labile multi-enzyme complexes that include specific isoforms of PAL (Hrazdina and Wagner, 1985; Rasmussen and Dixon, 1999).
Fruit ripening in raspberry requires coordinated expression of genes encoding enzymes in the core phenylpropanoid pathway, such as PAL and 4-coumarate CoA ligase, and enzymes in branch pathways such as chalcone synthase, to assure accumulation of the essential flavor metabolites and pigments. To determine how this biosynthetic activity is regulated and integrated within the ripening program, we have characterized each of these key genes in raspberry. In the present study, we describe the PAL gene family structure in this species, as well as the developmental expression pattern of the raspberry PAL genes.
RESULTS
PCR-Based Search for the Raspberry PAL Gene Family
We took advantage of the observation that PAL sequences from divergent taxa share a few short stretches of highly conserved residues, whose presence probably reflects a stringent requirement that the PAL protein undergo a novel posttranslational modification to form an unusual active site electrophile (Schuster and Retey, 1995). Degenerate PCR primers were designed to target these signature motifs, and with raspberry genomic DNA as a template, various combinations of the primers were used for amplification of PAL genes.
Because all angiosperm PAL genes characterized to date have a single intron in a conserved position between the binding sites of primers P5-(P2/P6), these primer combinations were expected to amplify fragments longer than 1.1 kb (Fig. 1). In fact, amplifications with combinations P5-P2 or P5-P6 each yielded an intense band of approximately 1.8 kb. Amplification reactions with the primer pair P7-(P2/P6), on the other hand, yielded the predicted size fragment of about 0.9 kb. Multiple colonies obtained from subcloning the products of two independent PCR reactions derived from each primer combination were analyzed for the presence of inserts by digestion with EcoRI and XbaI. We sequenced the 3′ end of >20 randomly chosen clones from each of the primer pair combinations P7-(P2/P6), and five additional clones from each of the primer pair combinations P5-(P2/P6).
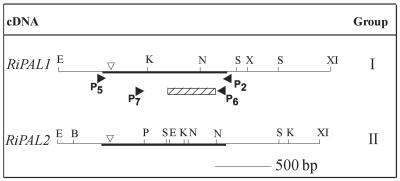
Figure 1.
Summary of the partial and full-length raspberry PAL clones identified. The dark line represents the partial fragment isolated and characterized during the PCR-based homology search. Closed arrows indicate the target positions of the degenerate primers. The 366-bp (122 amino acids) region compared and analyzed in detail for the PCR-based homology screening is indicated by the hatched box. Inverted open arrows ( 209 ) indicate the location of the intron found within the genomic fragments of the two raspberry PAL genes. Restriction enzymes distinguishing the two groups of cDNA clones are: E, EcoRI; K, KpnI; N, NcoI; S, SmaI; X, XbaI; XI, XhoI; B, BamHI; and P, PstI.
Comparison of a stretch of 366 nucleotides (122 amino acids) sequenced from all 50 clones revealed two different classes of PAL sequences, arbitrarily named Ripal1 (GenBank accession no. AF304366) and Ripal2 (GenBank accession no. AF304367) The Ripal1-like sequence class was represented by 43 clones: 16 clones obtained from primer-pair P7-P2, 18 clones from P7-P6, five clones from P5-P2, and four clones from P5-P6. A closer analysis of these sequences revealed that the clones could be grouped into two subcategories that varied from each other at fewer than 10 nucleotide positions. Twenty clones were identical in the 366-nucleotide positions and this sequence therefore was designated Ripal1. Each of the 15 additional clones had single base substitutions at nucleotide positions 75 (C→T), 114 (A→G), 141 (C→T), 144 (G→A), 162 (C→T), 228 (T→C), 267 (A→T), and 351 (C→G). It is interesting that none of these substitutions introduced a change in the amino acid sequence. Because these specific substitutions were seen in multiple clones obtained from two independent PCR reactions, and because the target DNA used for amplification in this analysis was diploid (and derived from multiple non-clonal parents), the observed differences presumably reflect allelic polymorphism at the Ripal1 locus. Eight additional clones representing the Ripal1 locus were found to contain single base substitutions at fewer than three nucleotide positions, each within the 366 nucleotides compared. These substitutions were not seen in clones obtained from another PCR reaction with the same or a different primer set, and thus likely reflect synthesis errors introduced by Taq DNA polymerase.
Ripal2 was represented by eight clones obtained from primer set P7-P2, four clones from P7-P6, two clones from P5-P2, and one clone from P5–6. Eight of these clones were identical over the 366 nucleotides compared and this sequence therefore was designated Ripal2. An additional four clones contained substitutions at both nucleotide positions 234 (T→C) and 246 (T→C). These sequences most likely represent allelic variation because they were detected in clones obtained from two independent PCR reactions with the same or different primer combinations. Three of the 15 Ripal2-like clones had single nucleotide substitutions at positions that were not represented in other clones, and thus most likely arose from the error introduced by Taq DNA polymerase. Ripal2 has an internal EcoRI site 554 bp downstream of primer P5 (281 bp downstream of primer P7; Fig. 1) that accounted for clones with inserts of 0.7 kb obtained with all the primer combinations used in this analysis.
Amplifications of Ripal1 and Ripal2 with primer combinations P5-(P2/P6) had revealed the likely presence of intron(s) within the regions amplified, based on the size of the PCR amplification products (1.8 kb). Because Ripal2 had an internal EcoRI site (Fig. 1), we cloned the 1.8-kb PCR-amplified products of P5-(P2/P6) into a dideoxy T-tailed vector (Holton and Graham, 1991). The two classes of raspberry PAL genes could be distinguished by digesting the cloned inserts with EcoRI. Sequencing the full 1.8-kb insert from each of the two classes confirmed the presence of introns 778 and 687 bp in size in RiPAL1 and RiPAL2, respectively. The two introns share 57% nucleotide sequence identity.
The sizes of PAL introns sequenced thus far vary from 90 bp in pea (Pisum sativum) PAL2 (Yamada et al., 1992) to approximately 1,900 bp in Nicotiana tabacum PALs. The raspberry intron is inserted between the second and third bases of a conserved Arg codon at a position that is identical to that observed in other angiosperm PAL genes (Arg-121 in Arabidopsis PAL3; Fig. 1). In PAL genes from potato (Solanum tuberosum; Joos and Hahlbrock, 1992) and orchid (Bromheadia finlaysoniana; Liew et al., 1996), this Arg is replaced by a Lys but the position of the intron is conserved. The sequence around the exon-intron junctions in the Ripal genes is similar to the consensus for dicot genes (Brown, 1986) and conforms to the “GT-AG” rule for donor/acceptor sites (Breathnach and Chambon, 1981). Within the amplified regions, the Ripals do not contain the second intron found only in Arabidopsis PAL3 and Arabidopsis PAL4 (GenBank accession no. Ac009400) genes to date.
Isolation and Characterization of Rubus PAL cDNAs
Three rounds of screening the cDNA library with a mixed population of Ripal1 and Ripal2 yielded 25 clones with potentially full-length PAL gene inserts, which RFLP analysis grouped into two discrete groups (Fig. 1). Group I clones had a similar RFLP pattern and an exact sequence match to Ripal1 locus. Group II clones had a similar RFLP pattern and exact sequence match with the overlapping regions of genomic fragments of Ripal2 locus. Because group I and group II cDNA clones were the full-length expressed versions of the two PAL loci in raspberry, we designated these clones as RiPAL1 and RiPAL2.
The open reading frame of RiPAL1 encodes a polypeptide of 710 amino acids with a predicted molecular mass and theoretical pI of 77,580 D and 6.2, respectively. The RiPAL2 open reading frame encodes a polypeptide of 730 amino acids with a predicted molecular mass and theoretical pI of 79,356 D and 5.9, respectively. The calculated molecular masses of the two deduced open reading frames are consistent with the size determined for PAL polypeptides (72–83 kD) from other plants using SDS/PAGE (Schomburg and Salzmann, 1990).
Sequence analysis of the cDNAs demonstrated that the introns detected in each of the two genes were spliced at the expected positions. The deduced amino acid sequences of RiPAL1 and RiPAL2 are 81% identical (88% similar) to each other. As has been noted in other PAL protein sequences (Cramer et al., 1989), the greatest divergence is found in the N-terminal region. This is reflected in the sequences of the exon I portions of the two raspberry PAL genes, which share 73% amino acid sequence identity, whereas the sequences representing the exon II regions shared 83% amino acid sequence identity.
In amino acid sequence comparisons between the RiPALs and the PAL gene family members from other species, the sequences of RiPAL1 and RiPAL2 were more closely related to predicted amino acid sequences of PAL genes isolated from dicot plant species than to those from monocot plants, as expected. The polypeptides encoded by rice (Oryza sativa) PAL genes, for example, share only 66% to 72% amino acid sequence identity with RiPALs. A posttranslational phosphorylation site (Thr-545) detected in the bean (Phaseolus vulgaris) PAL sequence (Allwood et al., 1999) is conserved in the two raspberry PALs, suggesting that phosphorylation might play a role in modulating the activity of these PALs. Other potential posttranslational modification sites are also predicted for both RiPALs, but their relevance is unknown.
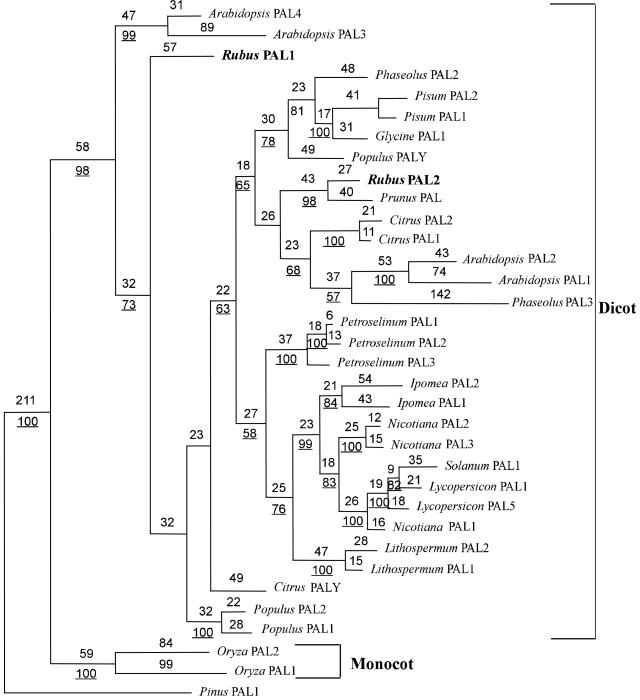
To study the evolutionary relationship among PAL genes, sequences of a set of PAL gene family members from other plant species, including raspberry, were analyzed using the maximum parsimony method. The heuristic search resulted in a single tree that we designate as the most parsimonious tree. The gymnosperm PAL sequence from Pinus taeda was selected as the outgroup because gymnosperms are considered on the basis of both morphological characters and 18s RNA sequences (Chaw et al., 1997) to be ancestral to the angiosperms. With the pine (Pinus taeda) PAL sequence as the outgroup, the high bootstrap values (100% of the 1,000 bootstrap replicates) place dicot and monocot PALs in separate monophyletic groups (Fig. 2). Among the dicot PALs, RiPAL1 formed a distinct subgroup, whereas RiPAL2 clustered with the cherry PAL sequence, the only other full-length PAL gene characterized from a Rosaceae species.
Figure 2.
A phylogenetic tree depicting the relationships among PALs. PALs identified from other species were used subject to the following restrictions: (a) Only species in which full-length PAL gene families have been characterized were used in the analysis; and (b) within a gene family, only sequences that differed by more than 5% in primary nucleotide sequence were incorporated in the analysis to reduce the inclusion of allelic sequences. Amino acid sequences were aligned and the parsimonious tree was constructed using the program PAUP 4.0.b2. Characters were reweighted for maximum retention indices. The tree shown has a consistency index of 0.788. Branch lengths are indicated above the branch lines and bootstrap support values are underlined. Support values below 50% are not indicated. The P. taeda PAL sequence (accession no. T09777) was used to root the tree.
Developmental Regulation of RiPAL Genes
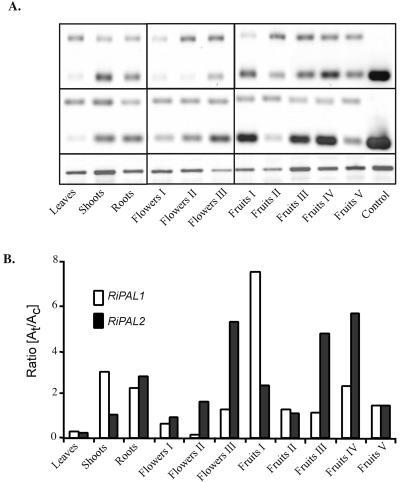
Because we were interested in the relationship between PAL gene expression and the process of raspberry fruit ripening, we examined the expression patterns of the two RiPAL gene family members both during fruit development and in other tissues. Using a semiquantitative competitive reverse transcriptase (RT)-PCR method based on an internal DNA competitor, we were able to demonstrate that both members of the RiPAL gene family were expressed in all the tissues examined (Fig. 4). Among the tissues examined, the two genes were expressed most actively in fertilized flowers or in fruits. Moderate expression was also detected in shoots and roots, whereas very low levels of expression were observed in the young leaves. Low expression of PAL genes in leaves accompanied by a higher level of expression in roots has also been seen in other plant species (Liang et al., 1989; Joos and Hahlbrock, 1992; Yamada et al., 1992; Subramaniam et al., 1993).
Figure 4.
Semiquantitative RT-cPCR estimation of the accumulation of specific RiPAL transcripts in different organs of raspberry. A, RT-cPCR was performed using 100 ng of total RNA isolated from: young leaves, shoots, roots, and different developmental stages of flowers and fruits. Following 32 cycles of amplification, the products (50% of each amplification mix) were resolved in a 3% (w/v) Tris-acetate-EDTA-agarose/ethidium bromide gel. B, The relative amounts of target (t; 217/218 bp) and competitor (c; 370 bp) amplification products were calculated, and the ratio of the two products graphed. Similar results were obtained in two independent experiments. The expression level of a given RiPAL gene can be compared between tissues, but expression of the two RiPAL genes cannot be compared within a tissue. The intensity of RiPAL bands was normalized to the intensity of the RiHis3 product (395 bp) as a control for equal amounts of starting RNA. No amplification products of this size were obtained in the absence of the RiHis3 primers. M, 100-bp DNA size marker; C, positive control cDNA.
The expression profile of RiPAL2 could be readily mapped onto the different stages of reproductive tissue development, with the highest expression of RiPAL2 occurring in fully fertilized flowers (stage III). As the flowers developed into fruits, levels of RiPAL2 were low during early stages of fruit development and then peaked as the fruits ripened through the pink (stage III) and then red (stage IV) stages. In contrast, RiPAL1 was most highly expressed in green immature fruits (stage I), and its expression declined as the fruits approached maturity. The expression of RiPAL2 followed the expression profile of the orthologous cherry PAL gene, which is maximally induced in “early pink” stages of fruit development (Wiersma and Wu, 1998).
Quantitation of the Expression of RiPAL Genes in Different Organs of Raspberry
Although the previous analysis showed that the two genes were each expressed in all tissues, their expression levels within a single tissue could not be directly compared using this method. Therefore, we included a positive standard in the RT-PCR assay to quantify the absolute amounts of each RiPAL gene in various tissue. As determined by this analysis, the absolute amounts of the two PAL gene transcripts in raspberry fruits (stage III) were essentially equal and equivalent in each case to 65 ng mg−1 total RNA. This result is consistent with the class distribution of the RiPAL clones isolated from the raspberry cDNA library, where the 26 full-length characterized cDNAs fell into two RFLP classes that contained 12 RiPAL1 and 14 RiPAL2 clones, respectively.
The quantitative analysis revealed that RiPAL1 mRNA is more abundant than RiPAL2 in most of the tissues examined (Table I). RiPAL1 is about 12-fold more abundant in shoots and about 5-fold higher in leaves and young fruit (stage I and II). Only in mature flowers (stage III) and maturing fruits (stage III) did transcripts of RiPAL1 and RiPAL2 appear at similar levels.
Table I.
Levels of specific RiPAL transcripts in different organs
| Organ |
RiPAL Transcript
Levels
|
Ratio RiPAL1:RiPAL2 | |
|---|---|---|---|
| RiPAL1 | RiPAL2 | ||
| ng mg−1 total cellular RNA | |||
| Leaves | 14 | 3 | 4.4 |
| Shoots | 169 | 15 | 11.5 |
| Roots | 126 | 39 | 3.3 |
| Flowers I | 37 | 13 | 2.9 |
| Flowers II | 8 | 23 | 0.3 |
| Flowers III | 76 | 73 | 1.0 |
| Fruits I | 426 | 75 | 5.7 |
| Fruits II | 73 | 15 | 4.9 |
| Fruits III | 65 | 65 | 1.0 |
| Fruits IV | 133 | 78 | 1.7 |
| Fruits V | 84 | 20 | 4.2 |
The absolute levels of the two RiPAL transcripts were first determined for stage III fruits. The levels of transcripts of each cDNA in other organs were then determined based on the relative ratio (target:competitor) of expression of each cDNA in those tissues compared with that seen in stage III fruits (Fig. 4B).
DISCUSSION
We have used a combination of approaches to identify the repertoire of PAL genes in raspberry and to study the tissue-specific and developmental expression of each gene family member during ripening of raspberry fruits. To our knowledge, this is the first comprehensive study of the PAL genes from a fruit crop.
To identify the PAL genes in raspberry, we initially used degenerate PCR primers that target evolutionarily conserved sequences in PAL. Such a PCR-based homology approach was previously successful in identifying multiple divergent PAL genes in pine (Butland et al., 1998), where an earlier cDNA library screening had led to the erroneous conclusion that PAL might be encoded by a single gene in gymnosperms (Whetten and Sederoff, 1992). By sequencing multiple PCR-derived clones, we identified partial fragments of two distinct raspberry PAL genes (Ripal1 and Ripal2), distinguishable by a polymorphic EcoRI restriction endonuclease site. We also isolated putative alleles of Ripal1 and Ripal2. Screening of a cDNA library derived from partially ripe fruits of raspberry resulted in the isolation of two classes of cDNA clones that represented the full-length sequences corresponding to the PCR-generated genomic fragments. The presence of both sequences in the cDNA library demonstrates that the genomic sequences do not represent pseudogenes.
The two genomic fragments designated Ripal1 and Ripal2 shared 71% nucleotide sequence identity, confirming that the PCR primers used in this study are capable of efficiently amplifying divergent PAL genes. Using the same sets of degenerate PCR primers, Butland et al. (1998) identified five pal loci in Pinus banskiana that share 69% to 94% nucleotide sequence identity within a 366-bp region similar to that used for comparison of the raspberry PAL genes. Likewise, we have used the primer pair combination P7-P2 to successfully sample the presence of PAL genes in various plant species, including those belonging to taxa as remote as the Hepatophyta, Bryophyta, and Lycophyta (M. McQuoid and B. Ellis, unpublished data). This emphasizes the conserved nature of the PAL primer target sequences across great evolutionary distance. In fact, the peptide sequences targeted by the PAL primers are also conserved in a catalytically similar enzyme, His ammonia-lyase (EC 4.3.1.3). The recently reported crystal structure of His ammonia-lyase reveals that the residues targeted by primers P5, P7, P2, and P6 are all required for the formation of the active sites in these tetrameric ammonia-lyase enzyme (Schwede et al., 1999).
Several lines of evidence support the conclusion that the RiPAL gene family consists of just two members. First, amplification of the raspberry genome with different sets of degenerate PCR primers amplified the same two classes of genes in each case. Second, screening of a cDNA library under moderate stringency conditions recovered an array of clones that could all be assigned to two groups, corresponding to the genomic PCR products. Finally, in a Southern-blot analysis with each of the two RiPAL cDNAs, single genomic restriction fragments generated by each of six restriction endonucleases hybridized to each cDNA (data not shown). Nevertheless, we cannot with certainty exclude the existence of additional, even more divergent members.
Gene duplication followed by divergence in regulatory and/or protein coding sequences has long been recognized as a potential source of genes with novel functional capabilities (Ohno, 1970). Our analysis of the clustering relationships of Arabidopsis PAL clones (Fig. 2) suggests that duplication has been an important theme in the evolution of PAL genes. As was observed earlier (Wanner et al., 1995), such duplication events apparently have occurred both before and after the divergence of monocots and dicots that is proposed to have taken place between 200 and 100 million years ago (Stewart and Rothwell, 1993). Events that occurred later in the evolution of angiosperms, after the split between monocots and dicots, resulted in PAL genes within a single species that cluster together, rather than with PAL genes from other species. For example, Petroselinum crispum PAL is encoded by a family of four genes that cluster closely together, as do PAL gene family members characterized from N. tabacum, potato, Lycopersicon esculentum, and Lithospermum erythrorhizon.
Consistent with this model of relatively recent divergence, the biochemical and molecular data for the Petroselinum sp. PALs expressed as recombinant proteins show that they have nearly indistinguishable catalytic properties (Appert et al., 1994). The four genes also share considerable sequence similarity in their promoter elements and are induced in a similar manner (Logemann et al., 1995). In contrast to these recently diverged PALs, however, Phaseolus vulgaris PAL genes do not cluster together. They also encode unique PAL isoforms (Liang et al., 1989), and are differentially induced by different stimuli (Liang et al., 1989). Because the two raspberry PAL sequences do not cluster together, they likewise may have evolved to encode distinct PAL isoforms that play different metabolic roles.
Although the expression profiles for RiPAL1 and RiPAL2 in raspberry are broadly similar (Fig. 4 and Table I), there are clearly both qualitative and quantitative differences in their regulation, implying that their expression is controlled by distinct regulatory signals. During fruit ripening, the strongest signal for RiPAL1 was detected in stage I fruits, whereas the strongest signal for RiPAL2 was observed in stage IV fruits. Although fruits at these two stages clearly differ in their chemistry, determination of the exact role played by each RiPAL isoform in supporting accumulation of specific phenylpropanoid products in fruits would require detailed metabolite profiling. It is perhaps noteworthy that two peaks of PAL activity have been reported in ripening strawberry. The first peak, observed in green fruits, has been suggested to be involved in the synthesis of flavonoids (e.g. condensed tannins) and phenolics during early fruit development, whereas the second peak, in nearly ripe fruits, is correlated with the anthocyanin accumulation that is the hallmark of ripe fruits (Cheng and Breen, 1991).
Although the flowering and fruiting developmental program clearly induced both RiPAL transcripts, their overall abundance is not uniquely correlated with the process of fruit ripening. RiPAL1 is strongly expressed in raspberry shoots and roots, whereas RiPAL2 is expressed at generally lower levels throughout the plant, a pattern that is reminiscent of the behavior of the three Phaseolus sp. PAL genes (Liang et al., 1989). All three members of the Phaseolus sp. PAL gene family are expressed in roots, whereas PAL1 and PAL2 are expressed in shoots. PAL1 is also weakly expressed in Phaseolus sp. petals, whereas PAL2 is strongly expressed in that tissue and PAL3 transcripts could not be detected. Only PAL1 is expressed in leaves (Liang et al., 1989). Thus, as in Phaseolus sp., the RiPAL1 and RiPAL2 genes are both phylogenetically separated and perhaps regulated by different developmental control mechanisms. We have recently observed a similar complexity in the differential regulation of members of a family of 4-coumarate:CoA ligase genes, and a family of polyketide synthase genes, in raspberry (A. Kumar and B. Ellis, unpublished data).
The identification and characterization of two PAL genes from a fruit cDNA library creates an opportunity to explore the possible functions of multiple PAL genes during fruit development. To resolve their respective roles, it would be informative to selectively silence each of the two raspberry PAL genes and monitor the resulting transgenic phenotypes. Transformation of this species has been accomplished (Hassan et al., 1993), and the presence of gene-specific sequences within the N-terminal regions of RiPAL1 and RiPAL2 should make it possible to deploy posttranscriptional gene silencing procedures (Waterhouse et al., 1998). Therefore, it should be feasible to dissect the functions of the raspberry PAL gene family, and the cis-acting elements involved in selective expression during fruit development.
MATERIALS AND METHODS
Plant Growth Conditions and Materials
Raspberry (Rubus idaeus L. cv Meeker) plants were grown in the experimental plots of the Agriculture and Agri-Food Canada Research Station (Abbotsford, BC) or in the greenhouse (Faculty of Agricultural Sciences, University of British Columbia) under ambient conditions. All harvested plant tissues were immediately frozen in liquid nitrogen and stored at −80°C.
Raspberry leaf, shoot, and root tissues were pooled from several plants grown under greenhouse conditions. Flowers and fruits at different developmental stages were pooled from multiple plants grown in an experimental field trial by Agriculture and Agri-Food Canada. Flowers I consisted of closed inflorescence buds, flowers II consisted of fully open flowers, and flowers III consisted of fertilized flowers. Fruits I were green, hard, and still undergoing cell expansion. Fruits II were still green but had almost reached mature size. Fruits III were yellow, starting to “blush,” and had reached full size. Fruits IV were fully ripe, with the color and aroma fully developed, whereas fruits V were slightly overripe and somewhat dehydrated.
Gene Amplification and Characterization
Degenerate PCR primers capable of amplifying segments of the PAL genes are: P5, 5′-CGGAATTCTACGG(T/C) GTCACIAC(T/C) GGITT(T/C) GG-3′; P7, 5′-CGGAATTCATC(T/A) CIGCITCIGGIGA(C/T)(T/C) T-3′; P2, 5′-GCTCTAGATG(T/C) TCIGCI(G/C)(A/T)(T/C) TGIAC(A/G) TG-3′; and P6, 5′-GCTCTAGATTIAC(G/A) TC (C/T) G(G/A) TT(A/G) TG(C/T) TC-3′. Each primer carried a restriction endonuclease site (underlined) to facilitate directional cloning of the PCR products. The potential target binding sites of these primers have been graphically represented in Figure 1.
Genomic DNA from young leaves was isolated using the method described by Doyle and Doyle (1990). Amplification reactions contained 100 ng of genomic DNA, 1× Appligene buffer (providing a final concentration of 1.5 mm MgCl2), 0.5 μm each 2′-deoxynucleoside-5′-triphosphate, 2 μm each primer, and 2.5 units Taq DNA polymerase. The reaction mixture was incubated at 95°C for 10 min and then subjected to 35 cycles of amplification (95°C for 50 s, 55°C for 50 s, and 72°C for 1 min), and completed by a final 10-min extension at 72°C in a Techne PHC-3 thermal cycler (Mandel Scientific, Guelph, ON). The PCR reaction products were analyzed by 1% (w/v) Tris-acetate-EDTA-agarose gel electrophoresis.
Amplified products of two independent PCR reactions from each primer combination were subcloned into EcoRI/XbaI-digested pUC19. Multiple clones from each subcloning experiment were amplified with vector-specific primers M13R and M13F to confirm the presence of inserts.
Construction and Screening of the Raspberry cDNA Library
Total RNA was isolated from raspberry fruits (stage III) using the RNeasy Maxi Kit (Qiagen, Valencia, CA) following the manufacturer's protocol. Poly(A+) RNA was isolated from 1.5 mg of total RNA using Dynabeads Oligo (dT25; Dynal, Lake Success, NY) following the manufacturer's instructions. A cDNA library was constructed from 5 μg of poly(A+) RNA using a Uni-ZAP XR Library Construction Kit (Stratagene, La Jolla, CA). The cDNA library, consisting of approximately 107 independent clones, was amplified once to obtain high-titer stock.
Approximately 5 × 105 plaques of the amplified cDNA library were blotted in duplicate on Hybond N+ nylon membrane (Amersham, Piscataway, NJ). The membranes were screened with a mixed population of Ripal1 and Ripal2 radiolabeled to a high specific activity with [α-32P]dATP using a Random Primer Labeling kit (Life Technologies, Burlington, ON). The nylon membranes were hybridized at 50°C for 16 h in hybridization buffer consisting of 6× SSC buffer, 20 mm NaH2PO4, 0.1% (v/v) SDS, 5× Denhardt's reagent, and 500 μg mL−1 denatured salmon sperm DNA. The membranes were washed three times (20 min each) at 50°C with 6× SSC buffer and 0.1% (v/v) SDS. After the tertiary screen, the insert of each positive plaque was amplified with vector-specific primer T3 and PAL gene-specific primer P6, employing the protocol used for amplifications of the genomic fragments. Clones with potential full-length genes were re-amplified with vector-specific primers T3 and T7 to obtain the full-length inserts. Amplified full-length inserts (5 μL) were digested with restriction enzymes (BamHI, HindIII, EcoRI, EcoRV, KpnI, NcoI, SalI, SmaI, and XhoI) to distinguish clones based on RFLP. One plaque representative of each RFLP class harboring the largest size insert was rescued as a pBluescript II SK(-) phagemid using the ExAssist (Stratagene) helper phage.
Sequence Analysis
Plasmid DNA from selected clones was isolated for sequencing following a mini-alkaline lysis/polyethylene glycol precipitation procedure (Ausubel et al., 1995). Both strands of the insert were sequenced with the M13 universal primer and/or synthetic oligonucleotide primers as needed to extend the sequence. Sequencing reactions were carried out at the Nucleic Acid Protein Service Unit (University of British Columbia) in an ABI 373 DNA sequencer (Applied Biosystems, Foster City, CA). All sequences were edited and analyzed using the PC/GENE Software (Intelligenetics, Mountain View, CA). Database searches for sequence homology and comparisons were performed using various web-based analytical tools. Sequences for the RiPAL1 and RiPAL2 cDNAs were submitted to GenBank (accession nos. AF237954 and AF237955, respectively).
RT-cPCR
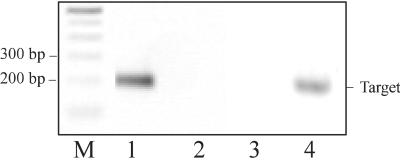
Gene-specific primers capable of amplifying a segment between nucleotides 2,009 and 2,227 of RiPAL1 cDNA are: P15 (5′-CGACAATGCCAGGATCGAAT) and antisense primer P13 (5′-TCCTTCAAACACTCCAGCAGA-3′). Gene-specific primers capable of amplifying a segment between nucleotides 2,009 and 2,225 of RiPAL2 cDNA are: P25 (5′-TGAGAGCGCTAGGGCTGCG-3′) and antisense primer P23 (5′-GCTGAGGCAGCTGAGAATG-3′). Gene-specific primers for amplification of a fragment of the Rubus sp. HistoneH3 gene (GenBank accession no. AF304365) had the nucleotide sequences: H15 (5′-ATGGCGCGGACGAAGGA-3′) and H13 (5′-GCCTACGCCGCCCGCTCAACCTA-3′). The specificity of the gene-specific primers was tested by control amplifications of the reciprocal plasmid cDNAs (Fig. 3).
Figure 3.
Specificity of the gene-specific primers designed to amplify individual members of the RiPAL gene family. Lane 1, RiPAL1 cDNA with RiPAL1-specific primers P15 and P13; lane 2, RiPAL2 cDNA with RiPAL1-specific primers P15 and P13; lane 3, RiPAL1 cDNA with RiPAL2-specific primers P25 and P23; lane 4, RiPAL2 cDNA with RiPAL2-specific primers P25 and P23. M, 100-bp Mr marker.
To generate PCR competitors, a 330-bp fragment of a spruce coniferin-β-glucosidase gene (M. Gray-Mitsumune and B.E. Ellis, unpublished data) was first amplified with primers C15 (5′-CCCCTAACAGGAATTCTGCG-3′) and C13 (5′-ACCATCGCAGATTGAAGGAC-3′). The PCR product was then re-amplified with composite primers containing both coniferin-β-glucosidase and RiPAL1/RiPAL2 gene-specific sequences, and finally with RiPAL1/RiPAL2 gene-specific primers only. This procedure generated two 370-bp non-homologous DNA fragments (competitors) containing at their ends appropriate templates for RiPAL1/RiPAL2 primers.
A constant amount of total RNA (100 ng) from various raspberry tissues was reverse-transcribed into cDNA using Omniscript Reverse Transcriptase (Qiagen) in a reaction volume (20 μL) containing 1× RT buffer, 0.5 mm each dNTP, 1 μm oligo-dT primer, and 10 units RNase inhibitor (Pharmacia, Piscataway, NJ). The reaction mixture was incubated at 37°C for 1 h followed by 5 min incubation at 95°C to destroy the RT enzyme. To normalize for equal amounts of total RNA and efficiency of cDNA synthesis from various tissue samples, the intensities of the bands were normalized with the average intensity of the RiHis3 product across the samples investigated. For competition assays, the first-strand cDNA reaction (1 μL) was amplified in a total volume of 20 μL containing 200 nm each PCR primer, 200 μm each dNTP, and 2.5 units of Taq DNA polymerase in 1× PCR buffer (Qiagen), 1× Q solution (Qiagen), and 1 attomol of competitor. The thermal cycling conditions were 94°C for 5 min, followed by 25 cycles for RiHis3 or 32 cycles for RiPAL of 94°C for 20 s, 59°C for 50 s, and 72°C for 50 s, followed by a final extension for 5 min at 72°C. The PCR product (10 μL) was analyzed on a 3% (w/v) TAE-agarose gel and stained with ethidium bromide. The staining intensity was digitally quantified using the Scion Image (Scion Corporations, Frederick, MD). Relative intensities of the target and competitor bands were expressed as arbitrary units after correction for band size differences between the competitor and target. This analysis was repeated twice with consistent results and representative data from one analysis have been presented.
To determine the absolute levels of the two transcripts across different tissues, the absolute levels were first determined in fruits (stage III) because it had been established by cDNA screening that both genes are expressed in this tissue. Constant aliquots of cDNA (2 μL) were used as PCR template with RiPAL1/RiPAL2 gene-specific primer pairs in the presence of a series of dilutions of each competitor ranging from 100 to 3 × 10−3 attomoles. The two targets, the 217-/218-bp fragment from RiPAL1/RiPAL2 mRNA and the control competitor (370-bp) fragments, are simultaneously amplified by the polymerase in proportions that reflect the relative abundance of each target species. As decreasing amounts of competitor are added, the amount of RiPALs cDNA being amplified increases. Hence, the ratio of the two bands (target:competitor) observed after agarose gel electrophoresis changes across the dilution series. After densitometric scanning of the stained gel, the logarithm of the molar ratio of the target to competitor was plotted as a function of the logarithm of the amount of competitor added, to establish the point at which the amount of target is equal to the amount of competitor. Once the absolute amounts of RiPAL1/RiPAL2 genes in developing fruits (stage III) were known, it was possible to calculate the amounts of each transcript in other tissues by comparing the ratio (target:competitor) obtained for each tissue with the ratio (target:competitor) established for expression in fruits stage III.
Phylogenetic Analysis
PAL amino acid sequences were downloaded from the GenBank database and aligned using Clustal W (Altschul et al., 1990). The alignment was manually optimized and used to find the most parsimonious tree, employing the heuristic search option algorithm in the program PAUP 4.0b2 (Sinauer Associates, Sunderland, MA). For statistical analysis, 1,000 bootstrap replications (Felsenstein, 1995) were analyzed. The following PAL amino acid sequences (accession nos. within parentheses) were used for phylogenetic analysis: Arabidopsis (P35510, P45724, P45725, and AC009400), Citrus limon (Q42667, AJ238753, and AJ238754), Glycine max (P27991 and JQ1070), Ipomoea batatas (P14166 and Q428588), Lithospermum erythrorhizon (O49835 and 049836), Lycopersicon esculentum (P35511 and P26600), Nicotiana tabacum (P25872, P35513, and P45733), Phaseolus vulgaris (P19142 and P19143), Pisum sativum (Q01861 and Q04593), Petroselinum crispum (P24481 and P45728), Populus kitakamiensis (P45731, P45730, Q43052, and Q40910), raspberry (AF237954 and AF237955), Prunus avium (AF036948), Solanum tuberosum (P31425 and P31426), Oryza sativa (P14717 and P53443), and Pinus taeda (T09777).
ACKNOWLEDGMENTS
The authors are greatly indebted to Hugh Daubney and Chaim Kempler (Agriculture and Agri-Food Canada, Summerland, BC) for their generous supply of plant materials, and to Stefanie Butland (Centre for Molecular Medicine and Therapeutics, Vancouver, BC) for her professional assistance with several aspects of this study.
Footnotes
This work was supported by the Natural Sciences and Engineering Research Council of Canada (grant to B.E.E.) and by the University of British Columbia (Graduate Fellowship to A.K.).
LITERATURE CITED
- Allwood EG, Davies DR, Gerrish C, Ellis BE, Bolwell GP. Phosphorylation of phenylalanine ammonia-lyase: evidence for a novel protein kinase and identification of the phosphorylated residue. FEBS Lett. 1999;457:47–52. doi: 10.1016/s0014-5793(99)00998-9. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Appert C, Logemann E, Hahlbrock K, Schmid J, Amrhein N. Structural and catalytic properties of the four phenylalanine ammonia-lyase isoenzymes from parsley (Petroselinum crispumNym.) Eur J Biochem. 1994;225:491–499. doi: 10.1111/j.1432-1033.1994.00491.x. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley; 1995. [Google Scholar]
- Barrit BH, Torre LC. Fruit anthocyanin pigments of red raspberry cultivars. J Amer Soc Hortic Sci. 1975;100:98–100. [Google Scholar]
- Borejsza-Wysocki W, Hrazdina G. Biosynthesis of p-hydroxyphenylbutan-2-one in raspberry fruits and tissue cultures. Phytochemistry. 1994;35:623–628. [Google Scholar]
- Brady CJ. Fruit ripening. Annu Rev Plant Physiol. 1987;38:155–178. [Google Scholar]
- Breathnach R, Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Brodelius PE. Phenylpropanoid metabolism in Vanilla planifolisAndr. (V): high performance liquid chromatographic analysis of phenolic glycosides and aglycone in developing fruits. Phytochem Anal. 1994;5:27–31. [Google Scholar]
- Brown JWS. A catalogue of splice function and putative branch point sequences from plant introns. Nucleic Acids Res. 1986;14:9549–9559. doi: 10.1093/nar/14.24.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butland SL, Chow ML, Ellis BE. A diverse family of phenylalanine ammonia-lyase genes in pine tree and cell cultures. Plant Mol Biol. 1998;37:15–24. doi: 10.1023/a:1005941228246. [DOI] [PubMed] [Google Scholar]
- Chaw S-M, Zhaekikh A, Sung HM, Lau T-C, Li W-H. Molecular phylogeny of extant gymnosperms and seed plant evolution: analysis of nuclear 18S rRNA sequences. Mol Biol Evol. 1997;14:56–68. doi: 10.1093/oxfordjournals.molbev.a025702. [DOI] [PubMed] [Google Scholar]
- Cheng GW, Breen PJ. Activity of phenylalanine ammonia-lyase (PAL) and concentrations of anthocyanins and phenolics in developing strawberry fruit. J Am Soc Hortic Sci. 1991;116:865–869. [Google Scholar]
- Cramer CL, Edwards K, Dron M, Liang X, Deldine SL, Bolwell GP, Dixon RA, Lamb CJ, Schuch WW. Phenylalanine ammonia-lyase gene organization and structure. Plant Mol Biol. 1989;12:367–383. doi: 10.1007/BF00017577. [DOI] [PubMed] [Google Scholar]
- Diallinas G, Kanellis AK. A phenylalanine ammonia-lyase gene from melon fruit: cDNA cloning, sequence and expression in response to development and wounding. Plant Mol Biol. 1994;26:473–479. doi: 10.1007/BF00039557. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Dubery IA, Schabort JC. Phenylalanine ammonia-lyase from Citrus sinensis: purification hydrophobic interaction chromatography and physical characterization. Biochem Int. 1986;13:579–589. [Google Scholar]
- Faragher JD, Brohier RL. Anthocyanin accumulation in apples skin during ripening: Regulation by ethylene and phenylalanine ammonia-lyase. Sci Hortic. 1984;22:89–96. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using bootstrap. Evolution. 1995;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Given NK, Venis MA, Grierson D. Purification and properties of phenylalanine ammonia-lyase from strawberry fruit and its synthesis during ripening. J Plant Physiol. 1988a;133:31–37. [Google Scholar]
- Given NK, Venis MA, Grierson D. Phenylalanine ammonia-lyase activity and anthocyanin synthesis in ripening strawberry fruit. J Plant Physiol. 1988b;133:25–30. [Google Scholar]
- Hassan MA, Swartz HJ, Inamine G, Mullineaux P. Agrobacterium tumefaciens-mediated transformations of several Rubusgenotypes and recovery of transformed plants. Plant Cell Tissue Organ Cult. 1993;33:9–17. [Google Scholar]
- Holton TA, Graham MW. A simple and efficient method for direct cloning of PCR products using ddT-tailed vectors. Nucleic Acids Res. 1991;19:1156–1157. doi: 10.1093/nar/19.5.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrazdina G, Parsons GF, Mattick LR. Physiological and biochemical events during development and maturation of grape berries. Am J Enol Vitic. 1984;35:220–227. [Google Scholar]
- Hrazdina G, Wagner G. Metabolic pathways as enzyme complexes: Evidence for the synthesis of phenylpropanoids and flavonoids in membrane-associated enzyme complexes. Arch Biochem Biophys. 1985;237:88–100. doi: 10.1016/0003-9861(85)90257-7. [DOI] [PubMed] [Google Scholar]
- Joos HJ, Hahlbrock K. Phenylalanine ammonia-lyase in potato (Solanum tuberosumL.) Eur J Biochem. 1992;204:621–629. doi: 10.1111/j.1432-1033.1992.tb16675.x. [DOI] [PubMed] [Google Scholar]
- Latza S, Dietmar G, Berger RG. Identification and accumulation of 1–0-traans-cinnamoyl-beta-D-glucopyra-nose in developing strawberry fruit (Fragaria ananassa Duch.Cv. Kent) J Agric Food Chem. 1996;44:1367–1370. [Google Scholar]
- Liang X, Dron M, Schmid J, Dixon RA, Lamb CJ. Developmental and environmental regulation of a phenylalanine ammonia-lyase-β-glucuronidase gene fusion in transgenic tobacco plants. Proc Natl Acad Sci USA. 1989;86:9284–9288. doi: 10.1073/pnas.86.23.9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew CF, Goh CJ, Loh CS, Lim SH. Cloning and nucleotide sequence of a cDNA encoding phenylalanine ammonia-lyase from Bromheadia finlaysoniana (Lindl.) Rchb.f. (accession no. X99997) (PGR 96-087) Plant Physiol. 1996;112:863. [Google Scholar]
- Logemann E, Parniske M, Hahlbrock K. Modes of expression and common structural features of the complete phenylalanine ammonia-lyase gene family in parsley. Proc Natl Acad Sci USA. 1995;92:5905–5909. doi: 10.1073/pnas.92.13.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Why gene duplications? In: Ohno S, editor. Evolution by Gene Duplication. New York: Springer-Verlag; 1970. pp. 59–65. [Google Scholar]
- Perkins-Veazie P, Nonnecke G. Physiological changes during ripening of raspberry fruits. Hortic Sci. 1992;27:331–333. [Google Scholar]
- Rasmussen S, Dixon RA. Transgene-mediated and elicitor-induced perturbations of metabolic channeling at the entry point into the phenylpropanpoid pathway. Plant Cell. 1999;11:1537–1551. doi: 10.1105/tpc.11.8.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Lopez J, Ordorica-Falomir C, Wesche-Ebeling P. Changes in anthocyanin concentration in Lychee (Litchi chinensisSonn.) pericarp during maturation. Food Chem. 1999;65:195–200. [Google Scholar]
- Roubelakis-Angelakis KA, Kliewer WM. Phenylalanine ammonia-lyase in berries of Vitis viniferaL.: extraction and possible sources of error during assay. Am J Enol Vitic. 1985;36:314–315. [Google Scholar]
- Schomburg D, Salzmann M. Class 4: lyases, phenylalanine ammonia-lyase. In: Schomburg D, Salzmann M, editors. Enzyme Handbook 1. Berlin: Springer-Verlag; 1990. pp. 1–5. [Google Scholar]
- Schuster B, Retey J. The mechanism of action of phenylalanine ammonia-lyase: the role of prosthetic dehydroalanine. Proc Natl Acad Sci USA. 1995;92:8433–8437. doi: 10.1073/pnas.92.18.8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede TF, Retey J, Schulz GE. Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile. Biochemistry. 1999;38:5355–5361. doi: 10.1021/bi982929q. [DOI] [PubMed] [Google Scholar]
- Stewart WN, Rothwell GW. Paleobotany and the Evolution of Plants. Cambridge, UK: Cambridge University Press; 1993. [Google Scholar]
- Subramaniam R, Reinold S, Molitor EK, Douglas CJ. Structure, inheritance, and expression of hybrid poplar (Populus trichocarpa × Populus deltoides) phenylalanine ammonia-lyase genes. Plant Physiol. 1993;102:71–83. doi: 10.1104/pp.102.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukrasno N, Yeoman MM. Phenylpropanoid metabolism during growth and development of Capsicum frutescensfruits. Phytochemistry. 1993;32:839–844. [Google Scholar]
- Wanner LA, Guoqing L, Ware D, Somssich IE, Davis K. The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol Biol. 1995;27:327–338. doi: 10.1007/BF00020187. [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang MB. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetten RW, Sederoff RR. Phenylalanine ammonia-lyase from loblolly pine: purification of the enzyme and isolation of complementary DNA clones. Plant Physiol. 1992;98:380–386. doi: 10.1104/pp.98.1.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersma PA, Wu Z. A full length cDNA for phenylalanine ammonia-lyase cloned from ripe sweet cherry fruit (Prunus avium; accession no. AF036948) (PGR 98-184) Plant Physiol. 1998;118:1102. [Google Scholar]
- Yamada T, Tanaka Y, Sriprasertsak P, Kato H, Hashimoto T, Kawamata S, Ichinose Y, Kato H, Shiraishi T, Oku H. Phenylalanine ammonia-lyase genes from Pisum sativum: structure, organ-specific expression and regulation by fungal elcicitor and suppressor. Plant Cell Physiol. 1992;33:715–725. [Google Scholar]