Abstract
Parasitic plants in the Scrophulariaceae use chemicals released by host plant roots to signal developmental processes critical for heterotrophy. Haustoria, parasitic plant structures that attach to and invade host roots, develop on roots of the hemiparasitic plant Triphysaria versicolor within a few hours of exposure to either maize (Zea mays) root exudate or purified haustoria-inducing factors. We prepared a normalized, subtractive cDNA library enriched for transcripts differentially abundant in T. versicolor root tips treated with the allelopathic quinone 2,6-dimethoxybenzoquinone (DMBQ). Northern analyses estimated that about 10% of the cDNAs represent transcripts strongly up-regulated in roots exposed to DMBQ. Northern and reverse northern analyses demonstrated that most DMBQ-responsive messages were similarly up-regulated in T. versicolor roots exposed to maize root exudates. From the cDNA sequences we assembled a unigene set of 137 distinct transcripts and assigned functions by homology comparisons. Many of the proteins encoded by the transcripts are predicted to function in quinone detoxification, whereas others are more likely associated with haustorium development. The identification of genes transcriptionally regulated by haustorium-inducing factors provides a framework for dissecting genetic pathways recruited by parasitic plants during the transition to heterotrophic growth.
Parasitic plants in the Scrophulariaceae invade the roots of neighboring plants and rob them of water, minerals, and nutrients (Kuijt, 1969; Press and Graves, 1995). The consequence to the parasitized plants can be debilitating and some of the world's most destructive agricultural pests are parasitic weeds (Parker and Riches, 1993). Parasitic Scrophulariaceae use allelopathic chemicals that are released by host plan roots to trigger developmental programs critical to the parasitic lifestyle. One such program results in the development of haustoria, specialized root structures that attach to and invade host roots. Once a haustorium has invaded the host root, it serves as the physiological conduit between the parasite and host roots through which the parasite robs host plant resources (Riopel and Timko, 1995). Parasitic Scrophulariaceae typically develop haustoria only in the presence of neighboring plants. Haustorium development can be readily monitored in vitro by applying host factors to parasite roots (Atsatt et al., 1978; Riopel and Musselman, 1979).
The first natural haustorium-inducing factor identified from host roots was 2,6-dimethoxybenzoquinone (DMBQ; Chang and Lynn, 1986). DMBQ induces haustorium development in both obligate and facultative parasites within hours of applying it to their roots (Baird and Riopel, 1984; Chang and Lynn, 1986; Albrecht et al., 1999). DMBQ and related quinones are secondary metabolites widespread in diverse plant families (Handa et al., 1983). These molecules are released into the rhizosphere as components of root exudates and as byproducts of fungal-mediated lignin degradation. Quinones are also produced through the oxidation of phenolic acids, another major constituent of root exudates (Thomson, 1987; Siqueira et al., 1991).
Several naturally occurring quinones and phenolics induce haustoria when applied to parasite roots (Steffens et al., 1982; Riopel and Timko, 1995; Albrecht et al., 1999). An important cue as to how structurally different molecules induce haustoria was obtained from the observation that the redox potentials of active factors fall within a relatively narrow window (Smith et al., 1990, 1996). The design of chemical inhibitors that trap radical intermediates established that haustorium induction is associated with redox cycling between quinone and semiquinone states (Zeng et al., 1996). Redox cycling also generates reactive oxygen intermediates that are largely responsible for quinone toxicity (Obrien, 1991). In fact, DMBQ has been investigated as a possible anticancer agent because of its mammalian cytotoxicity (Brambilla et al., 1988). Parasitic plants have recruited these potentially harmful intermediates to signal haustorium organogenesis.
We would like to understand how parasitic plants perceive and process allelopathic quinones. We are using Triphysaria, previously Orthocarpus (Chuang and Heckard, 1991), a small parasitic Scrophulariaceae that grows as a spring-time annual in coastal bluffs and inland grasslands throughout the Pacific coast, for these studies. Triphysaria is a facultative parasite that can be grown to maturity in the absence of host plants but will parasitize a broad spectrum of hosts, including Arabidopsis and maize (Zea mays; Thurman, 1966; Yoder, 1997). Haustorium development is rapid and can be monitored in vitro by applying host root exudates or haustoria inducing factors to Triphysaria roots (Atsatt et al., 1978; Albrecht et al., 1999). Triphysaria is a simple diploid whose perfect flowers are amenable to genetic manipulations (Chuang and Heckard, 1991; Yoder, 1998). Triphysaria is closely related to the parasitic weeds Striga and Orobanche and can serve as a useful model for these pernicious agricultural pests (Nickrent et al., 1998).
To begin the dissection of genetic pathways governing haustorium development, we characterized cDNAs homologous to transcripts up-regulated in Triphysaria versicolor roots exposed to DMBQ. We asked whether these transcripts were similarly regulated by host root factors by northern and reverse northern analyses. These represent plant genes regulated by subterranean signals produced by other plants. By categorizing the putative functions of DMBQ responsive genes, we can generate models that highlight potential pathways associated with quinone signaling and haustorium morphogenesis.
RESULTS AND DISCUSSION
Early Morphological Responses of T. versicolor Roots to DMBQ
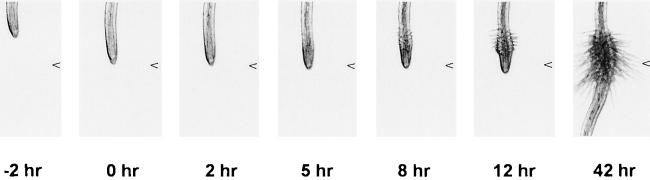
Morphological changes associated with early haustorium development were observed in T. versicolor root tips shortly after exposure to either 10 μm DMBQ or root exudates from hydroponically grown maize. Haustorium ontogeny in T. versicolor was similar to that described for other secondary haustoria (Atsatt et al., 1978; Riopel and Baird, 1987). The first change observed was a termination of root elongation within 30 min of DMBQ treatment (Fig. 1; data not shown). In Striga ariatica, this period is also hallmarked by a cessation of DNA synthesis (O'Malley and Lynn, 2000). Within 5 h of treatment, epidermal hairs could be seen to elongate and proliferate in the region just behind the root tip (Fig. 1). These hairs function to attach the developing haustorium to the host tissue (Baird and Riopel, 1985; Heide-Jorgensen and Kuijt, 1995). At about this same time, cortical cells underlying the proliferating epidermal hairs began to swell. This resulted in a swelling near the root tip noticeable within 8 h of treatment. The swelling and proliferation of haustorial hairs continued for about 24 h during which time the haustorium was competent to attach to a host root (J.I. Yoder, unpublished data). Under our induction conditions, approximately 70% to 80% of the root tips formed haustoria after DMBQ treatment, whereas virtually no haustoria developed when treated with water.
Figure 1.
Time course of haustorium development. T. versicolor were grown on agar media in vertically oriented petri dishes so that their roots grew along the surface of the agar. The roots were then exposed to 10 μm DMBQ and their tips photographed at different times under 2× magnification with a dissecting microscope. The pictures in the figure are a time lapse of the same root. The arrowhead to the right in each picture represents the position of the root tip when the treatment was applied.
Haustorium development in T. versicolor roots was transitory and after about 12 h the root tips reverted to their typical developmental program. This resulted in a normal appearing root distal to the haustorium. The reversion to normal root growth occurred even in the presence of active inducing factors because fresh T. versicolor sown onto these plates formed haustoria. Also, the same root made multiple haustoria if the root came out of contact with the media for a short period of time and then retouched the surface. Similar observations have been made for Striga root cultures (Wolf and Timko, 1992).
Early Transcriptional Responses to DMBQ and Host Exudate Exposure
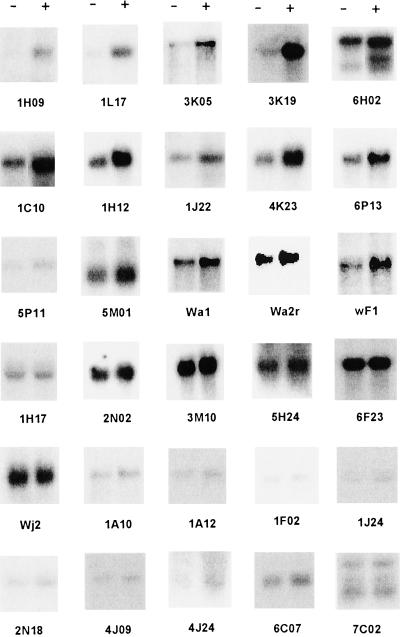
We made a normalized, subtracted cDNA library enriched for transcripts up-regulated in T. versicolor root tips 2 to 5 h after treatment with DMBQ. Following two rounds of colony hybridization with forward- and reverse-subtracted cDNA probes (see “Materials and Methods”), several hundred clones were selected for further characterization. Inserts from 30 cDNAs were PCR amplified and used as probes on northern blots containing RNA from T. versicolor roots exposed to either DMBQ or water. As seen in Figure 2 and summarized in Table I, five transcripts that were in low abundance in water- treated roots, were strongly up-regulated by DMBQ treatment. Ten cDNAs hybridized to transcripts with basal levels in untreated roots that increased upon DMBQ treatment; these are labeled as “moderate” in Table I. The steady-state levels of transcripts hybridizing to six cDNA clones were unchanged by DMBQ. These may be false positives that were carried through the screening. Hybridization to nine cDNA probes was very weak and it was not possible to conclude whether these transcripts were regulated. From these results we predict that between 50% and 80% of the clones in the subtractive library were up-regulated by DMBQ, about 10% of these strongly.
Figure 2.
Northern analysis using early DMBQ-induced transcript (EDIT) probes. Northern filters were prepared from mRNA extracted from T. versicolor root tips exposed to either water (left lane [−]) or DMBQ (right lane [+]) for 5 h. Inserts of 30 cDNAs were PCR amplified, labeled with 32P, and used as hybridization probes. The putative functions of these clones as predicted from virtual translations are given in Table I.
Table I.
Transcripts regulated by DMBQa
| Probeb | Regulation | EDITc | Function | Functional Class |
|---|---|---|---|---|
| 1H09 | High | 0173 | Quinone oxidoreductase | Metabolism |
| 1L17 | High | 0017 | Pirin | Transcription |
| 3K05 | High | 0120 | Multispanning membrane protein p76 | Membrane transport |
| 3K19 | High | 0010 | 1,4-Benzoquinone oxidoreductase | Metabolism |
| 6H02 | High | 0089 | Strictosidine synthase | 2 Metabolism |
| 1C10 | Moderate | 0012 | 40S ribosomal protein p40 | Translation |
| 1H12 | Moderate | 0171 | Ser-O-acetyltransferase | Metabolism |
| 1J22 | Moderate | 0005 | Alpha-soluble N-ethylmaleimide-sensitive fusion attachment protein | Membrane transport |
| 4K23 | Moderate | 0030 | 60S Ribosomal protein L7 | Translation |
| 5M01 | Moderate | 0026 | 60S Ribosomal protein L15 | Translation |
| 5P11 | Moderate | 0093 | Pyrrolidone-carboxylate peptidase | 2 Metabolism |
| 6P13 | Moderate | 0061 | 40S Ribosomal protein S15 | Translation |
| Wa1 | Moderate | 0077 | S-Adenosyl-Met decarboxylase | Metabolism |
| Wa2 | Moderate | 0075 | p450, CYP72 family | Metabolism |
| Wf1 | Moderate | 0071 | Hydroxy-Pro-rich glycoprotein | Cell wall |
| 1H17 | None | 0169 | No homology | Unknown |
| 2N02 | None | 0006 | 60S Ribosomal protein L10 | Translation |
| 3M10 | None | 0115 | No homology | Unknown |
| 5H24 | None | 0096 | GTP-binding protein | Signal transduction |
| 6F23 | None | 0090 | Unknown function | Unknown |
| Wj2 | None | 0074 | 60S Ribosomal protein L26 | Translation |
| 1A10 | Low abundance | 0189 | Transaldolase | Metabolism |
| 1A12 | Low abundance | 0188 | Cell cycle regulator p21 | Cell cycle |
| 1F02 | Low abundance | 0020 | Transmembrane protein | Membrane transport |
| 1J24 | Low abundance | 0013 | dTDP-Glu 4-6-dehydratase | Metabolism |
| 2N18 | Low abundance | 0137 | Reticuline oxidoreductase | 2 Metabolism |
| 4J09 | Low abundance | 0106 | GTP-binding protein | Signal transduction |
| 4J24 | Low abundance | 0105 | Unknown function | Unknown |
| 6C07 | Low abundance | 0035 | Transcription factor BTF3 | Transcription |
| 7C02 | Low abundance | 0082 | Protein kinase | Signal transduction |
Table derived from Figure 2.
Identification number of the cDNA probe.
Identification number of assembled transcript in unigene set.
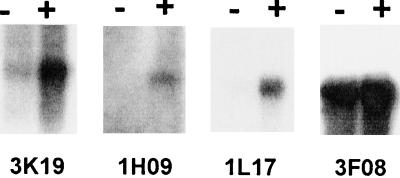
Host root exudates were made by growing maize seedlings in culture for 2 weeks and recovering the supernatant after centrifuging the agar. Maize root exudates induced haustoria in T. versicolor roots with the same frequency and time course as DMBQ. RNA was extracted from exudate-treated roots and analyzed by northern hybridizations. Three cDNAs, 3K19, 1H09, and 1L17, hybridized to transcripts strongly up-regulated by maize root exudate; these were similarly up-regulated in T. versicolor by DMBQ (Fig. 2). A fourth cDNA (3F08) was not up-regulated by either treatment (Fig. 3; data not shown).
Figure 3.
Transcription regulation by maize root exudate. A northern blot was prepared containing RNA isolated from T. versicolor roots mock treated with water (−) or with maize root exudate (+) for 4 h. The blot was sequentially probed with clones 3K19, 1H09, 1L17, and 3F08.
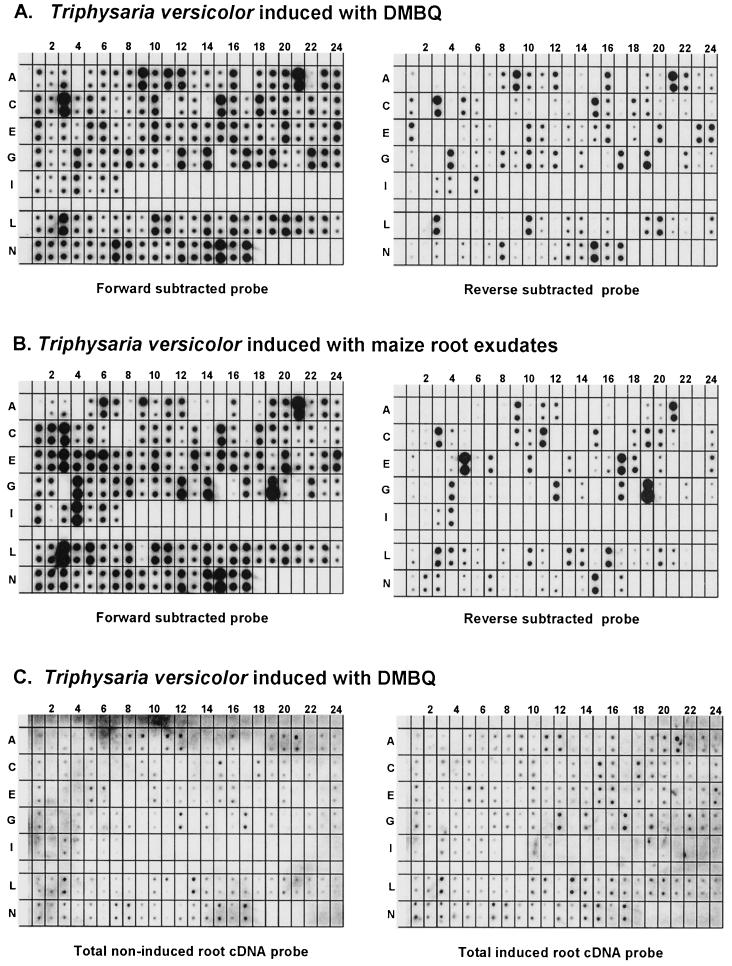
We used reverse northerns to obtain an independent estimate of the proportion of DMBQ regulated clones also up-regulated by host root exudates. Inserts from 141 clones were PCR amplified and arrayed in duplicate on a nylon membrane. The filter was sequentially hybridized with radioactive cDNA probes made from mRNA isolated from DMBQ-treated and untreated roots. Most of the inserts did not hybridize with these probes at levels over significantly over background. This is consistent with previous observations that 32P-labeled probes synthesized from complex cDNA pools generally detect only highly abundant transcripts (Wang and Brown, 1991). Even when hybridization was detected there were relatively few differences between induced and noninduced conditions (Fig. 4C). This can result if the cDNA probes cross hybridize to nonregulated members of a gene family.
Figure 4.
Probe interrogation of EDIT arrays. Inserts from 141 cDNA clones were spotted onto nylon membranes in duplicate. The same filter was sequentially hybridized with forward- and reverse-subtracted probes (A and B), and with non-subtracted probes made from complex root mRNA (C). In A and C, the probes were made from T. versicolor root tips exposed to DMBQ for 2 to 5 h. In B, the probes were made from T. versicolor root tips exposed to maize root exudate. The cDNAs spotted at each position is found in supplemental data online at www.plantphysiol.org.
To reduce the proportion of highly abundant transcripts in the probe populations, we normalized the cDNA probes by forward and reverse hybridizations (Gurskaya et al., 1996). Because this procedure employed restriction digests prior to subtractive hybridization, the probes were further enriched for low abundant gene domains (Yang et al., 1999). Forward-subtracted probes, enriched for transcripts more abundant after DMBQ treatment, and reverse-subtracted probes, enriched for sequences less abundant after DMBQ treatment, were hybridized to the filters (Fig. 4A). In this case, most of the probes hybridized to inserts on the filter and there was considerable variation between induced and noninduced treatments. We standardized the autoradiograph exposures using clones analyzed by northern blots. A visual examination of the standardized autoradiographs led to the estimate that about 80% of the cDNAs hybridized stronger to forward-subtracted than reverse-subtracted probes, a result consistent with the northern analyses. Because hybridization intensities cannot be quantified using normalized probes, we did not attempt to refine this estimate further.
We then made forward- and reverse-subtracted probes from RNA isolated from T. versicolor roots treated with maize exudates. When these probes were hybridized to the cDNA arrays, the hybridization pattern was very similar to that obtained following DMBQ treatments (Fig. 4B). Combining these results from those of the northern blots described above, we conclude that most of the transcripts up-regulated in T. versicolor by DMBQ are similarly up-regulated by maize root exudates.
Putative Functions of DMBQ-Responsive Transcripts
Based on the relative strength of signals in colony hybridizations, we selected 220 of the most differentially regulated cDNAs to sequence. The sequences clustered into 191 nonoverlapping assemblies using The Institute for Genomic Research Assembler program (Sutton et al., 1995). The virtual translations of the assembled sequences were analyzed by BLASTX.
Transcripts predicted to encode distinct proteins were assembled into a unigene set of 137 members with each assembly assigned its own EDIT number. Putative functions could be assigned to 117 EDIT proteins by homology searches of public data banks. An additional 12 EDIT translations had significant homology to proteins predicted from genome projects, typically Arabidopsis, but for which there were no well-annotated functions. Eight EDITs did not share significant homology with any protein sequences in the databases. The proteins predicted to be encoded by the EDITs were assigned to the functional classes shown in Table II. GenBank accession numbers and putative functions of these sequences and assemblies can be found in the supplemental data online at www.plantphysiol.org. Links of each sequence to FASTA files and BLAST reports can be found at http://veghome.ucdavis.edu/Faculty/Yoder/lab/index.html. The potential relevance of some of these clones to allelochemical signaling is discussed below.
Table II.
Functional classes of EDITs
| Functional Class | No. of EDITs |
|---|---|
| Cell cycle | 2 |
| Cell wall | 5 |
| Cytoskeleton | 2 |
| DNA binding | 1 |
| Membrane transport | 13 |
| Metabolism | 31 |
| Metabolism, secondary | 5 |
| Signal transduction | 11 |
| Transcription | 9 |
| Translation | 38 |
| Unknown | 12 |
| No homology | 8 |
| Total | 137 |
Quinone Detoxification
Several EDITs are homologous to proteins that function in quinone detoxification. Quinones are biologically reactive molecules that are used medically as anticancer, -fungal, and -malarial agents. The toxicity of pharmacological quinones is associated with reactive oxygen intermediates formed during redox transformations between quinone and hydroquinone forms (Obrien, 1991). Semiquinone radicals formed as intermediates during redox cycling readily donate their electrons to molecular oxygen forming superoxide anions (O2·−; Testa, 1995). Superoxides dismutate to hydrogen peroxide (H2O2), which undergoes reactions to form hydroxyl (OH·) and hydroperoxyl radicals (HO2·; Hammondkosack and Jones, 1996). These reactive oxygen intermediates are enormously destructive to membranes, proteins, and DNA (Smith, 1985).
Genetic mechanisms to defend organisms from oxidative stress evolved early in evolutionary history, presumably in defense against accumulating atmospheric oxygen (Testa, 1995). As such, proteins associated with oxygen stress tolerance tend to be similar in extant aerobic organisms. Several proteins predicted from the virtual translations of EDITs were homologous to proteins that confer oxidative stress tolerance in other organisms. Two EDITs, 0013and 0173, were homologous to Arabidopsis proteins that confer tolerance to diamide, an active oxidizer of cellular thiols and inducer of oxidative stress, in transformed yeast (Saccharomyces cerevisiae; Babiychuk et al., 1995; Kushnir et al., 1995). Another clone, EDIT 0010, was highly homologous to quinone oxidoreductase in the lignin-degrading fungus Phanerochaete chrysosporium. P. chrysosporium produces DMBQ and other quinones as byproducts of lignin degradation. It has been postulated that these quinones are detoxified by the quinone reductase homolog of EDIT 0010 (Brock and Gold, 1996).
Oxidative stress responses have been particularly well studied in yeasts. Twenty-five genes associated with oxidative stress tolerance in yeast are up-regulated by overproducing the YAP1 transcription factor (Toone and Jones, 1999). We compared the virtual translations of these YAP1 regulated transcripts with the 137 EDITs using a local copy of BLAST and found that 11 EDITs proteins shared homologies with at least one YAP1 regulated message. The putative proteins encoded by EDITs 0146, 0173, and 0189 had homology to more than one YAP1 regulated transcript because of the conservation of functional domains, such as the ATP-binding cassette (Table III).
Table III.
EDIT homologs in yeast that are regulated by YAP1
| Yeast Functiona | Open Reading Frameb | Pc | EDIT No. | EDIT Functiond |
|---|---|---|---|---|
| Membrane Transport | ||||
| Cadmium resistance factor | YDR135C | 3 × 10−5 | 0146 | ABC transporter |
| ABC transporter | YDR011W | 2 × 10−2 | 0146 | ABC transporter |
| Pleitropic drug resistance | YOR153W | 1 × 10−1 | 0146 | ABC transporter |
| Cell wall maintenance | YAL059W | 7 × 10−1 | 0163 | Ring canal protein |
| Flucanozole resistance | YBR008C | 5 × 10−1 | 0131 | ATP translocator |
| Nitroquinoline-N-oxide | YML116W | NS | – | – |
| Detoxification | ||||
| Malate dehydrogenase | YOL126C | 3 × 10−17 | 0058 | Malate dehydrogenase |
| Homology with YCR102C | YLR460C | 5 × 10−10 | 0173 | Quinone oxidoreductase |
| Arabidopsis zeta crystalin | YML131W | 6 × 10−1 | 0173 | Quinone oxidoreductase |
| NADPH oxidoreductase | YHR179W | 5.4 | 0173 | Quinone oxidoreductase |
| NADPH oxidoreductase | YPL171C | 3.2 | 0161 | Cinnamate 4-hydroxylase |
| Aryl-alcohol dehydrogenase | YFLO56C | 3.7 | 0189 | Transaldolase |
| Aryl-alcohol dehydrogenase | YJR155W | 2.2 | 0189 | Transaldolase |
| Aryl-alcohol reductase | YNL331C | 2.3 | 0189 | Transaldolase |
| Aryl-alcohol reductase | YCR107W | 2.1 | 0189 | Transaldolase |
| Aryl-alcohol dehydrogenase | YOL165C | NSe | – | – |
| Thioredoxin | YGR209C | NS | – | – |
| Thioredoxin | YDR353W | NS | – | – |
| Thioredoxin | YDR098C | 1 × 10−111 | 0156 | Yeast thioredoxin |
| Glutathione reductase | YPL091W | NS | – | – |
| Glutathione reductase | YLL060C | NS | – | – |
| Glutamylcysteine synthase | YJL101C | NS | – | – |
| Other | ||||
| HSP70 | YAL005C | 1 × 10−46 | 0178 | Chaperon-binding protein |
| Transcription factor | YML007C | NS | – | – |
| Putative protein | YCL08C | NS | – | – |
| Similar to bacterial csgA | YKL071W | NS | – | – |
Annotated function of the yeast YAP1 regulated transcript.
Yeast open reading frames up-regulated in YAP1 overproducing lines (De Risi et al., 1997).
Probability score obtained by the BLAST algorithm between the yeast and T. versicolor virtual translations.
The function ascribed to the EDITs by BLASTX searches.
NS, No significant homology.
Xenobiotic detoxification genes typically function by transporting the toxin out of the cells or by inactivating them chemically. Genes for both these mechanisms were found in the EDIT collection. In addition to the homologs of Yap regulated transcripts, 15 more EDIT genes were predicted to function in membrane transport. EDIT 0120 was a highly DMBQ-regulated transcript with homology to a human 76-kD membrane protein localized predominantly to endosomes (Schimmoller et al., 1998). This family of proteins contains nine potential membrane-spanning domains and is thought to function as a small molecule transporter in intracellular compartments.
From these results, we hypothesize that a significant number of messages regulated in T. versicolor roots by DMBQ fulfill xenobiotic detoxification functions. This is being tested by examining the expression of these genes in response to non-haustorial forming quinones and in parasitic and nonparasitic plants.
Signal Transduction and Transcript Regulation
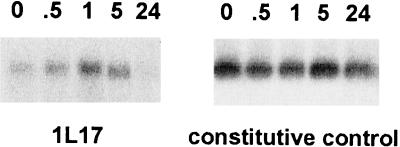
Twenty EDIT proteins (15%) were predicted to function in signal transduction and transcriptional regulation. One of these that we examined in detail was homologous to the human transcription factor pirin (0017). The human pirin was identified in a yeast two-hybrid system as binding to the NFI/CTF1 transcription factor (Wendler et al., 1997). Pharmacological studies suggest that pirin is associated with the protein kinase Raf-MEK-ERK signal pathway and important for oncogene-induced phenotypic changes (Bergman et al., 1999). Pirin abundance was up-regulated in T. versicolor roots within an hour of DMBQ treatment (Fig. 5). The level of pirin steady- state mRNA returned to that of noninduced roots by 24 h posttreatment.
Figure 5.
Regulation of TvPirin by DMBQ. A northern blot containing RNA isolated from T. versicolor root tips 0, 0.5, 1, 5, and 24 h after exposure to DMBQ was probed sequentially probed with TvPirin (1L17) and an unsequenced, constitutively expressed cDNA.
Among the weakly expressed EDITs was the cell cycle regulator p21 (0188). In Schizosaccharomyces pombe, p21 interacts with cdc2, a protein kinase controlling cell division at the M-G1 interface (Gould and Nurse, 1989). Treatment of Striga radicals with DMBQ leads to an almost immediate cessation of DNA synthesis (O'Malley and Lynn, 2000). This observation is consistent with the modification of cell cycle regulation, perhaps associated with a p21-like protein.
Haustorium Ontogeny
Although many of the EDIT clones are predicted to function in many plants for quinone detoxification, others more likely function specifically in haustorium development. A good candidate for such a gene was the GTP-binding protein (0106). This EDIT was homologous to an Arabidopsis protein that functions in epidermal cell elongation. Mutation of this gene in Arabidopsis results in the root hair-defective mutation rhd3 (Wang et al., 1997). It is likely that parasitic plants have recruited this gene to function in haustorial hair development.
The induction of tubulin (0042) and actin (0175) were possibly involved in establishing the cytoskeletal structure of the haustoria. It was suggested previously that changes in actin transcript levels might be associated with haustorium development in Striga sp. (Wolf and Timko, 1994). Other EDITs that may function in cell wall expansion included a xyloglucan endotransglycosylase (0100) and a beta-d-glucan exohydrolase (0182; Hrmova et al., 1996; Rose et al., 1996).
Mitochondrial Genes and Maternal Factors in DMBQ Recognition
At least nine EDIT genes encoded nuclear proteins that become localized in mitochondria. It was most notable that five transcripts encoded proteins functioning in the citric acid cycle. The citric acid cycle moves electrons from organic acids to NAD+ and FAD, forming NADH and FADH2 (Buchanan et al., 2000). These reduced molecules provide the reducing energy for quinone detoxification and induction of the citric acid cycle may be associated with these functions. It is interesting that several allelopathic quinones inhibit respiration and oxidative phosphorylation and DMBQ in particular has been shown to inhibit the citric acid system of mammalian mitochondria (Redfearn and Whittaker, 1962; Rabbani and Duhaiman, 1998).
CONCLUSIONS
The transition of parasitic plants from autotrophic to heterotrophic growth is coupled to transcriptional changes in many genes. Some of the proteins predicted to be activated during this period, such as those functioning in quinone detoxification, are probably not associated with parasitism. Comparing the expression of these genes in closely related parasitic and nonparasitic Scrophulariaceae can test this hypothesis (Matvienko et al., 2001). One of the long-standing questions in parasitic plant biology has been whether the genes that encode developmental pathways associated with parasitism are of an endogenous or exogenous origin (Atsatt, 1973). Most of the genes we examined have closely related homologs in other plants, so we can conclude that at least many of the genes responsive during haustorium development also function in autotrophic plants. Our experiments do not, however, rule out the involvement of extrinsic genes introduced into the parasitic lineage of Scrophulariaceae through endosymbiosis or horizontal gene transfer. In either case, the cDNAs described here represent a class of plant genes that are responsive to signal molecules released by other plants. As such they should be useful markers for monitoring plant-plant interactions and their regulatory elements may facilitate the engineering of allelopathy to improve crop performance.
MATERIALS AND METHODS
Materials
Triphysaria versicolor seeds were harvested from mature capsules on plants growing in a grassland stand near Napa California. Seeds were collected and pooled from hundreds of different plants growing within about 1 ha.
DMBQ was obtained from Pfalz and Bauer Inc. (Waterbury, CT). Maize (Zea mays) root exudates were prepared from in vitro-grown seedlings. Maize seeds (no. 3245IR, Pioneer Hi-Bred International, Johnston, IA) were surface sterilized with bleach and placed into 100 mL of 0.6% (w/v) phytoagar in Magenta culture boxes, four seeds per box (112 plants total). After 2 weeks growth at 25°C, the plants and their roots were removed from the agar, which was centrifuged at 10,000 rpm for 30 min and the supernatant filter sterilized and dried under a vacuum. The resulting powder was resuspended in 100 mL water and added to the parasite roots without further dilution. Approximately 1 mL of exudate corresponds to 1 g fresh root weight.
Haustorium Induction
T. versicolor seeds were surface sterilized and plated at 16°C in petri dishes containing 0.25× Hoagland solution [1.25 mm Ca(NO3)2, 1.25 mm KNO3, 0.25 mm KH2PO4, and 0.5 mm MgSO4], 1% (w/v) Suc, and 0.6% (w/v) phytoagar as previously described (Delavault et al., 1998). Two weeks after germination, seedlings were transplanted along one edge of square petri dishes containing 0.25× Hoagland media in 1% (w/v) agar, 10 seedlings to a dish. The dishes were then incubated for 10 d at 25°C at a near vertical orientation so that the roots grew down along the surface of the agar.
To induce haustoria, 2 mL of either maize root exudate or 10 μm DMBQ (dissolved in water) was applied to T. versicolor roots. An equal number of plants were mock treated with 2 mL water. For optimal induction, the plates needed to be kept horizontal for 2 h after exposure before returning them to the vertical orientation.
Construction of the Subtractive cDNA Library
The roots of 500 T. versicolor seedlings were treated with either 10 μm DMBQ or water for 2 to 5 h at which time 5-mm pieces were cut off the root tips and frozen in liquid nitrogen. The root tips were ground in liquid nitrogen and resuspended in a 1:1 mixture of RNA extraction buffer (0.1 m Tris-HCl, pH 9.0, 0.1 m LiCl, 10 mm EDTA, and 1% [w/v] SDS) and saturated phenol at 70°C. After vortexing and centrifugation (15 min, 6,000g), the water phase was collected and RNA extracted as described by Pawlowski et al. (1994).
The PCR-Select Subtraction Kit (CLONTECH, Palo Alto, CA) was used to make a cDNA library enriched for transcripts differentially abundant in T. versicolor root tips after exposure to DMBQ. This protocol uses suppression PCR to enrich for transcripts differentially expressed in the tester material (Diatchenko et al., 1996). The procedure also contains a hybridization step that equalizes transcript abundance (Gurskaya et al., 1996). We performed the subtractive hybridization using 2 μg poly(A+) RNA isolated from DMBQ-treated root tips as tester, and 2 μg poly(A+) RNA from water-treated root tips as driver (CLONTECH, 2000). Following PCR amplification, subtracted cDNAs were cloned into pCR2.1 using the TA Cloning System (Invitrogen, Carlsbad, CA). Recombinant clones were identified on X-Gal-containing plates. The average insert size was 650 bp.
Approximately 2,500 single colonies were picked into individual wells of 384-well trays (Nalge Nunc International, Rochester, NY) containing 0.2 mL of Luria broth. Replica trays containing 4.4% (w/v) glycerol were frozen at −80°C. Colonies were then replica plated from the multiwell trays onto seven, 8- × 12-cm nylon filters using a 384-pin replicator (V & P Scientific, Inc., San Diego). Each clone was spotted twice for total of 768 colonies per filter. The colonies were grown overnight on the filters, lysed, DNA fixed to the membrane, and prehybridized in 6.7% (w/v) SDS and 6.7× SSPE at 68°C (Sambrook et al., 1989).
The filters were hybridized sequentially with forward- and reverse-subtracted probes to identify false positives (CLONTECH, 1999). Forward-subtracted probes were made using mRNA obtained from DMBQ-induced root tips as tester and water-treated roots as driver. These probes were enriched for transcripts induced in DMBQ treated roots. Reverse-subtracted probes were similarly prepared except that the tester mRNA was derived from water-treated root tips, whereas the driver mRNA from DMBQ-treated roots. Reverse-subtracted probes were enriched for transcripts less abundant after DMBQ treatment.
About 2% of the 2,500 clones strongly hybridized to both forward- and reverse-subtracted probes. These clones were picked, the inserts amplified by PCR, pooled, and added as cold blocking agent in subsequent hybridizations. Clones giving the greatest differential in hybridization signal strength between the forward- and reverse-subtracted clones were selected for sequencing.
Sequence Analysis
Selected recombinant clones were picked into 0.2 mL of Luria broth in individual wells of a 384-well tray. Insert cDNA was PCR amplified from the intact clones using the primers 1 and 2R (CLONTECH, 2000). One side of the PCR-amplified inserts were sequenced using an Applied Biosystems 377 DNA sequencer (PE Applied Biosystems, Foster City, CA) and the insert sequences trimmed of vector sequences using Sequencher software (Gene Codes Co., Ann Arbor, MI). To identify redundancies, each sequence was searched against the entire database of sequences using a local BLAST. Some cDNAs were also sequenced from the other end. The sequences were clustered into 191 nonoverlapping assemblies using The Institute for Genomic Research Assembler program (Sutton et al., 1995). The virtual translations of the assembled sequences were analyzed by BLASTX to identify homologous sequences in the GenBank database (Benson et al., 1998).
cDNA Array Preparation and Probing
PCR-amplified insert DNA (100 ng μL) from 141 clones was put into individual wells of a 384-well tray. The cDNA inserts were then arrayed in duplicate onto an 8- × 12-cm nylon filter using a 384-pin replicator (V & P Scientific, Inc.) and the DNA fixed to the membrane. The filters were sequentially hybridized to radioactive cDNA probes and visualized by autoradiography. Filters were washed of probe between hybridizations by immersing into boiling 0.5% (w/v) SDS.
Northern Hybridizations
Total RNA was denatured in dimethyl sulfoxide/glyoxal, separated on 1.4% (w/v) agarose gels (Sambrook et al., 1989), and blotted onto Hybond N+ nylon membrane according to the recommendations of the manufacturer (Amersham, Arlington Heights, IL). The blots were hybridized in 6.7% (w/v) SDS ande 6.7× SSPE at 65°C (Sambrook et al., 1989). The cDNA inserts used as probes were radiolabeled by random priming using (α-32P) dCTP (3,000 Ci mmol−1; ICN Pharmaceuticals, Costa Mesa, CA).
Supplementary Material
ACKNOWLEDGMENTS
We wish to thank Paul Richnavsky and Kendra Kongkadee for their excellent technical assistance and Alexander Kozik for his help with database management. We also thank Russell Wrobel for unpublished results and Michael Lassner for invaluable discussions. DNA sequencing and analysis was done at the Plant Genetics Facility (University of California, Davis).
Footnotes
This work was funded by the National Science Foundation (grant no. 99–83053), by the U.S. Department of Agriculture National Research Initiative (grant no. 97–01934), and by the Univeristy of California, Davis (Biotechnology Fellowship to M.J.T.).
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
LITERATURE CITED
- Albrecht H, Yoder JI, Phillips DA. Flavonoids promote haustoria formation in the root parasite Triphysaria. Plant Physiol. 1999;119:585–591. doi: 10.1104/pp.119.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsatt PR. Parasitic flowering plants: how did they evolve? Am Nat. 1973;107:502–510. [Google Scholar]
- Atsatt PR, Hearn TF, Nelson RL, Heineman RT. Chemical induction and repression of haustoria in Orthocarpus purpurascens (Scophulariaceae) Ann Bot. 1978;42:1177–1184. [Google Scholar]
- Babiychuk E, Kushnir S, Bellesboix E, Van Montagu M, Inze D. Arabidopsis thaliana NADPH oxidoreductase homologs confer tolerance of yeasts toward the thiol-oxidizing drug diamide. J Biol Chem. 1995;270:26224–26231. doi: 10.1074/jbc.270.44.26224. [DOI] [PubMed] [Google Scholar]
- Baird WV, Riopel JL. Experimental studies of haustorium initiation and early development in Agalinis purpurea (L.) Raf. (Scrophulariaceae) Am J Bot. 1984;71:803–814. [Google Scholar]
- Baird WV, Riopel JL. Surface characteristics of root haustorial hairs of parasitic Scrophulariaceae. Bot Gaz. 1985;146:63–69. [Google Scholar]
- Benson D, Boguski M, Lipman D, Ostell J, Ouellette B. GenBank. Nucleic Acids Res. 1998;26:1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman AC, Alaiya AA, Wendler W, Binetruy B, Shoshan M, Sakaguchi K, Bergman T, Kronenwett U, Auer G, Appella E. Protein kinase-dependent overexpression of the nuclear protein pirin in c-JUN and RAS transformed fibroblasts. Cell Mol Life Sci. 1999;55:467–471. doi: 10.1007/s000180050303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla G, Robbiano L, Cajelli E, Martelli A, Turmolini F, Mazzei M. DNA-damaging and mutagenic properites of 2,6-dimethoxy-1,4-benzoquinone, formed by dimethoprine-nitrite interaction. J Pharmacol Exp Ther. 1988;244:1011–1015. [PubMed] [Google Scholar]
- Brock BJ, Gold MH. 1,4-Benzoquinone reductase from the basidiomycete Phanerochaete chrysosporium: spectral and kinetic analysis. Arch Biochem Biophys. 1996;331:31–40. doi: 10.1006/abbi.1996.0279. [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Gruissem W, Jones RL. Biochemistry & Molecular Biology of Plants. Rockville, MD: American Society of Plant Physiologists; 2000. [Google Scholar]
- Chang M, Lynn DG. The haustorium and the chemistry of host recognition in parasitic angiosperms. J Chem Ecol. 1986;12:561–579. doi: 10.1007/BF01020572. [DOI] [PubMed] [Google Scholar]
- Chuang TI, Heckard LR. Generic realignment and synopsis of subtribe Castillejinae (Scrophulariaceae: tribe Pediculareae) Syst Bot. 1991;16:644–666. [Google Scholar]
- CLONTECH. PCR-select differential screening kit, users manual PT 3138–1. Clonetech Labs: Technical Information; 1999. http://www.clontech.com/techinfo/manuals/PDF/PT3138-1.pdf [Google Scholar]
- CLONTECH. PCR-select cDNA subtraction kit, users manual PT 1117–1. Clonetech Labs: Technical Information; 2000. http://www.clontech.com/techinfo/manuals/PDF/PT1117-1.pdf [Google Scholar]
- Delavault P, Estabrook E, Albrecht H, Wrobel R, Yoder JI. Host-root exudates increase gene expression of asparagine synthetase in the roots of a hemiparasitic plant Triphysaria versicolor (Scrophulariaceae) Gene. 1998;222:155–162. doi: 10.1016/s0378-1119(98)00502-2. [DOI] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KL, Nurse P. Tyrosine phosphorylation of the fission yeast Cdc2-positive protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- Gurskaya NG, Diatchenko L, Chenchik A, Siebert PD, Khaspekov GL, Lukyanov KA, Vagner LL, Ermolaeva OD, Lukyanov SA, Sverdlov ED. Equalizing cDNA subtraction based on selective suppression of polymerase chain reaction: cloning of Jurkat cell transcripts induced by phytohemaglutinin and phorbol 12-myristate 13-acetate. Anal Biochem. 1996;240:90–97. doi: 10.1006/abio.1996.0334. [DOI] [PubMed] [Google Scholar]
- Hammondkosack KE, Jones JDG. Resistance gene-dependent plant defense responses. Plant Cell. 1996;8:1773–1791. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa S, Kinghorn A, Cordell G, Farnsorth N. Plant anticancer agents: XXVI. Constituents of Peddiea fischeri. J Nat Prod. 1983;46:248–250. doi: 10.1021/np50026a020. [DOI] [PubMed] [Google Scholar]
- Heide-Jorgensen HS, Kuijt J. The haustorium of the root parasite Triphysaria (Scrophulariaceae), with special reference to xylem bridge ultrastructure. Am J Bot. 1995;82:782–797. [Google Scholar]
- Hrmova M, Harvey AJ, Wang J, Shirley NJ, Jones GP, Stone BA, Hoj PB, Fincher GB. Barley β-d-glucan exohydrolases with β-d-glucosidase activity: purification, characterization, and determination of primary structure from a cDNA clone. J Biol Chem. 1996;271:5277–5286. doi: 10.1074/jbc.271.9.5277. [DOI] [PubMed] [Google Scholar]
- Kuijt J. The Biology of Parasitic Flowering Plants. Berkeley: University of California Press; 1969. [Google Scholar]
- Kushnir S, Babiychuk E, Kampfenkel K, Bellesboix E, Vanmontagu M, Inze D. Characterization of Arabidopsis thaliana cDNAs that render yeasts tolerant toward the thiol-oxidizing drug diamide. Proc Natl Acad Sci USA. 1995;92:10580–10584. doi: 10.1073/pnas.92.23.10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matvienko M, Wojtowicz A, Wrobel R, Jamison D, Goldwasser Y, Yoder JI. Quinone oxidoreductase message levels are differentially regulated in parasitic and non-parasitic plants exposed to allelopathic quinones. Plant J. 2001;25:375–387. doi: 10.1046/j.1365-313x.2001.00971.x. [DOI] [PubMed] [Google Scholar]
- Nickrent DL, Duff RJ, Colwell AE, Wolfe AD, Young ND, Steiner KE, dePamphilis CW. Molecular phylogenetic and evolutionary studies of parasitic plants. In: Soltis DE, Soltis PS, Doyle JJ, editors. Molecular Systematics of Plants II DNA Sequencing. Boston: Kluwer Academic Publishers; 1998. pp. 211–241. [Google Scholar]
- Obrien PJ. Molecular mechanisms of quinone cytotoxicity. Chem Biol Interact. 1991;80:1–41. doi: 10.1016/0009-2797(91)90029-7. [DOI] [PubMed] [Google Scholar]
- O'Malley RC, Lynn DG. Expansin message regulation in parasitic angiosperms: marking time in development. Plant Cell. 2000;12:1455–1465. doi: 10.1105/tpc.12.8.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C, Riches CR, editors. Parasitic Weeds of the World: Biology and Control. Wallingford, UK: CAB International; 1993. [Google Scholar]
- Pawlowski K, Kunze R, de Vries S, Bisseling T. Isolation of total, poly (A) and polysomal RNA from plant tissues. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Norwell, MA: Kluwer Academic Publishers; 1994. pp. 1–13. [Google Scholar]
- Press MC, Graves JD, editors. Parasitic Plants. London: Chapman and Hall; 1995. [Google Scholar]
- Rabbani N, Duhaiman AS. Inhibition of camel lens zeta-crystallin/NADPH:quinone oxidoreductase by pyridoxal-5′-phosphate. Biochim Biophis Acta-Protein Struct Mol Enzymes. 1998;1388:175–180. doi: 10.1016/s0167-4838(98)00185-x. [DOI] [PubMed] [Google Scholar]
- Redfearn E, Whittaker P. The inhibitory effects of quinones on the succinic oxidase system of the respiratory chain. Biochim Biophys Acta. 1962;56:440–444. doi: 10.1016/0006-3002(62)90595-4. [DOI] [PubMed] [Google Scholar]
- Riopel J, Musselman L. Experimental initiation of haustoria in Agalinis purpurea. Am J Bot. 1979;66:570–575. [Google Scholar]
- Riopel JL, Baird WV. Morphogenesis of the early development of primary haustoria in Striga asiatica. In: Musselman LJ, editor. Parasitic Weeds in Agriculture. Boca Raton, FL: CRC Press, Inc.; 1987. pp. 107–125. [Google Scholar]
- Riopel JL, Timko MP. Haustorial initiation and differentiation. In: Press MC, Graves JD, editors. Parasitic Plants. London: Chapman and Hall; 1995. pp. 39–79. [Google Scholar]
- Rose JKC, Brummell DA, Bennett AB. Two divergent xyloglucan endotransglycosylases exhibit mutually exclusive patterns of expression in Nasturtium. Plant Physiol. 1996;110:493–499. doi: 10.1104/pp.110.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Maniatis T, Fritsch EF. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schimmoller F, Diaz E, Muhlbauer B, Pfeffer SR. Characterization of a 76 kDa endosomal, multispanning membrane protein that is highly conserved throughout evolution. Gene. 1998;216:311–318. doi: 10.1016/s0378-1119(98)00349-7. [DOI] [PubMed] [Google Scholar]
- Siqueira JO, Nair MG, Hammerschmidt R, Safir GR. Significance of phenolic compounds in plant-soil-microbial systems. Crit Rev Plant Sci. 1991;10:63–121. [Google Scholar]
- Smith CE, Dudley MW, Lynn DG. Vegetative/parasitic transition: control and plasticity in Striga development. Plant Physiol. 1990;93:208–215. doi: 10.1104/pp.93.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Ruttledge T, Zeng Z, O'Malley RC, Lynn DG. A mechanism for inducing plant development- the genesis of a specific inhibitor. Proc Natl Acad Sci USA. 1996;93:6986–6991. doi: 10.1073/pnas.93.14.6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MT. Quinones as mutagens, carcinogens, and anticancer agents: introduction and overview. J Toxicol Environ Health. 1985;16:665–672. doi: 10.1080/15287398509530776. [DOI] [PubMed] [Google Scholar]
- Steffens JC, Lynn DG, Kamat VS, Riopel JL. Molecular specificity of haustorial induction in Agalinis purpurea (L.) Raf. (Scrophulariaceae) Ann Bot. 1982;50:1–7. [Google Scholar]
- Sutton G, White O, Adams M, Kerlavage A. TIGR Assembler: a new tool for assembling large shotgun sequencing projects. Genome Sci Technol. 1995;1:9–19. [Google Scholar]
- Testa B. The Metabolism of Drugs and Other Xenobiotics. New York: Academic Press; 1995. [Google Scholar]
- Thomson RH. Naturally Occurring Quinones III: Recent Advances. New York: Chapman and Hall; 1987. [Google Scholar]
- Thurman LD. Genecological studies in Orthocarpus subgenus Triphysaria. PhD thesis. Berkeley: University of California; 1966. [Google Scholar]
- Toone WM, Jones N. AP-1 transcription factors in yeast. Curr Opin Genet Dev. 1999;9:55–61. doi: 10.1016/s0959-437x(99)80008-2. [DOI] [PubMed] [Google Scholar]
- Wang HY, Lockwood SK, Hoeltzel MF, Schiefelbein JW. The root hair defective-3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev. 1997;11:799–811. doi: 10.1101/gad.11.6.799. [DOI] [PubMed] [Google Scholar]
- Wang Z, Brown DD. A gene expression screen. Proc Natl Acad Sci USA. 1991;88:11505–11509. doi: 10.1073/pnas.88.24.11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler WMF, Kremmer E, Forster R, Winnacker EL. Identification of pirin, a novel highly conserved nuclear protein. J Biol Chem. 1997;272:8482–8489. doi: 10.1074/jbc.272.13.8482. [DOI] [PubMed] [Google Scholar]
- Wolf SJ, Timko MP. Analysis of in vivo protein sythesis and histological studies of haustorial formation in root cultures of witchweed (Striga asiatica L. Kuntze) J Exp Bot. 1992;43:1339–1348. [Google Scholar]
- Wolf SJ, Timko MP. Characterization of actin-gene family members and their expression during development in witchweed (Striga asiatica L.) Planta. 1994;192:61–68. doi: 10.1007/BF00198693. [DOI] [PubMed] [Google Scholar]
- Yang GP, Ross DT, Kuang WW, Brown PO, Weigel RJ. Combining SSH and cDNA microarrays for rapid identification of differentially expressed genes. Nucleic Acids Res. 1999;27:1517–1523. doi: 10.1093/nar/27.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JI. A species-specific recognition system directs haustorium development in the parasitic plant Triphysaria (Scrophulariaceae) Planta. 1997;202:407–413. doi: 10.1007/s004250050144. [DOI] [PubMed] [Google Scholar]
- Yoder JI. Self and cross-compatibility in three species of the hemiparasite Triphysaria (Scrophulariaceae) Environ Exp Bot. 1998;39:77–83. [Google Scholar]
- Zeng ZX, Cartwright CH, Lynn DG. Cyclopropyl-p-benzoquinone: a specific organogenesis inhibitor in plants. J Am Chem Soc. 1996;118:1233–1234. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.