Abstract
A subtractive tomato (Lycopersicon esculentum) root cDNA library enriched in genes up-regulated by changes in plant mineral status was screened with labeled mRNA from roots of both nitrate-induced and mineral nutrient-deficient (−nitrogen [N], −phosphorus, −potassium [K], −sulfur, −magnesium, −calcium, −iron, −zinc, and −copper) tomato plants. A subset of cDNAs was selected from this library based on mineral nutrient-related changes in expression. Additional cDNAs were selected from a second mineral-deficient tomato root library based on sequence homology to known genes. These selection processes yielded a set of 1,280 mineral nutrition-related cDNAs that were arrayed on nylon membranes for further analysis. These high-density arrays were hybridized with mRNA from tomato plants exposed to nitrate at different time points after N was withheld for 48 h, for plants that were grown on nitrate/ammonium for 5 weeks prior to the withholding of N. One hundred-fifteen genes were found to be up-regulated by nitrate resupply. Among these genes were several previously identified as nitrate responsive, including nitrate transporters, nitrate and nitrite reductase, and metabolic enzymes such as transaldolase, transketolase, malate dehydrogenase, asparagine synthetase, and histidine decarboxylase. We also identified 14 novel nitrate-inducible genes, including: (a) water channels, (b) root phosphate and K+ transporters, (c) genes potentially involved in transcriptional regulation, (d) stress response genes, and (e) ribosomal protein genes. In addition, both families of nitrate transporters were also found to be inducible by phosphate, K, and iron deficiencies. The identification of these novel nitrate-inducible genes is providing avenues of research that will yield new insights into the molecular basis of plant N nutrition, as well as possible networking between the regulation of N, phosphorus, and K nutrition.
Nitrogen (N) is the essential mineral element required in the greatest amount in plants, comprising 1.5% to 2% of plant dry matter and approximately 16% of total plant protein (Frink et al., 1999). Thus, N availability is a major limiting factor for plant growth and crop production. Plant can utilize a wide range of N species including volatile ammonia (NH3), nitrogen oxides (NOx), mineral N (NO3− and NH4+), and organic N (amino acids, peptides, etc.; von Wirén et al., 1997). However, in most agricultural soils, nitrate (NO3−) is the most important source of N (Crawford and Glass, 1998; Hirsch and Sussman, 1999). Because of the high N requirements for crop plants, N fertilization is a major worldwide agricultural investment, with 80 million metric tons of N fertilizers (as nitrate and/or ammonium) applied annually (Frink et al., 1999). There are also negative environmental consequences for the extensive use of N fertilizers in crop production because agricultural crops only retain about two-thirds of the applied N, and the unabsorbed N can subsequently leach into and contaminate water supplies (Frink et al., 1999). Because of the high costs of N fertilizer to agricultural production, and the deleterious effect of N fertilizer pollution on the environment, it would be desirable to develop strategies to reduce N input while simultaneously maintaining productivity. A more complete understanding of the molecular and physiological basis of N uptake and metabolism in plants may reveal strategies for accomplishing these goals.
Nitrate uptake in plants is highly regulated and coordinated with other transport and metabolic pathways (Crawford, 1995), and a number of nitrate uptake and assimilation-related genes have been identified and characterized (Forde, 2000). Plants absorb nitrate via transporters localized to the root epidermal and cortical cell plasma membrane over a wide nitrate concentration range using several different transport mechanisms, including constitutive and nitrate-inducible high-affinity transport systems, as well as nitrate-inducible low-affinity transporters (von Wirén et al., 1997; Crawford and Glass, 1998; Daniel-Vedele et al., 1998; Forde and Clarkson, 1999; Hirsch and Sussman, 1999; Stitt, 1999). Once in the root cell cytoplasm, nitrate may be stored in the vacuole for later use, transported into the xylem and translocated to the shoot for assimilation and/or storage, released back into the rhizosphere, or reduced to nitrite and then ammonia via nitrate reductase (NR) and nitrite reductases (NiR; Crawford and Glass, 1998). The reduction of nitrate to nitrite and then ammonia generates N in a form that can be assimilated into amino acids via the GOGAT pathway (Stitt, 1999).
Nitrate is an interesting essential nutrient because unlike other mineral nutrients where deficiencies of the specific mineral induces genes involved in transport and assimilation, exposure of roots to nitrate has been shown to induce genes important for N assimilation. Thus, it is not surprising that this nutrient can induce a large number of genes including high (NRT2) and low (NRT1) affinity nitrate transporters, NR, NiR, and the enzymes of GOGAT pathway for ammonia assimilation (Wang et al., 2000). Nitrate also induces genes involved in different aspects of carbon metabolism, including the synthesis of organic acids used both for amino acid synthesis and the regulation of pH in response to OH− resulting from nitrate uptake and assimilation, and the oxidative pentose phosphate pathway, which provides high levels of NADPH needed for nitrate assimilation (Stitt, 1999).
Although nitrate-induced genes that are directly involved in nitrate transport and metabolism have been described and studied for a number of years; they may represent only a small fraction of the genes induced by nitrate. Identification of novel nitrate-inducible genes and elucidation of their function in plant N nutrition will be important to more fully understand the entire process of plant nitrate assimilation and utilization, which in turn should provide information for improving crop N use efficiency. Wang et al. (2000) recently used microarray analysis in Arabidopsis to identify at least 15 new genes induced by nitrate. Among these genes was a potential regulatory gene (a MYB transcription factor), a series of metabolic enzymes such as transaldolase, transketolase, malate dehydrogenase, Asn synthetase, and His decarboxylase; and several genes with unknown functions including hemoglobin, a senescence-associated protein, and two methyltransferases.
In this study, we employed a combination of mRNA subtraction and high-density cDNA arrays to conduct time course mRNA expression analysis, as a means of identifying additional nitrate-inducible genes. Recovery of genes previously reported to be induced by nitrate resupply validated the strategy we used. In addition to these genes, we identified a series of new genes that exhibit nitrate inducibility, including stress response genes, water transport genes, a transcription factor, and genes encoding known proteins whose functions have not yet been elucidated in plants. Nitrate was also discovered to induce phosphate (Pi) and potassium (K) transporters, suggesting potential coordination of nitrate, Pi, and K uptake in the plant.
RESULTS
Arraying Strategy
To identify novel genes induced by nitrate, a subtractive cDNA library was constructed from pooled roots of tomato (Lycopersicon esculentum) plants grown under a range of nutrient deficiencies (phosphorus [P], K, calcium [Ca], sulfur [S], magnesium [Mg], iron [Fe], zinc [Zn], and copper [Cu]) as well as under conditions of nitrate resupply. The plants for the nitrate resupply experiment were grown for 5 weeks on hydroponic media containing nitrate/ammonium, deprived of N for 48 h, and then 4.8 mm nitrate was resupplied for 12 h. We found that this 48-h period of N deprivation was sufficient to deplete nitrate pools in the roots without having significant effects on overall N nutrition, based on the lack of visual symptoms of N deficiency, no decrease in shoot growth, and no change in total shoot N content after the 48-h period of N deprivation. The library, consisting of approximately 14,000 clones, was initially screened with labeled mRNA from roots of plants resupplied with nitrate, and from roots of plants exposed to a spectrum of mineral deficiencies. Based on these preliminary expression studies, 716 cDNA clones that were up-regulated in response to changes in plant mineral nutrient status were chosen for array experiments. In addition, other clones were selected from a second mineral deficient tomato root library that was sequenced as part of a National Science Foundation-funded tomato genome project (in collaboration with Steve Tanksley, Cornell University), based on sequence homology to known genes (see “Materials and Methods”).
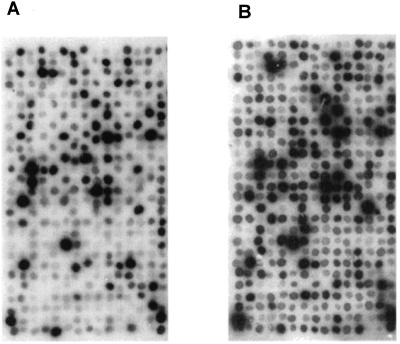
To identify genes among this set of 1,280 clones (see Supplementary Table I at www.plantphysiol. org) that were up-regulated by nitrate resupply, the amplified cDNAs were arrayed on nylon filters and were hybridized with 32P-labeled mRNA probes from roots of 5-week-old tomato seedlings grown without nitrate for 48 h and then exposed to nitrate for various times (0 = control, 1, 6, 12, 24, 48, and 96 h). For each time point, the particular array was compared with the 0-h time point array to identify clones whose expression was altered by exposure to nitrate. A representative comparison between the control and one such time point (1 h) is depicted in Figure 1. It can be seen that expression of a number of genes appears to be altered after the 1-h exposure to nitrate. After comparing the hybridization data for control versus each nitrate exposure time point, a total of 115 clones (Tables I and II) were identified that exhibited changes in expression. From these, a subset of clones exhibiting the greatest increase in expression was then chosen for more detailed RNA gel-blot analysis. The great majority of the overall pool of 115 cDNA clones exhibited increased expression, which should be expected because the original subtractive library was designed to be enriched in genes up-regulated by nitrate exposure. A number of nitrate-inducible genes were identified that participate in plant metabolic pathways, including those encoding transaldolase, transketolase, malate dehydrogenase, Asn synthetase, 6-phosphogluconate dehydrogenase, Glc-6-phosphate dehydrogenase, ferredoxin, ferredoxin NADP oxidoreductase, and His decarboxylase that were previously identified as being nitrate inducible in Arabidopsis (Wang et al., 2000), and will not be discussed in this report. However, the identification of a common set of metabolically related genes up-regulated by nitrate resupply in two different plant species, as well as genes for nitrate and NiR and nitrate transporters (for review, see Crawford, 1995; Stitt, 1999), validated our overall approach to identify novel genes induced by nitrate.
Figure 1.
Comparative hybridization profiles for the 1280 cDNA array hybridized with root mRNA either from plants grown without N for 48 h or −N-grown plants resupplied with nitrate for 1 h. A, Portion of the array hybridized with labeled mRNA at 0 h (−N grown plants). B, Same part of the array hybridized with labeled mRNA from roots resupplied with 48 mm potassium nitrate for 1 h. DNA was denatured on the filter with 0.4 m NaOH and neutralized with 2× SSC. The root mRNA probes were labeled with 32P-dATP via reverse transcription.
Table I.
Complete list of sequenced genes up-regulated by nitrate
| Clone ID | Accession No. | GenBank Hit | Gene/cDNA Clone | Expression (h after treatment) | 1-h/24-h Signal Ratio |
|---|---|---|---|---|---|
| B16-1 | AW979417 | X95098 | LeAMT2 | Up 6–12 h | 2.3/1.7 |
| B22-1 | AW979368 | A45772 | LeNRT1.2 | Up 1–24 h | 10/14.3 |
| H21-1 | AW219289 | U17987 | LeNRT1.2 | Up 1–24 h | 11/12 |
| D2-4 | AF092655 | AF092655 | LeNRT2.1 | Up 1–24 h | 12.1/11 |
| 11B22 | X14060 | X14060 | NR | Up 1–12 h | 4/3.8 |
| E20-4 | BG713767 | AF065616 | NiR | Up 1–24 h | 8/7.4 |
| E13-1 | AW979921 | AF129478 | K+ transporter HAK5 | Down after 3 h | 0.5/0.01 |
| G22-1 | AW218955 | AJ249962 | K channel (Kdc1 homolog) | Up 1–24 h | 4.5/6.8 |
| 23L8 | T07607 | T07607 | LePT2 (phosphate transporter) | Up 6–96 h | 1/2.8 |
| H10-1 | AW219193 | S49583 | Leu zipper transcription factor | Up 1–24 h | 3.9/3.4 |
| X12K3 | AW621495 | Y07563 | hin1 protein | Up 1–12 h | 5/1.8 |
| I4-1 | AW219480 | S22531 | Pathogenesis-related protein 1b | Up 1–24 h | 8.2/9.5 |
| L2-4 | AW624985 | U20809 | LeGST4 | Up 1–12 h | 4.4/2.8 |
| H9-2 | BG713784 | AF053076 | LEA5 protein | Up 1–24 h | 3/2.2 |
| H2-2 | BG713780 | A56555 | Trypsin inhibitor precursor | Up 1–96 h | 7/16 |
| J2-1 | AW219454 | S48032 | cim1 protein | Up 6–96 h | 0.8/3 |
| A15-2 | BG713769 | T03673 | pit1 protein | Up 6–96 h | 3/19 |
| I2-2 | BG713788 | T03673 | pit1 | Up 1–96 h | 1/10 |
| K12-2 | BG713797 | T03673 | pit1 | Up 1–48 h | 1.8/8 |
| K21-2 | BG713799 | T03673 | pit1 | Up 6–24 h | 2.1/15 |
| L11-2 | BG713802 | T03673 | pit1 | Up 6–48 h | 1.5/11 |
| D24-2 | BG713774 | T03673 | pit1 | Up 24–96 h | 4/12 |
| C5-3 | BG713810 | T03673 | pit1 | Up 1–96 h | 3.2/14 |
| C8-3 | BG713811 | T03673 | pit1 | Up 6–48 h | 1.3/7 |
| D9-3 | BG713814 | T03673 | pit1 | Up 6–96 h | 1.1/5 |
| F6-3 | BG713816 | T03673 | pit1 | Up 12–96 h | 2.4/6 |
| F22-3 | BG713817 | T03673 | pit1 | Up 12–48 h | 5.1/8 |
| H2-3 | BG713821 | T03673 | pit1 | Up 6–96 h | 1/6.8 |
| L4-3 | BG713833 | T03673 | pit1 | Up 6–48 h | 2.7/4 |
| M9-3 | BG713836 | T03673 | pit1 | Up 6–96 h | 3.5/13 |
| H9-1 | AW219182 | AL022023 | Plasma membrane intrinsic protein | Up 48–96 h | 1.2/3 |
| J21-4 | AW625013 | AB002149 | Water channel protein | Up 24–96 h | 1/3.2 |
| H3-2 | BG713781 | AF133530 | Water channel | Up 6–96 h | 0.8/2.8 |
| H15-2 | BG713786 | T06434 | Water channel | Up 6–96 h | 1.1/2.3 |
| G7-1 | AW218990 | JQ2288 | Soybean (Glycine max) nodulin 26 | Up 48–96 h | 1/1.6 |
| G10-1 | AW218928 | Q08451 | Tomato ripening-associated membrane protein | Up 48–96 h | 1.4/2.5 |
| L23-4 | AW625086 | U39486 | Delta tonoplast integral protein | Up 6–96 h | 1.4/2.5 |
| C5-2 | BG713771 | T02526 | Ribosomal protein L36 | Up 6 h | 1/1.2 |
| G10-2 | BG713779 | O23515 | Ribosomal protein L15 | Up 6–12 h | 2/1.2 |
| H6-2 | BG713783 | T02335 | Ribosomal protein L18a | Up 12 h | 0.9/1.6 |
| A4-3 | BG713807 | AF127042 | 60S ribosomal protein L37a | Up 6 h | 1.2/2 |
| I2-3 | BG713826 | X76714 | Ribosomal protein | Up 1–12 h | 1/1.8 |
| L12-3 | BG713834 | T48952 | Ribosomal protein S29 | Up 6 h | 1/1.9 |
| N1-1 | AW455251 | M90692 | Phe ammonia-lyase | Up 24–96 h | 0.5/2 |
| H13-1 | AW219196 | P23244 | LeHb1 (Glb1) | Up 6–24 h | 0.6/3 |
| O22-1 | AW218217 | X95905 | 14-3-3 Protein | Up 6–12 h | 1.2/1 |
| B4-4 | BG713766 | U88711 | Copper homeostasis factor | Up 1–12 h | 1.5/1.4 |
| A8-1 | AW979453 | Z73942 | RAB7C (ras-related protein) | Up 1–12 h | 1.4/0.8 |
| E5-4 | BG713768 | D86629 | NT16 polypeptide | Up 1–96 h | 3.5/4 |
| N19-1 | AW224533 | AF207621 | K transporter KUP3 | Down after 6 h | 0.6/1 |
| A18-1 | AW979462 | U83245 | ARF1 homolog | Up 1–12 h | 2/0.9 |
This table includes nitrate-induced genes that were subsequently subjected to northern analysis. All pit1 clones are either identical or highly homologous. One clone from each of the group of pit1 and ribosomal proteins, and two from the group of water channels, were used for northern analysis. Genes most strongly induced by nitrate resupply are shown in the Figures 2 through 8.
Table II.
A complete list of sequenced genes up-regulated by nitrate
| Clone ID | Accession No. | GenBank Hit | Gene/cDNA Clone | Expression (h after treatment) | 1-h/24-h Signal Ratios |
|---|---|---|---|---|---|
| B6-1 | AW979398 | D30719 | Dehydration-induced protein (ERD15) | Down after 6 h | 1.5/0.7 |
| E5-1 | AW979904 | L42814 | Acetyl coenzyme A carboxylase | Up 6–12 h | 0.9/1.4 |
| F4-1 | AW979983 | Z46944 | Non-photosynthetic ferredoxin | Up 1–96 h | 1.6/1.4 |
| H5-1 | AW219021 | Z97335 | Hydroxymethyltransferase | Up 1–96 h | 2/1.8 |
| H8-1 | AW219174 | X79770 | sti (stress-inducible protein) | Up 6–96 h | 1/2.4 |
| J3-1 | AW219553 | U55838 | Carbonic anhydrase | Up 1–12 h | 1.7/1 |
| J4-1 | AW219399 | O24661 | Asp synthetase | Up 1–96 h | 2.8/2 |
| J12-1 | AW219540 | A44133 | Phe ammonia-lyase | Down after 6 h | 1.2/0.7 |
| I13-1 | AW219330 | JA0177 | Salt-induced protein | Up after 1 h | 2/1 |
| D6-1 | AW224693 | AB004307 | Ferredoxin-NADP oxidoreductase | Up 1–12 h | 2/2.4 |
| I4-4 | AW624841 | X92975 | Xyloglucan endo-transglycosylase | Up 6–96 h | 1.6/1.8 |
| K6-4 | AW625018 | P08440 | Fru-bisphosphate aldolase | Up 1–6 h | 2/0.8 |
| L1-4 | AW218551 | AF104925 | 2-Oxoglutarate-dependent dioxygenase | Up 1–96 h | 1.4/1.6 |
| M5-4 | AW625127 | AF183891 | S-Adenosyl-l-methionine synthetase (sam2) | Up 1–96 h | 1.2/2 |
| A19-4 | AW624860 | AF022775 | Caffeoyl-coenzyme A 3-O-methyltransferase | Down after 6 h | 1.7/0.9 |
| B7-4 | AW154870 | AF007581 | Cytoplasmic malate dehydrogenase | Up 1–6 h | 2.2/1.1 |
| B18-4 | AW979983 | Z46944 | Non-photosynthetic ferredoxin | Up 1–12 h | 1.3/1 |
| B22-4 | AW219992 | U64924 | NTGP3 (small GTP-binding protein) | Down after 6 h | 0.7/1 |
| E4-4 | AW218294 | AF092547 | Profilin | Up 6–48 h | 1.1/1.7 |
| E9-4 | Y18931 | Y18931 | Tomato subtilisin-like protease | Up after 6 h | 1/1.8 |
| E16-4 | AW224694 | O04397 | Ferredoxin-NADP reductase | Up 1–6 h | 1.4/0.9 |
| F20-1 | AW218899 | D88156 | Tropinone reductase-I | Down after 12 h | 1.6/0.8 |
| D18-2 | BG713773 | S20925 | Polyubiquitin | Down 6–12 h | 1.3/1.1 |
| H5-2 | BG713782 | X99100 | Protein kinase | Up 6–12 h | 1/1.4 |
| F5-2 | BG713776 | Z48976 | Phosphoglycerate kinase | Up 6–96 h | 0.9/2 |
| G4-2 | BG713778 | AF112440 | Ribosomal protein S26 | Up 1–48 h | 1.5/1.8 |
| H12-2 | BG713785 | U17005 | Glyceraldehyde-3-phosphate dehydrogenase | Up 1–48 h | 0.8/1.6 |
| H23-2 | BG713787 | T04367 | Water channel | Down 6–48 h | 1.4/0.6 |
| J8-2 | BG713790 | U90212 | DNA-binding protein ACBF | Up 12 h | 1/1.4 |
| J9-2 | BG713791 | AC016662 | Enolase | Up 1–96 h | 2.2/3 |
| J10-2 | BG713792 | AC016662 | Enolase | Up 48 h | 1.1/1.4 |
| J11-2 | BG713793 | AC016662 | Enolase | Up 6–48 h | 1/1.8 |
| K13-2 | BG713798 | AB001891 | Ribosomal protein | Up 6–48 h | 0.8/2 |
| L5-2 | BG713800 | X98063 | Polyubiquitin | Up 6–48 h | 1.2/1.5 |
| M7-2 | BG713803 | T06434 | Water channel | Down 6–12 h | 1/1.3 |
| N5-2 | BG713805 | A48767 | Glu decarboxylase | Up 1–48 h | 1.7/1 |
| N6-2 | BG713806 | L44142 | Auxin-repressed protein | Up 1–48 h | 1.4/1.8 |
| A10-3 | BG713808 | T02215 | Ferrodoxin reductase | Up 1–6 h | 1.5/1 |
| C23-3 | BG713812 | T01984 | LEA 5 protein | Down 6–48 h | 4.2/1.3 |
| D2-3 | BG713813 | S51584 | Peroxidase | Up 12 h | 1.1/1.8 |
| E4-3 | BG713815 | T03673 | Enolase | Up 1–96 h | 1/1.7 |
| G3-3 | BG713818 | U44386 | LEA 5 protein | Up 1–12 h | 2/1.5 |
| H3-3 | BG713822 | AL163816 | Glutaredoxin-like protein | Up 6 h | 1.8/0.9 |
| H5-3 | BG713824 | Y10024 | Ubiquitin extension protein | Up 12–48 h | 1.1/1.6 |
| J10-3 | BG713829 | Y15371 | Induced by nod factor | Up 48 h | 0.9/1.4 |
| L13-3 | BG713835 | M94135 | Chloroplast carbonic anhydrase | Up 1 h | 1.8/1 |
This table includes nitrate-induced genes that were not subjected to northern analysis. For 18 of the sequenced nitrate-induced clones, we did not find sequence homology with any known genes in GenBank and thus these were not included in the Table. Genes with a clone ID ending in “−1” were identified from the Cornell tomato root cDNA libraries that were generated in collaboration with Dr. Steve Tanksley and sequenced by the Institute for Genomic Research. All other clones were obtained from the subtractive DNA libraries. The column labeled “1-h/24-h Signal Ratios” shows the quantified change in gene expression for plants exposed to nitrate for either 1 or 24 h compared with control plants (N withheld for 48 h).
Nitrate Uptake and Assimilation
Both families of nitrate transporters (NRT1 [low-affinity transporters] and NRT2 [high-affinity transporters]) were represented in the array. As shown in Figure 2, LeNrt1.2, a member of NRT1 family, was up-regulated rapidly after nitrate resupply (by 1 h) and this up-regulation persisted for the first 24 h of nitrate exposure. In contrast, the expression on another member of the NRT1 gene family, LeNrt1.1, was not altered by nitrate exposure (data not shown), in agreement with a previous report for tomato (Lauter et al., 1996). Expression of LeNrt2.1 (accession no. AF092655, a member of the NRT2 family) in response to nitrate was nearly identical to that seen for LeNrt 1.2 (rapid up-regulation that persisted through 24 h of nitrate exposure). This rapid up-regulation by nitrate of nitrate transporter genes from the NRT 2 gene family was also recently reported in barley roots (Vidmar et al., 2000). Genes encoding the other major immediate components of the nitrate assimilation pathway, NR and NiR, also exhibited a pattern of nitrate induction similar to the NRTs (Fig. 2). The similar expression patterns for nitrate transporters and nitrate and NiR is expected, given the tight coupling between nitrate provision to the cytoplasm and subsequent assimilation (Crawford, 1995).
Figure 2.
Expression profiles of genes encoding the nitrate transporters LeNRT12 and LeNRT21, NR, and NiR in response to nitrate resupply from 1 to 96 h. A, RNA gel blot. B, Quantitative expression data for the RNA gel blot depicted in A. In Figure 2B and the subsequent figures, the autoradiograph of the RNA gel blot was scanned and the image computer digitized. The gel blot intensities were quantified using the National Institutes of Health Image program. Changes in gene expression were quantified as a relative signal ratio, which was determined by dividing the quantified gel blot intensity at each nitrate exposure time point by the RNA blot intensity for the control plants (grown for 48 h without nitrate). LeNRT12, LeNRT21, and NR are tomato genes deposited in GenBank, whereas NiR is a homolog of a pepper NiR (GenBank accession no. AF065616) with a score of 291 and an E value of 6e-78. Approximately 1 μg of mRNA was loaded for each lane and α-tubulin was the loading control. Quantitated signals were weighted against the control in all the graphs.
Novel Nitrate-Induced Genes
In addition to the structural genes encoding the early components of the nitrate assimilation pathway described above, as well as the genes involved in plant metabolism previously identified by Wang et al. (2000), a number of genes not previously reported to be inducible by nitrate were identified in our study (Tables I and II). These included genes encoding water channels, other mineral nutrient transporters, stress response genes, signaling and regulatory genes, and genes encoding ribosomal proteins. Expression of these genes was subsequently analyzed using RNA gel blots.
Water Channel Proteins
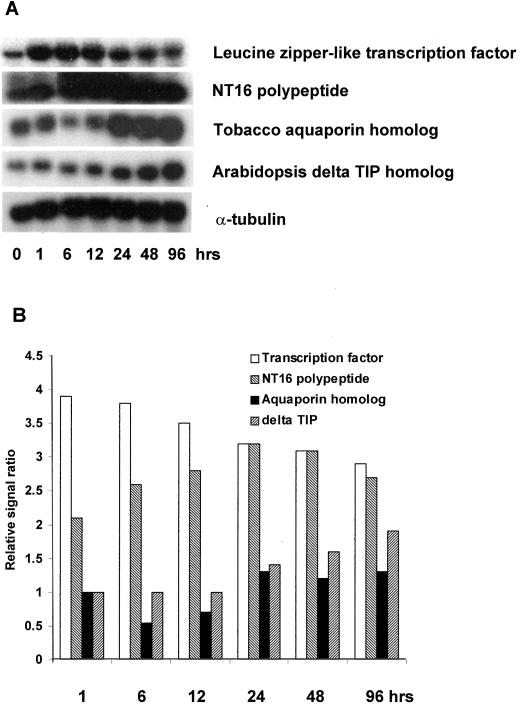
Water channel proteins (aquaporins) reside in the plasma membrane and tonoplast membranes and facilitate cellular water transport and help regulate turgor pressure in plants (Maurel, 1997). A wide range of possible roles for aquaporins have been suggested, including inhibition of self-pollination, closure of leaf guard cells, and root water uptake. This is supported by the fact that expression of certain aquaporins have been shown to be regulated by light, gibberellic acid, abscisic acid, drought, salt stress, and nematodal infestation (Maurel, 1997; Mariaux et al., 1998; Johansson et al., 2000). In this study, we found that at least seven different water channel genes were up-regulated by nitrate (Table I). The majority (five) of the putative water channel genes were not up-regulated until late (48 h) in the nitrate exposure time course (Fig. 3 and Table I). This up-regulation at 48 h coincided with the time at which the expression of nitrate uptake and assimilation genes decreased (see Figs. 2 and 3), which would be consistent with stimulated water uptake in response to an increase in symplasmic solute concentrations due to the nitrate uptake.
Figure 3.
Up-regulation of a transcription factor, and NT 16 and aquaporin gene homologs induced by nitrate resupply between 1 and 96 h. The tomato transcription factor EST is homologous to the tobacco (Nicotiana tabacum) TGA1a transcription factor with a score/E value of 238/4e-62. See Table I for additional details about all seven aquaporin genes induced by nitrate exposure. A, RNA gel blot. B, Quantitative expression data for the RNA gel blot depicted in A.
Ammonium, Pi, and K Transporters
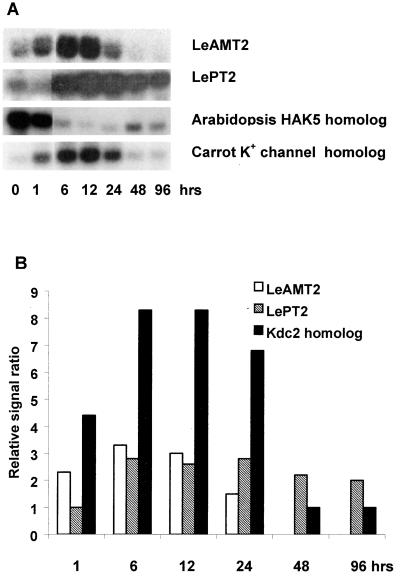
In addition to inducing root nitrate transporters, it was surprising to find that nitrate exposure rapidly altered expression of other mineral ion transporters, including ammonium, Pi, and K transporters. As seen in Figure 4, the ammonium transporter LeAMT2 was up-regulated rapidly (1 h) and strongly up to 24 h after nitrate resupply. Nitrate exposure had differential effects on two tomato Pi transporters. LePT2 was rapidly (within 3 h) up-regulated by nitrate exposure and remained highly expressed through the 96-h treatment (Fig. 4). A homolog of the Arabidopsis high-affinity K+ transporter, HAK5, was strongly down-regulated in response to nitrate, whereas a homolog of a novel carrot K+ channel, Kdc1 (Downey et al., 2000) was strongly up-regulated at 1 h and remained up-regulated until 24 h after nitrate exposure (Fig. 4). On the other hand, expression of the tomato Fe transporter LeIRT2 was not significantly changed in response to nitrate (data not shown).
Figure 4.
Changes in expression of ammonium, Pi, and K+ transporter genes induced by nitrate. HAK5 was not graphed. Time course used in the deficiency experiments was 1, 3, 6, 12, 24, and 48 h. A, RNA gel blot. B, Quantitative expression data for the RNA gel blot depicted in A.
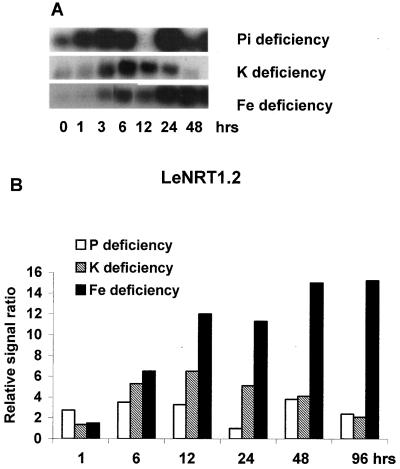
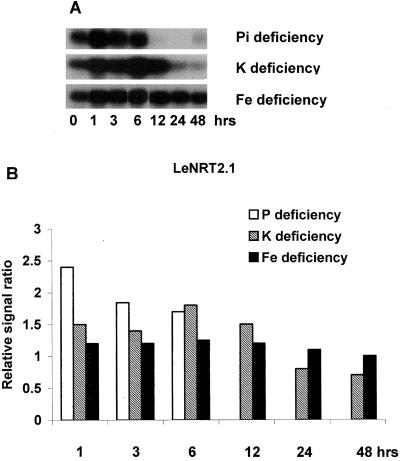
Because nitrate exposure altered expression of transporters for other mineral nutrients, we also looked at the influence of other mineral deficiencies on expression of the tomato nitrate transporters. As seen in Figures 5 and 6, LeNRT1.2 and LeNRT2.1 expression were rapidly increased after exposure to P and K deficiencies. Transcript abundance for both transporters increased rapidly (within 1 h) and strongly in response to exposure to P deficiency, whereas exposure to K deficiency increased expression within 1 to 6 h. A different pattern of nitrate transporter expression was seen in response to withholding Fe. LeNRT1.2 expression increased at 3 h of exposure and this was maintained for the entire 96-h deficiency period, whereas LeNRT2.1 expression was only slightly increased by Fe-deficient conditions.
Figure 5.
Changes in expression of nitrate transporter, LeNRT12, induced by Pi, K, and Fe deficiencies. Time course used in the deficiency experiments was 1, 3, 6, 12, 24, and 48 h. A, RNA gel blot. B, Quantitative expression data for the RNA gel blot depicted in A.
Figure 6.
Response of nitrate transporter, LeNRT21, to Pi, K, and Fe deficiencies. Time course used in the deficiency experiments was 1, 3, 6, 12, 24, and 48 h. A, RNA gel blot. B, Quantitative expression data for the RNA gel blot depicted in A.
Regulatory and Signaling Genes
Nitrate is unique as a mineral nutrient because it is the only one where exogenous exposure to the nutrient induces transport and assimilatory processes in the root. Thus, there is considerable interest in elucidating potential nitrate signaling elements in roots. A MADS box transcription factor, ANR1, previously was shown to respond to nitrate in N-starved Arabidopsis roots (Zhang and Forde, 1998). Our tomato cDNA library contained a homolog of ANR1, but it was not induced by nitrate exposure in tomato roots, which was also the case in the study by Wang et al. (2000). We identified here another transcription factor gene that was highly up-regulated within 1 h after nitrate resupply (Fig. 3). This increased expression was transient, and decreased after 24 h of nitrate exposure. It is interesting that this transcription factor was also rapidly and transiently induced by P, K, and Fe deficiencies (Y.-H. Wang, D.F. Garvin, and L.V. Kochian, unpublished data). Comparison of the amino acid sequence for this tomato transcription factor with homologs from other plant species indicated it was identical to a putative potato Leu zipper transcription factor and 80% identical to the tobacco bZIP transcription factor, TGA1a (Katagiri et al., 1989). We found that expression of this tomato transcription factor was not altered by other environmental stresses including salt and low temperature (data not shown), suggesting that it may be specific for plant responses to changes in mineral nutrient status.
A second putative transcription factor was also transiently induced by nitrate exposure. This gene, which is a homolog of ARF1 a transcription factor that binds to auxin response elements in Arabidopsis (Ulmasov et al., 1997), showed a 2-fold increase in expression 1 h after nitrate resupply (Table I). However, expression of this gene returned to control levels at subsequent time points after 1 h. Also, we found a gene highly similar to other tomato 14-3-3 proteins that was up-regulated 2- to 3-fold 6 to 12 hours after initiation of nitrate resupply (Table I). In plants, it has been suggested that 14-3-3 proteins regulate important N and carbon assimilatory enzymes such as NR and Suc phosphate synthase, in addition to possibly playing a role in responses to pathogen attack and activation of the plasma membrane H+-ATPase (Chung et al., 1999).
Finally, a homolog of the gene encoding the tobacco NT 16 polypeptide that has been implicated in root development, also was up-regulated (Fig. 3). In tobacco, NT 16 was found to be expressed almost exclusively in roots and was inducible by Agrobacterium tumefaciens infection that triggers root proliferation as well as tumorgenesis (Yasuda et al., 1997).
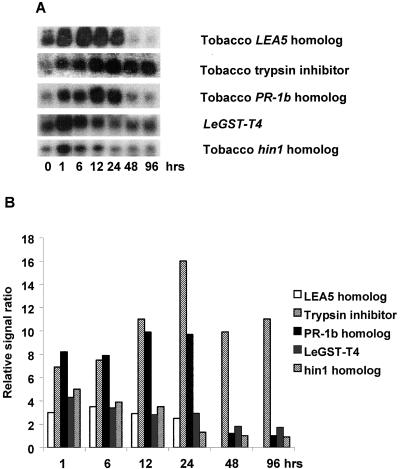
Stress Response Genes
A significant fraction of the novel nitrate-inducible genes we identified were similar to stress response genes reported in tomato and other plant species; expression of these clones is summarized in Figure 7. A homolog of the tobacco late embryogenesis-abundant protein 5 gene (LEA5) was found to be highly induced by nitrate. The induction was rapid and sustained between 1 and 24 h of nitrate resupply. It was previously reported that LEA5 is activated in response to salt, drought, and heat stress in citrus (Naot et al., 1995). Nitrate resupply also induced the expression of a homolog of the gene encoding a tobacco trypsin inhibitor type I precursor, and this homolog was not significantly induced by any of the mineral deficiency treatments (data not shown). In tobacco, trypsin inhibitor genes are coordinately expressed with PR-1b (a pathogenesis-related protein gene) and are inducible by a range of stresses, including tobacco mosaic virus infection, salycylic acid, ethylene, UV radiation, and wounding (Linthorst et al., 1993). We found that a homolog of the tobacco PR-1b was induced by nitrate resupply.
Figure 7.
Changes in expression of five different putative stress response genes to nitrate resupply. A, RNA gel blot. B, Quantitative expression data for the RNA gel blot depicted in A.
Several other stress response genes were also induced by nitrate exposure. A putative tomato glutathione-S-transferase T4 gene was transiently and rapidly induced between 1 and 3 h after nitrate resupply. Last, the tomato homolog of the tobacco hin1 gene (Gopalan et al., 1996) also was rapidly induced by nitrate resupply. The increase in hin 1 transcript level was only seen during the first 3 h after nitrate resupply. It was previously shown that hin1 is inducible by the bacterial hrp gene, which elicits a hypersensitive response in plants during plant infection by the bacterial pathogens Erwinia amylovora or Pseudomonas syringae (Gopalan et al., 1996).
Ribosomal Proteins
We found that a number of ribosomal protein genes (RPs) were up-regulated in response to nitrate exposure (Table I). In all cases, up-regulation occurred during the early stages of nitrate resupply (1–12 h). RNA gel-blot analysis was conducted on one of the RP clones (clone G10-2 that is homologous to RP L15) and this confirmed the time course for increased expression. In eukaryotes, four ribosomal RNAs and about 80 RPs contribute to the structure of the ribosomes. They are essential for ribosome assembly, which requires the coordinated expression of genes for all ribosomal constituents (Mager, 1988). However, the functional roles of different RPs in the translational process are not well understood in plants. Of the six RPs found to be up-regulated by nitrate resupply, a specific role for only one, L15, has been postulated (Lee et al., 1999). Nonetheless, the relatively early induction of these RP genes in response to nitrate resupply (occurring slightly after increased expression of the nitrate transporter genes) in this study may suggest a role in increased protein synthesis for N-depleted plants suddenly provided with N.
Other Genes Induced by Nitrate
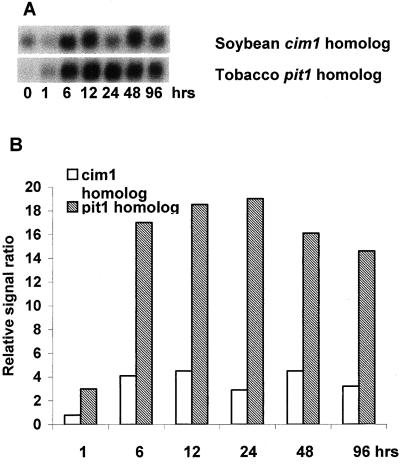
There were two novel genes up-regulated by nitrate for which a function with respect to N metabolism cannot be readily identified. A cDNA clone homologous to the soybean cim1 gene was up-regulated 3 h after nitrate resupply (Fig. 8). cim1 was shown previously to be induced strongly by cytokinin (20–60-fold) in cytokinin-starved soybean suspension cells. It was suggested that the cim1 protein is probably localized to the cell wall and may play a role in cytokinin-regulated cell wall expansion because Cim1 in soybean shares similarity with cell wall expansins (Downes and Crowell, 1998). Another cDNA clone partially homologous to the tobacco pit1 gene was strongly up-regulated after 6 h of nitrate exposure and this expression remained high throughout the 96 h of nitrate resupply (Fig. 8). In tobacco, pit1 is induced by aluminum toxicity and Pi deficiency, but in tomato, we found that this clone was only up-regulated by nitrate resupply. The homology between the tobacco pit1 and the tomato clone was only observed between the first one-half of the tomato cDNA and 80% of pit1 sequence, but we are interested in the function of this gene because it was highly abundant in our population of nitrate-inducible genes (14 clones among the 119 responding to nitrate). It was also found that a non-symbiotic hemoglobin gene, LeHB1, which exhibits more sequence similarity to Arabidopsis AHB1, was up-regulated by nitrate exposure. In Arabidopsis, Wang et al. (2000) found that AHB1 and not AHB2 was up-regulated by nitrate exposure. We also have found that other changes in plant mineral status, including P, K, and Fe deficiency, rapidly up-regulated expression of this gene that is detailed by Y.-H. Wang, L.V. Kochian, and D.F. Garvin (unpublished data).
Figure 8.
Changes in expression of homologs of the soybean Cim1 and tobacco pit1 genes in response to nitrate exposure for 1 to 96 h. pit1 was induced specifically by nitrate exposure and not by exposure to deficiencies of other mineral nutrients. A, RNA gel blot. B, Quantitative expression data for the RNA gel blot depicted in A.
DISCUSSION
In this study, we used array analysis to screen cDNA libraries enriched for genes responsive to changes in mineral nutrient status to successfully identify new genes induced by root nitrate exposure. Fourteen different novel nitrate-induced genes were identified, along with a set of genes previously reported to be induced by nitrate resupply. A number of putative orthologs of genes recently identified as responsive to nitrate resupply in Arabidopsis (Wang et al., 2000; including ferredoxin, ferredoxin-NADP oxidoreductase, Asn synthetase, and malate dehydrogenase) were also identified in this study, indicative of a general role of these genes in plant nitrate metabolism. We initially identified candidate nitrate-inducible genes visually, based on intensity of spots on the autoradiographs from arrayed membranes following hybridization of 32P-labeled mRNA probes to arrays containing 1,280 clones. The nitrate-inducible expression of selected genes subsequently was verified using RNA gel-blot assays.
The 14 new genes that were induced by nitrate were based on both cDNA array and RNA gel-blot assays, which have not been previously reported to be nitrate inducible. These genes can be loosely organized into several categories, including water transporters (aquaporins), mineral ion transporters (other than nitrate transporters), regulatory genes, stress response genes, ribosomal proteins, and genes with unknown functions. For a number of these genes, a possible role in nitrate assimilation and nutrition can be logically hypothesized; however, for other nitrate-inducible genes such as the pit1 homolog and stress genes, their function in this complex nutritional response is not obvious and will require more detailed investigation. The identification of new genes that could play a role in nitrate assimilation via pathways and processes that are not intuitively obvious based on the function of the gene product points out the major strength of a genomics-based analysis to understanding complex physiological processes at the molecular level.
One of the interesting features of this set of nitrate-induced genes is the large number of transport genes that were induced by nitrate exposure. Nitrate exposure increased expression of membrane proteins involved in both water transport (aquaporins), as well as the ammonium transporter LeAMT2, the Pi transporter LePT2 (Liu et al., 1998), and a root hair K+ channel homologous to the carrot Kdc1 gene (Downey et al., 2000), all of which are believed to play major roles in root mineral acquisition. This finding points to the possibility of an integrated aspect to root mineral acquisition, particularly for an abundantly required macronutrient such as N. It is not surprising that increased nitrate influx is associated with enhanced K+ transporter expression because it has been previously documented that stimulated nitrate provision and uptake is associated with increased K+ influx, which functions as the major charge-balancing cation (Marschner, 1995). It is presumable that the enhancement of Pi transporter expression is a more general response associated with increased plant metabolism and protein synthesis upon N resupply. There are observations in the literature from whole plant studies that support the general idea that N, P, and K nutrition are linked. For instance, field experiments have shown that cotton plants with adequate K fertilization had increased efficiency of N fertilizer use and N utilization (Pettigrew and Meredith, 1997). Furthermore, it has been reported that P deficiency led to severe inhibition of nitrate uptake, nitrate transport into the xylem, and nitrate reduction in castor bean roots (Jeschke et al., 1997), and decreases in plant N and K accumulation (Soraya et al., 1996). Finally, when N-starved barley plants were transferred to solutions containing 0.2 mm KNO3, K+ uptake was increased immediately; in contrast, uptake of nitrate increased more slowly, matching the rate of K+ uptake after 4 h (Tischner, 1990).
The induction of a root ammonium transporter, LeAMT2, by nitrate exposure seen in Figure 4 was similar to that recently reported by von Wirén et al. (2000) for LeAMT1.2 (which was not present in our mineral nutrition array). This is in contrast to LeAMT1.1 and its Arabidopsis homolog, AtAMT1.1, where expression of the gene encoding this protein was repressed by nitrate exposure and induced by ammonium treatment (von Wirén et al., 2000; Wang et al., 2000). von Wirén and colleagues speculated that nitrate induction of LeAMT1.2, a root-hair localized transporter, may play a role in retrieval of NH4+ lost from the roots during nitrate assimilation, thus compensating for the root NH4+ efflux usually seen during nitrate assimilation. The increased expression of water channel genes may be due to the fact that nitrate resupply should trigger large accumulations of nitrate and K+ from the nutrient solution, facilitating a significant increase in osmotically active solutes in the symplasm and large increases in water influx and turgor pressure. Thus, the nitrate induction of a large number of water channel genes presumably is to mediate and regulate the increased water influx into root cells.
Because nitrate exposure increased expression of Pi and K transporters, suggesting some common features in the regulation of N, P, and K nutrition, we also looked at the influence of the imposition of −P and −K conditions on expression of the nitrate transporters, LeNRT1.2 and LeNRT2.1. For both transporters, exposure of tomato roots to solutions lacking either P or K rapidly (within 1–3 h) and significantly increased the expression of the two nitrate transporters (Figs. 5 and 6). We also have found, in a companion study, that imposition of P or K deficiency also up-regulated Pi or K transporters within the same rapid time frame (Y.-H. Wang, D.F. Garvin, and L.V. Kochian, unpublished data). These findings yield two interesting observations. First, there appears to be “cross talk” at molecular level in the plant's systems for sensing changes in N, P, and K status that previously have not been recognized. At this time, it is difficult to provide a cogent explanation for the interrelationships between changes in plant status for these three essential nutrients and regulation of N, P, and K transporters, and this may be a focus of future research. Second, we were surprised to find that nitrate transporter gene expression responded so rapidly (within a few hours) to the withholding of P or K from the nutrient solution. It has been shown previously that expression of root Pi transporters and certain root K+ transporters is increased upon the imposition of −P or −K conditions, but these studies usually focus on gene expression changes after longer periods of nutrient deprivation (earliest time point of 12 h; Liu et al., 1998; Wang et al., 1998). The general assumption is that after the withholding of a nutrient such as P or K from the plant roots, the decline in mineral nutrient concentration in either the root and/or leaf symplasm is the signal triggering increased transporter expression. The findings presented here, suggests that plants respond very rapidly to the lack of a mineral nutrient in the solution bathing the root. This raises the possibility that like nitrate, roots may be able to “sense” changes in external availability of other mineral nutrients via currently unexplained mechanisms.
The concept that plants sense either their mineral nutrient status or external nutrient availability and have signaling and regulatory pathways to link plant nutrient status to nutrient assimilatory pathways is intriguing and is causing researchers to look for signaling and regulatory genes involved in mineral nutrient acquisition/assimilation. In this study, we found that a cim1 homolog was induced 4- to 5-fold by nitrate resupply in the root (Fig. 8). cim1 was found previously to be induced by cytokinin exposure 20- to 60-fold in cytokinin-starved soybean suspension cells (Downes and Crowell, 1998). Cytokinin has been suggested as signaling molecule for sensing N availability (Sakakibara et al., 2000); thus, these findings provide further circumstantial evidence for an involvement of cytokinins in N nutrition signaling.
One potentially interesting nitrate-inducible gene we identified was a Leu zipper transcription factor (Fig. 3) that was also found to respond to P, K, and Fe deficiencies in tomato roots (Y.-H. Wang, D.F. Garvin, and L.V. Kochian, unpublished data), and was highly homologous to the tobacco TGA1a transcription factor. TGA1a has been found to interact with PR-1, a group of pathogenesis-related proteins strongly induced in plants by pathogen attack, salicylic acid, developmental cues, and nutrient status changes (Strompen et al., 1998). In that study, TGA1a was shown to interact with as-l-like cis elements in the promoter region of the PR-1a gene. This suggests that PR-1a is controlled by an as-1-like sequence motif that also interacts with the transcription factor, TGA1a (Strompen et al., 1998). Although we have identified a similar pathogenesis-related protein, PR-1b, which was induced by nitrate in our study, we do not know if its promoter also contains an as-1-like sequence. Further experiments are under way to identify genes and DNA sequences that interact with this transcription factor.
Other transcription factors induced by nitrate that have recently been identified include the Arabidopsis MADS-box transcription factor, ANR1 (Zhang and Forde, 1998). In that study, ANR1 did not respond to P and K resupply and its response to −P or −K conditions was not studied (Zhang and Forde, 1998). Our tomato cDNA library contained a homolog of ANR1, but it was not induced by nitrate exposure in tomato roots, which was also the case in the study by Wang et al. (2000) in Arabidopsis. In that study, Wang et al. (2000) also identified an MYB transcription factor that was nitrate induced.
We have noted that some of genes induced by nitrate were actually stress response genes. These genes have been shown to be induced by a number of abiotic and biotic stresses and code for PR proteins, LEA proteins, trypsin inhibitors, hin1, and glutathione-S-transferase. Because crop plants in the field are never exposed to the abrupt changes in nitrate supply used in our experiments, it is possible that the plant is perceiving the rapid resupply of nitrate as some type of stress. One possibility is that rapid nitrate resupply causes a transiently increased production of nitrite, which can be toxic to plants. It alternatively could be that nitrate is directly inducing expression of these genes, which then play a currently unknown role in nitrate nutrition.
The possibility exists that at least some of the genes induced in this study were influenced by simple growth requirements of the plant that were promoted by N provision to plants that have been withheld N for 48 h, and not due to nitrate exposure per se. However, we feel it is unlikely that the many varied responses we found after nitrate resupply were merely due to a general response to N provision because it is highly unlikely that tomato plants grown on 2 mm nitrate and 0.2 mm ammonium for 5 weeks would experience a significant drop in total N status during a 48-h period of exposure to −N conditions. This is supported by the observations that there were no visual symptoms of N deficiency, no decrease in shoot growth, and no change in total shoot N content after the 48-h period of N deprivation.
Also, it is possible that genes found up-regulated after nitrate resupply in this study may also be inducible by other environmental stimuli, such as light and circadian responses. Based on transcript profiling with Arabidopsis microarrays, it has been shown recently that expression of a number of different genes can be influenced by circadian rhythms, including genes involved in N, S, and K nutrition (Harmer et al., 2000). In this study, the authors found that expression of both nitrate and ammonium transporters exhibited a circadian response. However, the patterns of nitrate and ammonium transporter gene expression influenced by the circadian clock were completely different than those found for nitrate resupply in our investigation. In the Harmer et al. (2000) study, these transporters showed maximal increases of gene expression of 1.5- to 2-fold approximately 20 h into the 24-h subjective day, for plants initially grown on a 12-h day-12-h night cycle and then transferred to constant light for the microarray experiments. For our experiments, we always initiated the withholding of N and the subsequent nitrate resupply at 9 am, which was 3 h into the light period. Thus, plant tissue was harvested at 3, 4, 9, and 15 h into the 16-h light period for the 0-, 1-, 6-, and 12-h time points, and then 3 h into the subsequent light periods for days 2 and 3 for the 24- and 48-h time points. This was done to minimize changes in gene expression due to either light versus dark exposure or circadian regulated responses. For the tomato nitrate transporters, we saw a 10- to 16-fold increase in expression at the 1-, 6-, 12-, and 24-h time points, and then gene expression declined dramatically after 24 and 48 h of nitrate resupply. For the ammonium transporter, nitrate resupply increased gene expression 2.5- to 3.5-fold over the first 12 h of exposure, and then transporter expression declined during the longer nitrate exposures.
In summary, subtraction approaches were used to generate a tomato cDNA library enriched in root genes up-regulated by changes in plant mineral status, including nitrate provision to nitrate-starved tomato plants. Genomic analysis using high density arrays of this library allowed us to identify 14 new nitrate-inducible genes, including water channels, root Pi and K+ transporters, genes potentially involved in transcriptional regulation, stress response genes, and genes encoding ribosomal proteins. The identification of these genes is providing new avenues of research into the molecular basis of plant mineral nutrition, including the possible linkages and networking between regulation of N, P, and K nutrition, processes and mechanisms by which plants sense and respond to changes in plant mineral status, and the connections between mineral ion transporters and plant metabolic pathways.
MATERIALS AND METHODS
Plant Materials
Tomato (Lycopersicon esculentum) plants were grown hydroponically in black plastic pots containing 2 L of modified one-fifth Johnson's solution (control), which consists of the following macronutrients: KNO3, 1.2 mm; Ca(NO3)2, 0.8 mm; NH4H2PO4, 0.2 mm; and MgSO4, 0.2 mm; and the following micronutrients: KCl, 50 μm; H3BO3, 12.5 μm; MnSO4, 1 μm; ZnSO4, 1 μm; CuSO4, 0.5 μm; H2MoO4, 0.1 μm; and NiSO4, 0.1 μm. The solutions were supplemented with 10 μm Fe-EDDHA. Five plants were grown in each pot in a controlled environment growth chamber with a 16-h (from 6 am–10 pm), 24°C d and an 8-h (from 10 PM-6 AM), 20°C night regime and a photon flux density of 350 μmol m−2 s−1. Modest aeration was provided. The nutrient solution was replaced with fresh solution after weeks 2, 3, and 4, and then every 2nd d after that.
After 5 weeks of growth, N was withheld for 48 h from the nutrient solution by replacing the 1.2 mm KNO3 and 0.8 mm Ca(NO3)2 with 0.6 mm K2SO4 and 0.8 mm CaSO4. After the 48 period of exposure to −N hydroponic media, nitrate was resupplied and plants were harvested after 0, 1, 6, 12, 24, 48, and 96 h of nitrate resupply. To minimize effects due to light/dark exposure and/or circadian regulated responses, the withholding of N and subsequent resupply of nitrate were always initiated at 9 am, which was 3 h into the light period. In addition, all plant tissues for the other time points were harvested during periods of light exposure. Roots and leaves were then separated, frozen, and stored in −80°C until use.
To obtain tissue for the other nutrient deficiencies, tomato seedlings were grown under nutrient-sufficient conditions for 5 weeks as described above and then the following deficiencies were imposed: Pi, K, S, Ca, Mg, Fe, Zn, and Cu. The deficiency solutions were based on the composition of the control solution with the following substitutions. For Pi, 0.2 mm NH4H2PO4 was replaced with 0.1 mm (NH4)2SO4; for K, 1.2 mm KNO3 was replaced with 0.6 mm Ca(NO3)2 and6 50 μm KCl was replaced with 25 μm CaCl2; for S, 0.2 mm MgSO4 replaced with 0.2 mm Mg(NO3)2 and all other sulfate salts were replaced by their respective chloride salts; for Ca, 0.8 mm Ca(NO3)2 was replaced with1.6 mm KNO3; and for Mg, 0.2 mm MgSO4 was replaced by 0.2 mm CaSO4. For the withholding of micronutrients (Fe, Zn, and Cu), the specific micronutrient salt simply was not added to the nutrient solution. Roots and leaves were harvested once the shoots exhibited visual deficiencies for the specific mineral nutrient. For the Pi, K, and Fe deficiency time course experiments, tomato plants were grown as described above with the deficiency treatments initiated 3 h into the light period and roots and shoots were harvested at the same time points as for the nitrate resupply experiments.
Construction of Subtractive cDNA Library
Two subtractions were performed. The first was between nitrate-induced and N-starved plants and the second was between pooled mineral-deficient and control plants. Root tissue mRNA was isolated using the Poly(A+) Pure Kit (Ambion, Austin, TX) following the protocols provided by the manufacturer (http://www.ambion.com/techlib/prot/bp_ppure.pdf). To provide a population enriched in nitrate-induced transcripts for the nitrate resupply library, the tester mRNA was isolated from roots resupplied with nitrate, whereas the driver mRNA was obtained from roots of plants grown without N for 48 h. For the second subtraction, the tester mRNA was isolated from roots of plants exposed to the specific mineral deficiencies, whereas driver mRNA was isolated from roots of nutrient-sufficient plants. First strand cDNA was synthesized from the tester mRNA using the SMART cDNA construction kit (CLONTECH, Palo Alto, CA; http://www.clontech.com/smart/pdf/SMARTBR.pdf), resulting in single-stranded, first strand cDNAs that contain a poly(A+) tail and a unique primer sequence incorporated at the 3′ end. Subtraction was then performed using the Subtractor kit (Invitrogen, Carlsbad, CA; http://www.invitrogen.com/pdf_manuals/sub_man.pdf). The residual first strand tester cDNA that did not formed duplexes with the driver mRNA was purified and second strand cDNA synthesis was completed with components of the CLONTECH kit by taking advantage of the presence of the unique primer sequences at both ends of these cDNAs for PCR amplification.
The resulting double-stranded cDNA was then digested with the restriction enzyme, SfiI, which produced SfiIA and SfiIB sites in both ends that permitted unidirectional insertion into vector. The vector used was pBluescript (Stratagene, La Jolla, CA) modified to harbor the two SfiI sties for unidirectional cloning. After transformation, colonies were picked into 384-well plates. A total of 3,456 clones were picked from the nitrate resupply library, and 10,368 clones were picked from the mineral deficiency subtraction library. For library screening, the combined library was transferred onto nylon membranes for hybridization using a 384-pin hand-held replicator (Nalge, Naperville, IL).
Sequence Identification of cDNA Clones
The arrayed mineral nutrient-related subtractive cDNA libraries were screened using hybridization techniques described in the next section. mRNA from roots of nitrate-exposed and mineral-deficient (−N, −P, −K, −Fe, and −Zn) plants was used to make 32P-labeled single stranded cDNA probes for hybridization to the arrayed libraries. From this initial screen, we identified 12,10 unsequenced cDNA clones that exhibited strong changes in expression due to the different manipulations of plant mineral status employed in this project. Of these 1,210 clones, 500 were sequenced using the Cornell DNA Sequencing Facility and 99 of the sequenced cDNAs were found to be highly similar (E value < 10−30) to known genes and were included on our 1,280-clone array. The other 610 unsequenced clones from the initial pool of 1,210 were also included on the final array. During the subsequent nitrate-induced expression profiling of the 1,280-clone array, the cDNAs in this subset of 610 unsequenced clones that exhibited significant changes in nitrate-induced expression were then sequenced and studied further.
Root cDNA libraries previously had also been constructed from tomato roots at different developmental stages and from roots of tomato plants where a number of mineral deficiencies were imposed (R.S. van der Hoeven, D. Garvin, A.L. Matern, I.L. Holt, F. Lian, J. Upton, T. Hansen, M.B. Craven, C.L. Bowman, S. Ahn et al., unpublished data). Approximately 6,000 clones from these libraries were sequenced by the Institute for Genomic Research and made available to us through a collaboration with Dr. Steven Tanksley (Cornell University). The DNA sequences for these clones were searched against GenBank using the BlastX algorithm (Altschul et al., 1997) for similarity to other potentially interesting genes associated with mineral nutrition, including mineral ion transporters, mineral nutrient assimilation genes, signaling genes such as kinases and phosphatases, and genes induced by abiotic stresses. As a consequence, 500 clones were selected based on sequence similarity to known mineral nutrition-related genes (E value < 10−30) for arraying. Finally, six additional MAP kinase clones were obtained from the Clemson University Genomics Institute (Clemson, SC) and one MAP kinase kinase (MEK1) was kindly provided by Dr. Donald Grieson (University of Nottingham, Loughborough, UK). All of these cDNAs from different sources were then spotted on a nylon filter to make up the final 1,280 cDNA mineral nutrition-related array.
Construction and Analysis of cDNA Arrays
The above 1,280 selected clones were amplified with Tfl DNA polymerase (Epicentre, Madison, WI). The 50-μL PCR reaction mix contained 2.5 μL of 20× buffer (supplied by the manufacturer), 2 units of Tfl polymerase, 0.2 mm each of T3 and T7 primers, 2.5 mm MgCl2, and 0.25 mm dNTPs. A portion of the individual colonies from the library plates was transferred into the reaction mix and amplified. The reaction ran for 45 cycles using the following temperature profile: 94°C for 2 min, 55°C for 1.5 min, and 72°C for 2 min. After the cycling, the temperature was maintained at 72°C for 10 min. Two microliters of the PCR reaction was run on 1% (w/v) agarose gel to ensure the proper amplification of a single product. The PCR products were arrayed directly on nylon membrane using a hand-held 0.5-μL 384 Floating Slot Pin Multi-Blot Replicator (V&P Scientific, San Diego). After spotting, the membrane was laid over filter paper saturated with 0.4 m NaOH for 7 min followed by 7 min on 2× SSC saturated paper.
mRNA was directly isolated from root tissue using the Poly(A+) Pure kit (Ambion). One gram of tissue was ground in liquid N to fine powder before 10 mL of the lysis solution was added. Two volumes of the diluent solution were then added to the mixture and centrifuged for 15 min at 6,000g in a 50-mL tube. The supernatant was mixed with 100 mg of oligo-dT cellulose and incubated for 90 min at room temperature with slow shaking. The mRNA-bound oligo-dT cellulose was washed three times each with binding and washing buffers. The mRNA-bound oligo-dT cellulose subsequently was transferred to a spin column and washed twice with washing buffer. Then, mRNA was eluted with elution solution and precipitated by adding 0.1 volume of 5 m sodium acetate, 2 μL of glycogen, and 2.5 volumes ethanol and frozen in dry ice at −80C for 30 min. Integrity of the sample was checked on a 1% (w/v) agarose gel and the concentration measured by spectrophotometer.
To generate probes to screen for nitrate induced genes on the array, root mRNA from each nitrate resupply time point was labeled with α-32P dCTP according to the following protocol. Approximately 2 μg of mRNA was mixed with 2 μg of oligo-dT primer and heated at 70°C for 10 min. The mixture was put on ice immediately and the following components were added: 6 μL of 5× SuperScript II first strand buffer, 1 μL each of 25 mm dNTPs (minus dCTP), RNase inhibitor and SuperScript II reverse transcriptase (Life Technologies, Rockville, MD), and finally 10 μL of α-32P dCTP (New England Nuclear, Boston; 3,000 mmol specific activity). The reaction was carried out at 42°C for 1 h and labeled probes purified using a Sephadex G50 column. Purified probe was denatured at 100°C for 10 min and hybridized to the array membrane overnight using PerfectHyb buffer (Sigma, St. Louis) at 65°C for overnight. The array membrane was washed with 1× and then 0.2× SSC each for 20 min before being exposed to film. Array results from each time point were visually compared with the control results (0-h exposure to nitrate).
RNA gel-blot analysis was performed for selected clones using the NorthernMax kit (Ambion) with α-tubulin as a loading control. RNA samples were run on 1% (w/v) agarose gel using glaxoal buffer after incubating at 50°C for 30 min. The gel was then photographed and transferred onto a nylon membrane. Probes were generated from PCR products that had been purified from agarose gels with QiaEx II (Qiagen, Valencia, CA). The cDNA probes were labeled with α-32P dATP (New England Nuclear) using a Strip-EZ DNA kit (Ambion). Hybridization was performed at 50°C overnight and membranes were washed at 50°C and 65°C in 0.2× SSC for 30 min each before exposing to film. Hybridized membranes were stripped according to Strip-EZ DNA protocol for reuse after autoradiography. Autoradiographs were scanned, and the gel images digitized and quantified using the National Institutes of Health Image program. Gene expression was quantified after adjusting the gel blot image intensities for variations in loading (using the loading control) by determining the relative signal ratio. This was done by dividing the gel blot intensity for each nitrate exposure time point by the intensity of the gel blot for the zero time point (determined from the tissue of seedlings grown without N for 48 h).
Supplementary Material
ACKNOWLEDGMENTS
We thank Holly Manslank and Jon Shaff (U.S. Plant Soil and Nutrition Lab, Ithaca, NY) for their expertise in setting up and maintaining the tomato hydroponic culture system. We also want to note that 6,000 tomato root cDNA clones and sequence information for these clones were provided via a collaborative effort with Dr. Steven Tanksley as part of his National Science Foundation-supported tomato genome project. One of the MEK1 clones was kindly provided by Dr. Donald Grieson (pLeMEK1). Six additional MAP kinase-related cDNA clones were provided by the Clemson University Genomics Institute.
Footnotes
This work was supported by the Agricultural Research Service Agricultural Genome Program (funding to L.V.K. and D.F.G.).
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
LITERATURE CITED
- Altschul SF, Madden TL, Schäffer A, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H-J, Sehnke PC, Ferl RJ. The 14-3-3 proteins: cellular regulators of plant metabolism. Trends Plant Sci. 1999;4:367–371. doi: 10.1016/s1360-1385(99)01462-4. [DOI] [PubMed] [Google Scholar]
- Crawford NM. Nitrate: nutrient and signal for plant growth. Plant Cell. 1995;7:859–868. doi: 10.1105/tpc.7.7.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM, Glass DMA. Molecular and physiological aspect of nitrate uptake in plants. Trends Plant Sci. 1998;3:389–395. [Google Scholar]
- Daniel-Vedele F, Filleur S, Caboche M. Nitrate transport: a key step in nitrate assimilation. Curr Opin Plant Biol. 1998;1:235–239. doi: 10.1016/s1369-5266(98)80110-6. [DOI] [PubMed] [Google Scholar]
- Downes BP, Crowell DN. Cytokinin regulates the expression of a soybean beta-expansin gene by a post-transcriptional mechanism. Plant Mol Biol. 1998;37:437–444. doi: 10.1023/a:1005920732211. [DOI] [PubMed] [Google Scholar]
- Downey P, Szabo I, Ivashikina N, Negro A, Guzzo F, Ache P, Hedrich R, Terzi M, Lo Schiavo F. Kdc1, a novel carrot root hair K+ channel: cloning, characterization and expression in mammalian cells. J Biol Chem. 2000;275:39420–39426. doi: 10.1074/jbc.M002962200. [DOI] [PubMed] [Google Scholar]
- Forde BG. Nitrate transporters in plants: structure, function and regulation. Biochim Biophys Acta. 2000;1465:219–235. doi: 10.1016/s0005-2736(00)00140-1. [DOI] [PubMed] [Google Scholar]
- Forde BG, Clarkson DT. Nitrate and ammonium nutrition of plants: physiological and molecular perspectives. Adv Bot Res. 1999;30:2–90. [Google Scholar]
- Frink CR, Waggoner PE, Ausubel JH. Nitrogen fertilizer: retrospect and prospect. Proc Natl Acad Sci USA. 1999;96:1175–1180. doi: 10.1073/pnas.96.4.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan S, Wei W, He SY. hrp gene-dependent induction of hin1: a plant gene activated rapidly by both harpins and the avrPto gene-mediated signal. Plant J. 1996;10:591–600. doi: 10.1046/j.1365-313x.1996.10040591.x. [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Hirsch RE, Sussman MR. Improving nutrient capture from soil by the genetic manipulation of crop plants. Trends Biotechnol. 1999;17:356–361. doi: 10.1016/s0167-7799(99)01332-3. [DOI] [PubMed] [Google Scholar]
- Jeschke DW, Kirkby EA, Peuke AD, Pate JS, Hartung W. Effects of P deficiency on assimilation and transport of nitrate and phosphate in intact plants of castor bean (Ricinus communis L) J Exp Bot. 1997;48:75–91. [Google Scholar]
- Johansson I, Karlsson M, Johanson U, Larsson C, Kjellbom P. The role of aquaporins in cellular and whole plant water balance. Biochim Biophys Acta. 2000;1465:324–342. doi: 10.1016/s0005-2736(00)00147-4. [DOI] [PubMed] [Google Scholar]
- Kitagiri F, Lam E, Chua NH. Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature. 1989;340:727–730. doi: 10.1038/340727a0. [DOI] [PubMed] [Google Scholar]
- Lauter F-R, Ninnemann O, Bucher M, Riesmeier JW, Frommer WB. Preferential expression of an ammonium transporter and of two putative nitrate transporters in root hairs of tomato. Proc Natl Acad Sci USA. 1996;93:8139–8144. doi: 10.1073/pnas.93.15.8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Mun JH, Kim SG. Characterization of cDNAs encoding cytoplasmic ribosomal protein L15 and L27a in petunia (Petunia hybrida): primary structures and coordinate expression. Gene. 1999;226:155–163. doi: 10.1016/s0378-1119(98)00582-4. [DOI] [PubMed] [Google Scholar]
- Linthorst HJ, Brederode FT, van der Does C, Bol JF. Tobacco proteinase inhibitor I genes are locally, but not systemically induced by stress. Plant Mol Biol. 1993;21:985–992. doi: 10.1007/BF00023597. [DOI] [PubMed] [Google Scholar]
- Liu C, Muchhal US, Uthappa M, Kononowicz AK, Raghothama KG. Tomato phosphate transporter genes are differentially regulated in plant tissues by phosphorus. Plant Physiol. 1998;116:91–99. doi: 10.1104/pp.116.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager WH. Control of ribosomal protein gene expression. Biochem Biophys Acta. 1988;949:1–15. doi: 10.1016/0167-4781(88)90048-6. [DOI] [PubMed] [Google Scholar]
- Mariaux JB, Bockel C, Salamini F, Bartels D. Desiccation- and abscisic acid-responsive genes encoding major intrinsic proteins (MIPs) from the resurrection plant Craterostigma plantagineum. Plant Mol Biol. 1998;38:1089–1099. doi: 10.1023/a:1006013130681. [DOI] [PubMed] [Google Scholar]
- Marschner H. Mineral Nutrition of Higher Plants. Ed 2. New York: Springer; 1995. [Google Scholar]
- Maurel C. Aquaporins and water permeability of plant membranes. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:399–429. doi: 10.1146/annurev.arplant.48.1.399. [DOI] [PubMed] [Google Scholar]
- Naot D, Ben-Hayyim G, Eshdat Y, Holland D. Drought, heat and salt stress induce the expression of a citrus homologue of an atypical late-embryogenesis Lea5 gene. Plant Mol Biol. 1995;27:619–622. doi: 10.1007/BF00019327. [DOI] [PubMed] [Google Scholar]
- Pettigrew WT, Meredith WR., Jr Dry matter production, nutrient uptake, and growth of cotton as affected by potassium fertilization. J Plant Nutr. 1997;20:531–548. [Google Scholar]
- Sakakibara H, Taniguchi M, Sugiyama T. His-Asp phosphorelay signaling: a communication avenue between plants and their environment. Plant Mol Biol. 2000;42:273–278. doi: 10.1023/a:1006334926388. [DOI] [PubMed] [Google Scholar]
- Soraya R, Takuji O, Takamitsu K, Taro I. Deficiency of N, P, K, Ca, Mg, or Fe mineral nutrients in Narcissus cv “Garden Giant.”. Soil Sci Plant Nutr. 1996;42:809–820. [Google Scholar]
- Stitt M. Nitrate regulation of metabolism and growth. Curr Opin Plant Biol. 1999;2:178–186. doi: 10.1016/S1369-5266(99)80033-8. [DOI] [PubMed] [Google Scholar]
- Strompen G, Gruner R, Pfitzner UM. An as-1-like motif controls the level of expression of the gene for the pathogenesis-related protein 1a from tobacco. Plant Mol Biol. 1998;37:871–883. doi: 10.1023/a:1006003916284. [DOI] [PubMed] [Google Scholar]
- Tischner R. New regulatory steps in nitrate assimilation of lower and higher plants. In: Ullrich WR, Rigano C, Fuggi A, Aparicio PJ, editors. Inorganic Nitrogen in Plants and Microorganisms. New York: Springer-Verlag; 1990. pp. 51–53. [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- Vidmar JJ, Zhuo D, Sddiqi MY, Glass ADM. Isolation and characterization of HvNRT23 and HvNRT24 cDNAs encoding high-affinity nitrate transporters fromm roots of barley. Plant Physiol. 2000;122:783–792. doi: 10.1104/pp.122.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wirén N, Gazzarrini S, Frommer WB. Regulation of mineral nitrogen uptake in plants. Plant and Soil. 1997;196:191–199. [Google Scholar]
- von Wirén N, Lauter F, Ninnemann O, Gillissen B, Walch-Liu P, Engels C, Jost W, Frommer WB. Differential regulation of three functional ammonium transporter genes by nitrogen in root hairs and by light in leaves of tomato. Plant J. 2000;21:167–175. doi: 10.1046/j.1365-313x.2000.00665.x. [DOI] [PubMed] [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM. Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell. 2000;12:1491–1509. doi: 10.1105/tpc.12.8.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T-B, Gassmann W, Rubio F, Schroeder JI, Glass ADM. Rapid upregulation of HKT1, a high-affinity potassium transporter gene, in roots of barley and wheat following withdrawal of potassium. Plant Physiol. 1998;118:651–659. doi: 10.1104/pp.118.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda E, Ebinuma H, Wabiko H. A novel glycine-rich/hydrophobic 16 kDa polypeptide gene from tobacco: similarity to proline-rich protein genes and its wound-inducible and developmentally regulated expression. Plant Mol Biol. 1997;33:667–678. doi: 10.1023/a:1005714119561. [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998;279:407–409. doi: 10.1126/science.279.5349.407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.