Abstract
The ABC superfamily of genes is one of the largest in the genomes of both bacteria and eukaryotes. The proteins encoded by these genes all carry a characteristic 200- to 250-amino-acid ATP-binding cassette that gives them their family name. In bacteria they are mostly involved in nutrient import, while in eukaryotes many are involved in export. Seven different families have been defined in eukaryotes based on sequence homology, domain topology, and function. While only 6 ABC genes in Dictyostelium discoideum have been studied in detail previously, sequences from the well-advanced Dictyostelium genome project have allowed us to recognize 68 members of this superfamily. They have been classified and compared to animal, plant, and fungal orthologs in order to gain some insight into the evolution of this superfamily. It appears that many of the genes inferred to have been present in the ancestor of the crown organisms duplicated extensively in some but not all phyla, while others were lost in one lineage or the other.
The common progenitor of plants, animals, fungi, and Dictyostelium discoideum appears to have inherited a large number of genes for transmembrane transporters with related ATP-binding domains from the bacterial ancestor. While bacteria use ABC (ATP-binding cassette) transporters for both import and export, eukaryotes are thought to use them only for export (30). Some of the inherited genes were retained and expanded in one phylum or another, while others were lost. ABC transporters are composed of two copies of the ABC domain with the conserved sequence LSGG recognizable between the Walker A and B motifs of the ATP-binding site (38) and two copies of a transmembrane (TM) domain, usually consisting of six TM helixes (18). These four domains may be present in a single polypeptide or may come from different proteins which associate to form a functional transporter. Many types of molecules, including small ions, lipids, and polypeptides, are transported by ABC transporters (20, 41). Eukaryotic ABCs were initially identified as the agents giving rise to multiple drug resistance of neoplastic cells, where they rapidly export compounds used in chemotherapy (14). Since these drugs are unlikely to be encountered naturally, the physiologically relevant functions of ABC transporters are mostly unknown. The ABC family also includes a few genes that carry the ABC domain but have lost the TM portion; their products no longer function as transporters but are likely to carry out other conserved functions.
Eukaryotic ABC genes have been classified in seven families, from ABCA to ABCG, based on gene organization and primary sequence homology (20). This classification was established to simplify the naming and identification of the ABC genes, since some had more than one name or had confusing names. At least 68 members of the ABC family can be recognized in the genomic sequences of Dictyostelium discoideum. The predicted products of these genes share a conserved ABC domain of about 200 amino acid residues, which includes an ATP-binding site. Most of these proteins also carry one or more TM domains either N-terminal or C-terminal to the ABC domain, and this topology has been used in their classification. We have adopted the recently revised nomenclature for human ABC families to facilitate comparisons. We have clustered these genes on the basis of their ABC domains as well as their full sequences, and we find that they form a robust tree. Detailed sequence comparisons have allowed additional grouping within each family and given some insight on the evolutionary history of these genes.
MATERIALS AND METHODS
Database searches and sequence analysis.
Although the complete sequences of the six Dictyostelium chromosomes have not yet been assembled, ongoing efforts by the Dictyostelium genome sequencing consortium have generated more than eightfold coverage of protein coding regions, and more than 30,000 cDNA reads have been generated by the Japanese project. The raw sequences are accessible through the links on a website containing supplementary materials related to this report (http://www.biology.ucsd.edu/labs/loomis/ABCwebsite/abcfamily.html). With this level of coverage, a given gene has more than a 95% chance of being represented. To identify sequences encoding one or more ABC domains, we performed a series of TBlastN searches of the Dictyostelium database using ABC proteins representative of each eukaryotic family. Besides the raw shotgun sequences, the database includes all Dictyostelium sequences deposited in GenBank, cDNAs from the Japanese EST project, and preliminary contigs generated by the Sanger Centre and the German Sequencing Group at Jena. Initially, 55 contigs were found to contain genes with ABC domains. Contigs were extended whenever possible from individual reads. A second series of Blast searches was performed using the consensus ABC sequence from pfam005 (see the website) using a cutoff of 10. In this way we were able to assemble a total of 68 genes and 2 pseudogenes (Table 1).
TABLE 1.
Numbers of genes in the ABC families
| Family | No. of ABC genesa in:
|
|||
|---|---|---|---|---|
| Dictyostelium discoideum | Saccharomyces cerevisiae | Homo sapiens | Arabidopsis thaliana | |
| ABCA | ||||
| Full | 7 (1 cluster) | None | 12 | 1 |
| Half | 5 (2 clusters) | None | None | 16 |
| ABCB | ||||
| Full | 2 (1 cluster) | 1 | 4 | 22 |
| Half | 7 (4 clusters) | 3 | 7 | 5 |
| ABCC | 14 (3 clusters) | 7 | 12 | 15 |
| ABCD | ||||
| Full | None | None | None | 1 |
| Half | 3 (2-3 clusters) | 2 | 4 | 1 |
| ABCE | 1 (1 cluster) | 1 | 1 | 2 |
| ABCF | 4 (3 clusters) | 5 | 3 | 5 |
| ABCG | ||||
| Full | 15 (2 clusters) | 10 | None | 13 |
| Half | 6 (3 clusters) | 1b | 5 | 29 |
| Otherc | 4 (3 clusters) | 2 | 2 | 16 |
| Total | 68 (25 clusters) | 32 | 50 | 127 |
Numbers of independent clusters are deduced from full sequence trees for each family. Numbers of genes in S. cerevisiae, H. sapiens, and A. thaliana are based on previous analyses of these organisms (3, 11, 29).
This protein corresponds to a half-transporter plus part of a TM domain.
ArsA homologs are included, but the SMC group is not.
Some of the gene sequences are incomplete, and in the case of ABCD.3, part of the ABC domain is missing. ABCA.10 and ABCA.11 may be two portions of the same gene, but the intervening sequence was not found. Each encodes a single ABC domain and a TM domain with sequences most closely related to those in the A family. However, at present they are considered separately (see below). The ABCA.12 partial sequence was too short to be used for proper analysis.
Comparisons of the ABC genes to homologs present in completed genomes of other eukaryotes were made by using BlastP on the National Center for Biotechnology Information (NCBI) website (with the filter option “Mask for lookup table only” to mask repetitive amino acid sequences) for seed recognition, followed by alignment of unmasked sequences.
Building trees.
Homologs of the Dictyostelium ABC proteins were identified in GenBank by multiple Blast searches. The closest homologs of the Dictyostelium members of each family from various species were aligned by using ClustalW as part of the MacVector program. Alignments were inspected for obvious errors, which were corrected manually. Trees were initially built by using the MacVector program for neighbor joining to obtain the best tree (28). The robustness of the trees was determined by bootstrap analysis with 1,000 replications (13). Bootstrap trees are presented with midpoint rooting.
RESULTS AND DISCUSSION
Classification of Dictyostelium ABC proteins.
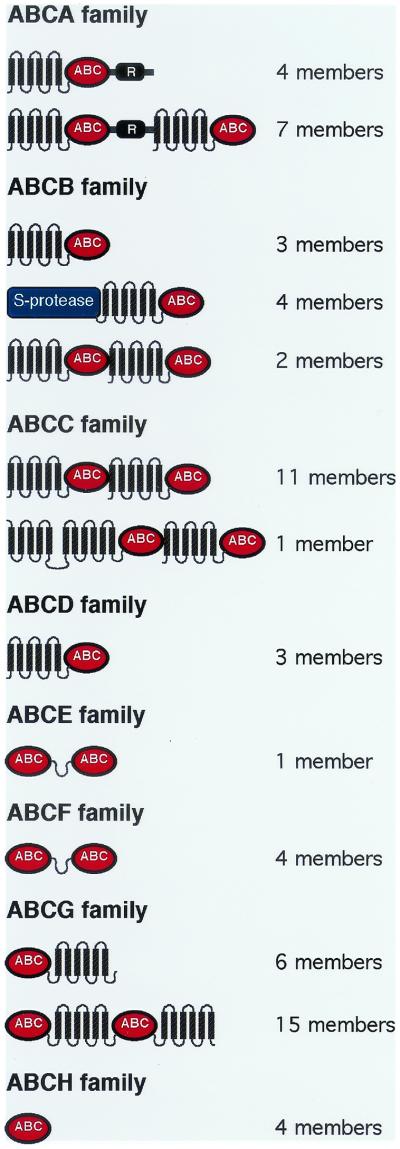
The 68 putative ABC proteins were compared both with each other and with sequences from other organisms. A simple Blast score obtained by comparison with protein sequences in GenBank was sufficient to sort the majority of the protein sequences into the seven major eukaryotic families, while a few sequences appeared more related to bacterial proteins. The positions of ABC and TM domains were identified in each coding sequence, allowing us to assign the genes to specific family groups (Fig. 1). Dictyostelium ABC genes follow the expected organization for each family, even though some additional subtypes were found. Those in which the TM domain is predicted to be N-terminal to the ABC domain fall into families A to D, while those in which the ABC domain precedes the TM domain are members of the ABCG family (Fig. 1). The ABC genes which do not encode a TM domain belong to the E, H, and F families. The H family includes members which do not fit into the seven standard families found in eukaryotes. While these genes might not be closely related to each other, they all share similar characteristics: a single ABC domain, absence of a TM domain, and a close homology to bacterial ABCs. Sequence comparisons with previously assigned ABCs of other organisms supported our assignments of the Dictyostelium ABC proteins to the seven major eukaryotic families (see below).
FIG. 1.
Domain arrangements in the ABC families. TM domains are represented by six connected hatched segments. Red ovals, ABCs; R, phosphorylatable regulatory region of ABCA members. The order of the domains in the various proteins is given, but the intervening distances are not drawn to scale. These proteins range from 300 to 2,000 amino acids.
The ABC domains were compared with each other in order to further refine the families. ABC domains were delineated by using the ABC sequence of pfam005 as a guide (see the website). Many proteins contain two ABC domains, numbered 1 and 2, resulting in a total of 103 complete ABC domains. Some proteins appear to contain a third, degenerate ABC domain of unknown function, which was not included in these analyses.
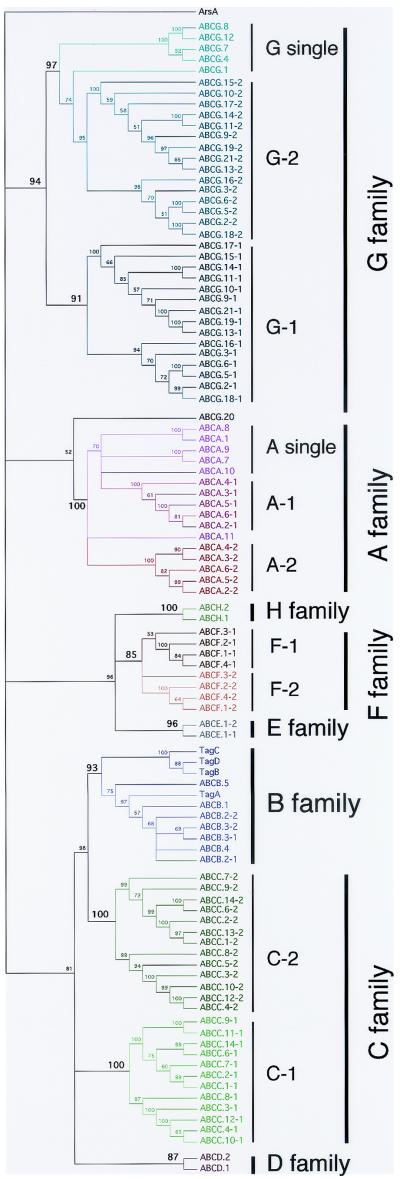
With a single exception, the individual ABC domains sorted out according to the known families (Fig. 2). It appears that the sequence of the ABC domain alone is sufficient to identify the family of a given eukaryotic ABC. Within families A, C, F, and G, the first and second ABC domains cluster together within their family but separately from each other. Very similar trees were obtained by using the ABC domain sequences from human and Drosophila ABCs, with the same subdivision between first and second ABC domains (11). If these genes arose by duplication and fusion of a region encoding a progenitor ABC domain and a TM domain, this must have happened long ago in the common progenitor of the crown organisms, since genes encoding two distinct ABC domains are found in all the eukaryotes. The ABCA and ABCG family proteins include proteins with a single ABC domain (half-transporters) as well as those with two ABC domains (full transporters). In both cases, the ABC domains from half-transporters cluster together, indicating that they share some common features that distinguish them from the ABC domains found in full transporters.
FIG.2.
Clustering of the ABC domains of Dictyostelium proteins. The amino acid sequences of 103 Dictyostelium ABC domains (see the website) were aligned and related to each other. An unrooted tree with bootstrap values is presented, with the values for separation of the families given in boldface. With a single exception (ABCG.20), domains from members of the same family clustered together. The first and second ABC domains in full transporters are marked with a bar to the right and clustered together. The ABC domains from half-transporters found in members of the A, B, and G families also clustered together within their respective families. ABCA.10 and ABCA.11 have only a single ABC domain but may be the first and second ABC domains of a full transporter, respectively. They cluster with the ABC domains of other full transporters of this family.
The only exception was the ABC domain of ABCG.20, which sorted with the ABC domains of the A family. Nevertheless, ABCG.20 has the typical organization of the G family, with the ABC domain preceding the TM domain, and thus belongs to this family (see below).
The most divergent ABC domain is found in ArsA. Its product is related to that of a Saccharomyces cerevisiae gene involved in arsenic resistance and shares more than 55% identity with human, Arabidopsis thaliana, and Schizosaccharomyces pombe ArsA proteins. It encodes a single ABC domain lacking the conserved LSGG motif between the Walker A and B motif, as do all its orthologs. Thus, it is not surprising that the ABC domain of Dictyostelium ArsA does not cluster with any of the other ABC domains.
One putative ABC gene, ABCH.3, was not included in this analysis because a 300-amino-acid insert between the Walker A and B motifs precluded its alignment with other ABC domains.
The 68 ABCs clustered into the same families when their complete sequences were used, indicating that each family has conserved amino acid sequences in addition to the ABC domain. These whole-sequence trees are further analyzed in the sections focused on the separate families. Supplementary materials including the trees and links to the sequences are available at http://www.biology.ucsd.edu/labs/loomis/ABCwebsite/abcfamily.html.
The ABCA family of Dictyostelium.
A few years ago it was suggested that the ABCA family first arose in animals, since members of this family were found in the human, Drosophila, and Caenorhabditis elegans genomes but not in those of yeast or fungi (20). However, clear members of the ABCA family were recently found in the genome sequences of Arabidopsis, indicating that this is an ancient family that was present in the common ancestor of animals and plants (29). It appears that it was subsequently lost from some lineages. Dictyostelium has 11 or 12 members of the A family, depending on whether one counts ABCA.10 and ABCA.11 as separate genes or two halves of the same gene (Table 1). One of the defining characteristics of the A family is the presence of a regulatory domain with multiple sites for phosphorylation by various protein kinases in the region after the first ABC domain. This region is present in all of the members of the ABCA family, including those in Dictyostelium. It is not found in other ABC genes.
Searches of GenBank uncovered ABCA genes in other protists, including trypanosomes and entamoebae. In addition to full transporters, half-transporters (TM-ABC) were identified in Dictyostelium, Arabidopsis, and Entamoeba histolytica. Half-transporters of the ABCA family are absent in animals. Clustering of the ABCA proteins from Dictyostelium, humans, Arabidopsis, and the protists showed that they fall into three distinct groups (Fig. 3 A). Genes in the first group encode full transporters with two ABC domains and two TM domains. The first five genes appear to have expanded from a single precursor gene, since they cluster together, separately from the human, plant, and protist homologs. The Arabidopsis ortholog is the only full transporter of the ABCA family in this plant (29).
FIG. 3.

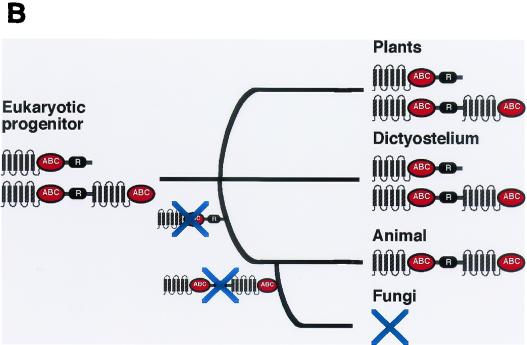
(A) Tree of the ABCA family. The complete amino acid sequences of the ABCA transporters were aligned and related to each other and homologs from other species. In this and other figures, the bootstrap values are presented for unrooted trees. Full transporters with two TM-ABC domains formed a single group that clustered with homologs in Trypanosoma cruzi (Tc), Homo sapiens (Hs), and A. thaliana (At). Two half-transporters clustered with other genes from these organisms as well as one from E. histolytica (Eh). Sequences of genes shown in this and other figures are available on the website (http://www.biology.ucsd.edu/labs/loomis/ABCwebsite/abcfamily.html).(B) Proposed order of gene loss in the ABCA family. The common ancestor of animals, plants, fungi, and Dictyostelium is presumed to have carried a gene encoding a half-transporter as well as one encoding a full transporter. The half-transporter was lost in the line leading to animals and fungi before these two kingdoms diverged. The full transporter was subsequently lost from the progenitor of fungi. Gene losses are indicated by an X.
There are multiple half-transporters of the A family in the Arabidopsis genome (29). They cluster into two groups that also contain the group 2 and group 3 genes of Dictyostelium, respectively, suggesting that both organisms inherited two genes encoding half-transporters.
The lack of ABCA half-transporters in animals and the complete lack of the family in fungi can be explained by two events in which genes were lost during the descent from a common ancestor which carried two genes encoding half-transporters and a single gene for a full transporter (Fig. 3B). All of these genes were retained and expanded in Dictyostelium and Arabidopsis, but the genes for the two half-transporters were lost in the progenitor of animals and fungi, leaving only the gene encoding a full transporter of the ABCA family. After animals and fungi diverged, the fungal ancestor lost the gene for the full transporter, although it was retained and duplicated in the animal lineage to form a large family. The alternate hypothesis of independent evolution of ABCA genes in plants, animals, protists, and Dictyostelium seems highly unlikely.
The ABCB family of Dictyostelium.
The B family includes three main subtypes: full transporters involved in multiple drug resistance (MDR), half-transporters targeted to mitochondria, and half-transporters involved in peptide transport (9, 17, 33). Dictyostelium ABCB.2 and ABCB.3 are full transporters homologous to human MDR1A (ABCB.1) protein. They cluster with similar full transporters from other organisms (see the ABCB tree on the website). Three other Dictyostelium genes of this family are most similar to mitochondrial half-transporters. Dictyostelium ABCB.1 clusters with human ABCB.10, and Dictyostelium ABCB.4 clusters with human ABCB.8. These two human proteins have been shown to be localized in the mitochondria, where they appear to form a heterodimeric full transporter (17). It is likely that the same situation occurs in Dictyostelium.
Dictyostelium ABCB.5 shares more than 50% amino acid sequence identity with human ABCB.7 and yeast ATM1, which have been shown to mediate transport of the Fe/S binding protein into mitochondria (21). Moreover, the Dictyostelium protein has a well-defined signal sequence targeting it to mitochondria. Since there is only one gene encoding such a protein in the yeast, human, Arabidopsis, and Dictyostelium genomes, there is little question that these genes are orthologous.
Four members of the ABCB family, TagA, -B, -C, and -D, are unusual in that they each have a serine protease domain N-terminal to the half-transporter (32; A. Kuspa, personal communication). Such fusion products have not been encountered in other organisms. The genes encoding TagB, -C, and -D are located next to each other on chromosome 4 and appear to have arisen by tandem duplications. They encode proteins that are more than 65% identical, with the differences located mostly in the terminal regions. In the central region, their nucleotide sequences are more than 95% identical, so that even synonymous mutations are rare. It is likely that they have recently undergone rectification. Mutations in either tagB or tagC block postaggregation morphogenesis (32) and affect the release of signaling peptides regulating terminal differentiation (2).
Although TagA carries both a half-transporter and a protease domain, it does not cluster closely with the other Tag proteins. TagA may have arisen from an independent fusion of a serine protease gene with a half-transporter gene. TagA has been implicated in the regulation of cell type proportioning in Dictyostelium (A. Kuspa, personal communication).
The ABCC family of Dictyostelium.
Transporters of the C family have been shown to transport glutathione conjugants as well as to export cadmium ions. They are always found as full transporters with two copies of the TM-ABC unit. Many of the genes of this family in humans and other animals have an additional TM domain at the N terminus (34). The Dictyostelium C family is composed of 14 members, only 1 of which, ABCC.8, has the extra TM domain. The closest homolog of ABCC.8 in humans is ABCC.2. Mutations in ABCC.2 result in the Dubin-Johnson syndrome (37). ABCC.8 is also the closest Dictyostelium homolog to the human protein CFTR. This protein has been extensively studied, since mutations in CFTR result in cystic fibrosis, one of the most common genetic diseases (10, 12, 40). CFTR acts not only as a transporter but also as a chloride channel (1). The closest homolog in yeast, Ycf1p, has been shown to be involved in cadmium resistance but is also able to transport glutathione-conjugated organic anions into vacuoles (39).
The remainder of the ABCC genes of Dictyostelium form two separate clusters, one related to the Arabidopsis gene MRP4 but to no genes found in animals or fungi and one related to Arabidopsis genes MRP.1 and MRP.2 as well as human ABCC.5 (see the ABCC tree on the website). Two of the group 1 genes, ABCC.9 and ABCC.11, encode proteins that are 92% identical to each other, suggesting that they arose from a recent duplication. It appears likely that the progenitor of the crown organisms carried two genes of the ABCC family that both expanded considerably in Dictyostelium and Arabidopsis. One of these genes, the group 1 homolog, appears to have been lost in both animals and fungi.
The ABCD family of Dictyostelium.
The ABCD family contains only half-transporters. Those that have been studied are all targeted to the peroxisome, where they regulate the transport of long-chain fatty acids (15). There are only two such proteins in yeast, three in Dictyostelium, and four in humans. Mutations in either of the yeast genes result in cells that are unable to grow on oleic acid, suggesting that they act as a heterodimer (31).
Dictyostelium ABCD.1 and ABCD.3 sequences are incomplete, but they are located near each other in the genome and may have arisen by duplication. However, they cluster separately. ABCD.1 clusters with a yeast ABCD gene, PXA2, while ABCD.3 is closer to human ABCD.4 (see the ABCD tree on the website). Dictyostelium ABCD.2 protein clusters with the human ABCD.3 protein and contains the motif for peroxisome localization. Thus, ABCD.2 may form a heterodimer with either ABCD.1 or ABCD.3 to transport fatty acids. ABCD.2 is the ortholog of human ABCD.3, which has been found to be mutated in Zellweger syndrome 2 (24).
The ABCE gene of Dictyostelium.
Sequenced eukaryotic genomes contain only one or at most two genes of the ABCE family. Only one has been recognized in Dictyostelium. They all have a conserved ferredoxin motif (pfam00037) at the N terminus, a motif found in nucleic acid binding proteins. They contain two ABC domains but no TM domains and so are unlikely to act as transporters. In animals, ABCE protein has been shown to inhibit RNase L, the double-stranded RNA nuclease, and is referred to as RLi (6). The Dictyostelium ABCE gene is more closely related to animal and plant homologs than to yeast genes (see the ABCE tree on the website). Less closely related homologs are also found in archaebacteria. It is unlikely that they act as RNase L inhibitors, since these bacteria do not have RNase L homologs. These genes may have retained an earlier function that has been either modified or supplanted in eukaryotes.
The ABCF family of Dictyostelium.
Like the ABCE family, the ABCF family is characterized by two ABC domains and no TM domains. One member of this family, GCN20 of yeast, has been shown to be involved in regulation of translation in amino acid-starved cells by interaction with eukaryotic initiation factor 2 (eIF2) and ribosomes (23, 36). The four Dictyostelium ABCF proteins sort into three separate clusters (see the ABCF tree on the website). ABCF.1 and ABCF.4 are the most closely related to GCN20 and so are candidates for translational regulators. ABCF.2 clusters with a yeast gene of unknown function as well as human ABCF.2. The closest homologs of ABCF.3 are found in bacterial genomes. Among eukaryotic genes it clusters with human ABCF.1, which has been shown to interact with eIF2 and ribosomes (35).
The ABCG family of Dictyostelium.
The ABCG family is characterized by the ABC domain preceding the TM domain (Fig. 1). All of the members of this family in animals are half-transporters, while all of the fungal members are full transporters with two ABC-TM units. No members of this family are found in bacteria. Dictyostelium and Arabidopsis have members of both types. Robust trees could be constructed only when half and full transporters were separately clustered.
Dictyostelium full transporters separate into two major groups (see the ABCG tree on the website). The first clusters with sequences from plants, while the second clusters with fungal sequences. Fungal ABCG proteins present an unusual variation in the Walker A consensus motif of the first (N-terminal) ABC domain (3). The highly conserved lysine in the sequence GXXXXGK(S/T) is replaced by a cysteine in the fungal ABCG protein. The Dictyostelium ABCG genes which cluster with fungal homologs also have replaced this lysine with a cysteine, suggesting that this mutation occurred in the common ancestor.
It seems likely that the ancestor of the crown organisms carried two closely related ABCG genes for full transporters, both of which were retained and amplified in Dictyostelium but only one of which was kept in plants while a different one was retained in fungi. Neither of the genes encoding full transporters were retained in the animal lineage.
Most of the ABCG half-transporters of Dictyostelium clustered together with each other, but ABCG.1 and ABCG.20 clustered with Drosophila, Arabidopsis, and human homologs (see the ABCG tree on the website). The closest homolog of ABCG.20 is the Drosophila protein CG9990, which, like the Dictyostelium protein, has the topology of the G family but clusters with the A family when the ABC superfamily is analyzed (11). It has been suggested that CG9990 and two related sequences in Drosophila should be considered a new family, since no related genes have been found in other organisms. However, since a gene with these properties is present in Dictyostelium, it does not seem sensible to make a new family.
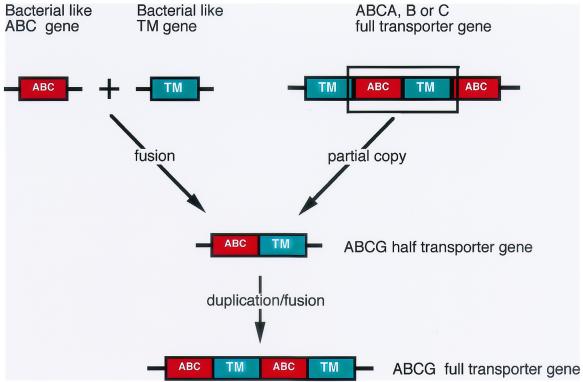
It has been assumed that the ABCG family arose from the fusion of independent ABC and TM domains, since it is the only ABC family in which the ABC domain precedes the TM domain. Alternatively, it may have arisen from the central portion of a member of the A, B, or C family that included only the first ABC domain and the second TM domain (Fig. 4). In the case of ABCG.20, CG9990, and related sequences, the second possibility appears more likely, considering the high similarity of their ABC domains with those of the A family. Tandem duplication and fusion of this gene could then have generated the full transporters of the ABCG family. The ABC domains of the G family cluster together on the branch that also carries the ABC domains of the A family (Fig. 2), making an ABCA gene the most likely source of the original ABCG gene. The ABC domains from ABCG proteins of Drosophila and humans also cluster on the same branch with the ABC domains of the A family (11).
FIG. 4.
Two possible routes by which the ABCG genes may have arisen. It has been assumed that the original gene in which the ABC domain precedes the TM domain was formed by the fusion of independent regions encoding such domains. Alternatively, a copy of the central region of a preexisting ABC gene could have generated a functional half-transporter. Tandem duplication and fusion of this gene could have generated the full transporters.
Among the full-transporters of the ABCG family, group 2 genes are surprisingly similar. Many appear to have arisen from recent duplications or replacements from other members of the family. For instance, there are 70 differences in the first 700 bases of the ABCG.19 and ABCG.13 genes. Furthermore, ABCG.19 has an additional intron in this area. However, for the next 3 kb, only 3 nucleotides differ between the nucleotide sequences of ABCG.19 and ABCG.13. At the 3′ ends of the genes, the sequences are more divergent and ABCG.19 has two introns not present in ABCG.13. It appears that a cDNA was generated from an ABCG.13 mRNA before it was processed and that this cDNA replaced the central region of ABCG.19 fairly recently. The complete ABCG.21 coding sequence is 97% identical with that of ABCG.13. One hundred forty-four of the 146 differences occur in the first 2.7 kb, while only two base pair differences can be found in the last 2 kb. The first two introns of ABCG.13 and ABCG.21 are similar but not identical, and ABCG.21 lacks the third intron present in ABCG.13 and ABCG.19. A cDNA generated from an ABCG.13 mRNA may have recently rectified the 3′ end of ABCG.21. Other members of this group seem to have been rectified in a similar manner.
Recently, a mutation affecting both ABCG.2 and ABCG.18 has been shown to impair endocytosis and affect endosomal pH (7). These genes are adjacent on a contig and were both disrupted by insertion mutagenesis. While ABCG.2 and ABCG.18 are more similar to each other than to any other ABCG (see the ABCG tree on the website), they show only 50% identity and so are unlikely to have arisen from a recent duplication. It is not yet clear whether only one of these two genes or both are required for regulation of endocytosis and endosomal pH.
Other Dictyostelium ABCs.
An ABC domain is found in the ArsA family of bacterial genes that are involved in the transport of arsenate, selenate, and other anionic compounds (9). Members of this family do not have TM domains but associate with another protein, ArsB, that has two potential TM domains. There are homologs of ArsA in plants, animals ,and fungi, although they show only about 30% identity to the bacterial gene products (4, 5, 19). Dictyostelium has a single ArsA gene with more than 55% identity with its eukaryotic counterparts. It clusters with the plant and animal homologs more closely than do the yeast homologs (see the ABCH tree on the website).
The Dictyostelium ABCH.1 and ABCH.2 genes have a single ABC domain and no TM domain. They are 45% identical to each other and share more than 40% identity with Escherichia coli ybbA, Methanococcus jannaschii glnQ, and Bacillus subtillis yvrO (see the ABCH tree on the website). All the identified ABC proteins related to ABCH.1 and ABCH.2 are importers rather than exporters (30). They cluster with ABC transporters involved in the uptake of polar amino acids. These proteins also lack TM domains and depend on a substrate binding protein as well as on a TM anchoring protein to carry out their function. There are no significant homologs to ABCH.1 and ABCH.2 in any eukaryotic genome. However, these genes do not appear to have arisen by a recent lateral transfer from a bacterium, since the Dictyostelium genes have the high A/T content typical of their genome and contain introns. It appears likely that the common eukaryotic ancestor carried either one or two of these ABC genes, but they were lost in most descendants.
Dictyostelium has another gene encoding an unusual ABC protein in which there is a 200-amino-acid insertion between the Walker A motif and the conserved LSGG sequence. The closest homolog of this gene, ABCH.3, is from the plague bacterium (Yersinia pestis) plasmid pMT1. We were unable to generate significant clusters with this gene.
Several Dictyostelium genes encode products related to the SMC family of proteins, which is involved in chromosomal maintenance and homologous recombination (16). Sequence analyses of the ATP-binding domains of the members of this family suggest that they are derived from the ABC family, but almost all have lost the ABC signature motif LSGG between the Walker A and B motifs and have acquired a conserved insert of about 800 amino acids. However, an Arabidopsis SMC family member has retained the LSGG motif (29). There are three SMC genes in the Dictyostelium genome, and like their Arabidopsis counterparts, all have retained the LSGG motif. However, the SMC family has diverged so much from the ABC family that these genes are not included in the ABC family trees or counted in the list of ABCs.
Conclusions.
Multigene families are subject to random birth-and-death processes, resulting in considerable variation in the number of genes (22, 25, 26). Duplications result in growth of the family, while deletions reduce, and can eliminate, the family. Members of multigene families gradually diverge by accumulation of point mutations and may eventually acquire different functions; alternatively, one of the copies may become a nonfunctional pseudogene and ultimately diverge so much that it cannot be recognized as belonging to the family. Members of the ABC superfamily appear to have been subject to all these forces.
Based on the number of clusters that we see for the Dictyostelium ABC genes (Table 1), the common progenitor of Dictyostelium, yeast, plants, and animals most likely had at least 25 genes encoding ABC proteins. Some encoded half-transporters, while others encoded full transporters. A few may have consisted only of the ABC domain. These founder genes gave rise to other ABC genes by duplication until the superfamily grew to have 68 members in D. discoideum. This is more than are present in either the yeast or the human genome but only half as many as are found in the plant A. thaliana.
Considering the number of related genes in the ABC superfamily, it is somewhat surprising that we were able to recognize only two pseudogenes. Regions adjacent to functional ABC genes were carefully scrutinized, but only ABCG.9 and ABCF.4 were found to have a linked pseudogene. In the ABCG.9 pseudogene, a large deletion removed the beginning of the gene and the end was found to be inverted. The nucleotide sequences of the remnants of this gene are still 95% identical to their comparable sequences in ABCG.9, suggesting that little time for random nucleotide mutations has passed since this gene became nonfunctional. The ABCF.4 pseudogene also suffered deletions while the remnants are nearly identical to the gene. It appears that the rate of deletion of dispensable regions is high in Dictyostelium, which may account for its relatively small genome size (22). Pseudogenes do not last long in this genome.
There is also clear evidence for rectification in the ABC superfamily. Reverse-transcribed copies of both processed and unprocessed mRNAs appear to have frequently replaced homologous regions in various members of the ABC families. There is no other plausible way to account for the observations that nucleotides at synonymous codon positions as well as the sequences of introns are conserved over long stretches of the genes and yet the nucleotide sequences flanking these regions show divergence levels indicating a far more ancient birth of the genes. Rectification among members of a gene family increases the degree of similarity and can lead to underestimation of the divergence time if these genes are used as a molecular clock.
Recent structural studies have indicated that active transport by ABC transporters requires that substrates enter a chamber formed by the paired TM domains in the membrane and partition into the aqueous environment following conformational changes of the transporter (8, 27). The energy of ATP binding to the ABC domains appears to provide the initial energy for translocation of the substrate across membranes. Subsequent hydrolysis of ATP and release of ADP and Pi allow the transporter to return to its original conformation by repacking the α-helices within the plane of the membrane using a combination of rotation and tilting. Although all ABC transporters are evolutionarily related, the details of their transport mechanisms may vary between members of different families.
Alignment of the proteins belonging to each of the ABC families allowed us to recognize identifier motifs that distinguish members of one family from members of all other ABC families (Table 2) (see also Fig. 8 in the supplementary figures on the website). Likewise, unique identifiers can distinguish among the different groups within a family. These short sequences are often sufficient to enable recognition of members of the family in other organisms when GenBank is queried.
TABLE 2.
Identifiers to discriminate between the different ABC families and domains
| ABC family | Sequencea | Blast searchb | IDc | Notes |
|---|---|---|---|---|
| ABCA | IGVCPQxDILW | ++ | + | DILW part is less conserved |
| DEPSTGxDPxxRR | ++ | ++ | Overlap with the Walker B motif | |
| IILTTMSMEEA | +++ | +++ | Gives a few hits with bacterial ABC | |
| ABCB | GKSTVLxLIxRFYDP | +++ | +++ | Overlap with the Walker A motif |
| ANAHEFI | ++ | +++ | Gives a few hits with bacterial ABC | |
| DEATSALDxESE | ++ | ++ | Overlap with the Walker B motif; gives a few hits with bacterial ABCs | |
| ABCC-1 | DLTEIGERGINLSGG | +++ | +++ | Overlap with the ABC signature |
| DDPLSAVDxHVGxHLF | +++ | +++ | Overlap with the Walker B motif | |
| ABCC-2 | EKIGIVGRTGAGKSS | +++ | +++ | Overlap with the Walker A motif |
| SVGQRQLLCL | +++ | +++ | Overlap with the ABC signature | |
| ABCD | GCGKSSLFRILGGLWP | ++ | +++ | Overlap with the Walker A motif; gives some hits with other ABCs |
| TLRDQIIYPDS | ++ | +++ | ||
| HRxSLWKYH | ++ | + | Gives some hits with unrelated proteins | |
| ABCE-1 | GKSTALKILAGKxKPNLGRY | +++ | +++ | Overlap with the Walker A motif |
| DEPSSYLDVKQRLKAAQVLRSLL | +++ | +++ | Overlap with the Walker B motif | |
| ABCE-2 | LNVSYKPQKIxPK | ++ | ++ | Gives some hits with unrelated proteins |
| HDFIMATYLAD | +++ | ++ | Gives some hits with unrelated proteins | |
| ABCF-1 | GRRYGLXGXNGXGKST | +++ | +++ | Overlap with the Walker A motif; gives some hits with bacterial ABCs |
| FLNxVCTxIIH | +++ | ++ | ||
| ABCF-2 | DSRIALVGPNGAGKST | +++ | +++ | Overlap with the Walker A motif; gives some hits with bacterial ABCs |
| DEPTNHLDIEXIDAL | +++ | +++ | Overlap with the Walker B motif; gives some hits with bacterial ABCs | |
| ABCG-1 | LVLGRPGxGCSTLLK | +++ | +++ | Detects only the ABCG-1 domain from the fungal group; overlap with the Walker A motif |
| DTxVGNExxRGxSGGERKR | +++ | +++ | Overlap with the ABC signature; specific for domain G-1 | |
| ABCG-2 | GKTTLLDVLAxRKTxG | +++ | +++ | Overlap with the Walker A motif; specific for domain G-2 |
| GLSVEQRKRLTIGVELVAKP | +++ | +++ | Overlap with the ABC signature; specific for domain G-2 | |
| ABCG half | IXTIHQPR | ++ | ++ | Also detects domain G2 from yeast ABCs; will not detect Dictyostelium ABCG20 |
| ABCH | PXQLSGGEQQRV | + (see note) | ++ | Overlap with the ABC signature; will detect bacterial permease. |
| ArsA | GGKGGVGKTTxSCS | +++ | +++ | Overlap with the Walker A motif |
| FDTAPTGHTLRLL | +++ | +++ | Overlap with an unusual Walker B motif |
Highly conserved amino-acids are boldfaced. The unusual cysteine in ABCG-1 is underlined.
Sequences can be used with “the search for short nearly exact matches” from NCBI, and the result was checked for preferentially sorting the specified ABC domain and given a rating from + to +++.
Effectiveness in identifying the family for a known ABC domain; rated from + to +++.
Dictyostelium carries genes encoding members of every ABC family that has been recognized so far. Many are closely related to transporters that can confer drug resistance. Dictyostelium appears to be equipped to rapidly export a wide variety of compounds. There may have been a strong selective advantage in the ability of the amoebae to protect themselves from toxic compounds they might have encountered in their soil habitat by pumping them out as soon as they entered. Other members of the superfamily may serve specialized developmental roles by releasing intercellular signals at appropriate stages of development.
Acknowledgments
We thank Negin Iranfar for preparation of the website containing supplementary materials.
Sequences searched in this study were generated as part of the Dictyostelium Genome Project by A. Kuspa and R. Gibbs (The Baylor Sequencing Center, Houston, Tex.; sequencing supported by the National Institutes of Health); G. Glöckner, A. Rosenthal, L. Eichinger, and A. Noegel (the Institute of Biochemistry, Cologne, Germany, together with the Institute of Molecular Biotechnology, Jena, Germany; sequencing supported by the Deutsche Forschungsgemeinschaft, grants 113/10-1 and 10-2); and M.-A. Rajandream and B. Barrell (the EUDICT consortium, supported by The European Union). This work was supported by a grant from the National Institutes of Health (GM60447).
REFERENCES
- 1.Anderson, M. P., D. P. Rich, R. J. Gregory, A. E. Smith, and M. J. Welsh. 1991. Generation of cAMP-activated chloride currents by expression of CFTR. Science 251:679-682. [DOI] [PubMed] [Google Scholar]
- 2.Anjard, C., C. Zeng, W. F. Loomis, and W. Nellen. 1998. Signal transduction pathways leading to spore differentiation in Dictyostelium discoideum. Dev. Biol. 193:146-155. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, B. E., H. Wolfger, and K. Kuchler. 1999. Inventory of yeast ABC proteins: about sex, stress, pleitropic drug and heavy metal resistance. Biochim. Biophys. Acta 1461:217-236. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharjee, H., and B. P. Rosen. 1996. Spatial proximity of Cys113, Cys172, and Cys422 in the metalloactivation domain of the ArsA ATPase. J. Biol. Chem. 271:24465-24470. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharjee, H., Y. S. Ho, and B. P. Rosen. 2001. Genomic organization and chromosomal localization of the Asna1 gene, a mouse homologue of a bacterial arsenic-translocating ATPase gene. Gene 272:291-299. [DOI] [PubMed] [Google Scholar]
- 6.Bisbal, C., C. Martinand, M. Silhol, B. Lebleu, and T. Salehzada. 1995. Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2-5A pathway. J. Biol. Chem. 270:13308-13317. [DOI] [PubMed] [Google Scholar]
- 7.Brazill, D. T., L. R. Meyer., R. D. Hatton, D. A. Brock, and R. H. Gomer. 2001. ABC transporters required for endocytosis and endosomal pH regulation in Dictyostelium. J. Cell Sci. 114:3923-3932. [DOI] [PubMed] [Google Scholar]
- 8.Chang, G., and C. B. Roth. 2001. Structure of MsbA from E. coli: a homolog of the multidrug resistance ATP binding cassette (ABC) transporters. Science 293:1793-1800. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. M., T. K. Misra, S. Silver, and B. P. Rosen. 1986. Nucleotide sequence of the structural genes for an anion pump. The plasmid-encoded arsenical resistance operon. J. Biol. Chem. 261:15030-15038. [PubMed] [Google Scholar]
- 10.Cutting, G. R., L. M. Kasch, B. J. Rosenstein, J. Zielenski, L. C. Tsui, S. E. Antonarakis, and H. H. Kazazian. 1990. A cluster of cystic fibrosis mutations in the first nucleotide-binding fold of the cystic fibrosis conductance regulator protein. Nature 346:366-369. [DOI] [PubMed] [Google Scholar]
- 11.Dean, M., A. Rzhetsky, and R. Allimets. 2001. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 11:1156-1166. [DOI] [PubMed] [Google Scholar]
- 12.Dean, M., M. B. White, J. Amos, B. Gerrard, C. Stewart, K. T. Khaw, and M. Leppert. 1990. Multiple mutations in highly conserved residues are found in mildly affected cystic fibrosis patients. Cell 61:863-870. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 14.Gros, P., Y. B. Ben Neriah, J. M. Croop, and D. E. Housman. 1986. Isolation and expression of a complementary DNA that confers multidrug resistance. Nature 323:728-731. [DOI] [PubMed] [Google Scholar]
- 15.Hettema, E. H., B. Distel, and H. F. Tabak. 1999. Import of proteins into peroxisomes. Biochim. Biophys. Acta 1451:17-34. [DOI] [PubMed] [Google Scholar]
- 16.Hirano, T., T. J. Mitchison, and J. R. Swedlow. 1995. The SMC family: from chromosome condensation to dosage compensation. Curr. Opin. Cell Biol. 7:329-336. [DOI] [PubMed] [Google Scholar]
- 17.Hogue, D. L., L. Liu, and V. Ling. 1999. Identification and characterization of a mammalian mitochondrial ATP-binding cassette membrane protein. J. Mol. Biol. 285:379-389. [DOI] [PubMed] [Google Scholar]
- 18.Holland, I. B., and M. A. Blight. 1999. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol. 293:381-399. [DOI] [PubMed] [Google Scholar]
- 19.Kaur, P., and B. P. Rosen. 1992. Plasmid-encoded resistance to arsenic and antimony. Plasmid 27:29-40. [DOI] [PubMed] [Google Scholar]
- 20.Klein, I., B. Sarkadi, and A. Varadi. 1999. An inventory of the human ABC proteins. Biochim. Biophys. Acta 1461:237-262. [DOI] [PubMed] [Google Scholar]
- 21.Leighton, J., and G. Schatz. 1995. An ABC transporter in the mitochondrial inner membrane is required for normal growth of yeast. EMBO J. 14:188-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loomis, W. F., and M. Gilpin. 1986. Multigene families and vestigial DNA. Proc. Natl. Acad. Sci. USA 83:2143-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marton, M. J., C. R. Vazquez de Aldana, H. Qiu, K. Chakraburtty, and A. G. Hinnebusch. 1997. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2α kinase GCN2. Mol. Cell. Biol. 17:4474-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mosser, J., Y. Lutz, M. E. Stoeckel, C. O. Sarde, C. Kretz, A. M. Douar, J. Lopez, P. Aubourg, and J. L. Mandel. 1994. The gene responsible for adrenoleukodystrophy encodes a peroxisomal membrane protein. Hum. Mol. Genet. 3:265-271. [DOI] [PubMed] [Google Scholar]
- 25.Nei, M., X. Gu, and T. Sitnikova. 1997. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc. Natl. Acad. Sci. USA 94:7799-7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ota, T., and M. Nei. 1994. Divergent evolution and evolution by the birth-and-death process in the immunoglobulin VH gene family. Mol. Biol. Evol. 11:469-482. [DOI] [PubMed] [Google Scholar]
- 27.Rosenberg, M. F., Q. Mao, A. Holzenburg, R. C. Ford, R. G. Deeley, and S. P. Cole. 2001. The structure of the multidrug resistance protein 1 (MRP1/ABCC1). Crystallization and single-particle analysis. J. Biol. Chem. 276:16076-16082. [DOI] [PubMed] [Google Scholar]
- 28.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 29.Sánchez-Fernández, R., T. G. Davies, J. O. Coleman, and P. A. Rea. 2001. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J. Biol. Chem. 276:30231-30244. [DOI] [PubMed] [Google Scholar]
- 30.Saurin, W., M. Hofnung, and E. Dassa. 1999. Getting in or out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J. Mol. Evol. 48:22-41. [DOI] [PubMed] [Google Scholar]
- 31.Shani, N., and D. Valle. 1996. A Saccharomyces cerevisiae homolog of the human adrenoleukodystrophy transporter is a heterodimer of two half ATP-binding cassette transporters. Proc. Natl. Acad. Sci. USA 93:11901-11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaulsky, G., A. Kuspa, and W. F. Loomis. 1995. A multidrug resistance transporter serine protease gene is required for prestalk specialization in Dictyostelium. Genes Dev. 9:1111-1122. [DOI] [PubMed] [Google Scholar]
- 33.Shepherd, J. C., T. N. Schumacher, P. G. Ashton-Rickardt, S. Imaeda, H. L. Ploegh, C. A. Janeway, Jr., and S. Tonegawa. 1993. TAP1-dependent peptide translocation in vitro is ATP dependent and peptide selective. Cell 74:577-584. [DOI] [PubMed] [Google Scholar]
- 34.Tusnady, G. E., E. Bakos, A. Varadi, and B. Sarkadi. 1997. Membrane topology distinguishes a subfamily of the ATP-binding cassette (ABC) transporters. FEBS Lett. 402:1-3. [DOI] [PubMed] [Google Scholar]
- 35.Tyzack, J. K., X. Wang, G. J. Belsham, and C. G. Proud. 2000. ABC50 interacts with eukaryotic initiation factor 2 and associates with the ribosome in an ATP-dependent manner. J. Biol. Chem. 275:34131-34139. [DOI] [PubMed] [Google Scholar]
- 36.Vazquez de Aldana, C. R., M. J. Marton, and A. G. Hinnebusch. 1995. GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2α kinase GCN2 in amino acid-starved cells. EMBO J. 270:3184-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wada, M., S. Toh, K. Taniguchi, T. Nakamura, T. Uchiumi, K. Kohno, I. Yoshida, A. Kimura, S. Sakisaka, Y. Adachi, and M. Kuwano. 1998. Mutations in the canilicular multispecific organic anion transporter (cMOAT) gene, a novel ABC transporter, in patients with hyperbilirubinemia II/Dubin-Johnson syndrome. Hum. Mol. Genet. 7:203-207. [DOI] [PubMed] [Google Scholar]
- 38.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wemmie, J. A., M. S. Szczypka, D. J. Thiele, and W. S. Moye-Rowley. 1994. Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCF1. J. Biol. Chem. 269:32592-32597. [PubMed] [Google Scholar]
- 40.White, M. B., J. Amos, J. M. Hsu, B. Gerrard, P. Finn, and M. Dean. 1990. A frame-shift mutation in the cystic fibrosis gene. Nature 344:665-667. [DOI] [PubMed] [Google Scholar]
- 41.Young, J., and I. B. Holland. 1999. ABC transporters: bacterial exporters-revisited five years on. Biochim. Biophys. Acta 1461:177-200. [DOI] [PubMed] [Google Scholar]