Abstract
Pathology plays a crucial role in diagnosing and evaluating patient tissue samples obtained via surgeries and biopsies. The advent of whole slide scanners and the development of deep learning technologies have considerably advanced this field, promoting extensive research and development in pathology artificial intelligence (AI). These advancements have contributed to reduced workload of pathologists and supported decision-making in treatment plans. Large-scale AI models, known as foundation models (FMs), are more accurate and applicable to various tasks than traditional AI. Such models have recently emerged and expanded their application scope in healthcare. Numerous FMs have been developed in pathology, with reported applications in various tasks, such as disease and rare cancer diagnoses, patient survival prognosis prediction, biomarker expression prediction, and scoring of the immunohistochemical expression intensity. However, several challenges persist in the clinical application of FMs, which healthcare professionals, as users, must be aware of. Research to address these challenges is ongoing. In the future, the development of generalist medical AI, which integrates pathology FMs with FMs from other medical domains, is expected to progress, effectively utilizing AI in real clinical settings to promote precision and personalized medicine.
Keywords: artificial intelligence, medicine, pathology, foundation model, generalist medical AI
1. Introduction for Digital Pathology
(a) Development of digital pathology
For more than 50 years, surgical pathology has played a vital role in diagnosing diseases, evaluating disease progression, and elucidating causes by observing patient tissue sections obtained from surgeries and biopsies through formalin fixation, embedding, and staining. Next, these sections were examined by trained pathologists under microscopes. During this process, several scoring systems, such as the Gleason score for prostate cancer and the Nottingham score for breast cancer, were utilized to grade tumors based on their morphology. These scores provide essential information for determining treatment plans; however, given that they are determined based on the pathologist’s subjective judgment, a notable variability exists between pathologists. Moreover, some indicators impose a burden on pathologists in routine practice (1). However, introducing whole slide scanners in the 1990s easily created digital images of entire specimens with the same resolution as microscopes. This development highlighted the application of image analysis and machine learning technologies to histopathology, thereby expanding digital pathology, wherein the interpretation and analysis of digitized images (whole slide images [WSI]) can be performed quantitatively using computers. Furthermore, the remarkable speed of technological innovation in deep learning developed artificial intelligence (AI) that reduces the workload of pathologists and aids in predicting patient prognosis and supporting treatment decisions based on WSI, further resulting in research and development of AI with high potential clinical utility (2).
(b) Current applications of AI in pathology
The applications of current pathology AI are broadly categorized into three types: i) improving the accuracy and efficiency of pathology diagnosis, ii) predicting patient prognosis and supporting treatment decisions, and iii) integrating data with genomic information.
Some items in ii) include iii) as elemental technology.
i) Improving the accuracy and efficiency of pathology diagnosis
AI enables accurate quantitative evaluation of various tissue features, such as immunohistochemical biomarker evaluation, cell counting, spatial cell arrangement, structural density, distribution patterns, and tissue structure, thereby developing diagnostic AI tools that diagnose with the same or higher accuracy obtained by general pathologists in specific tissues or provide information that is difficult to identify by pathologists. Examples include diagnostic support tools that evaluate the morphological features necessary for tumor assessment, such as tumor grade, histological type, and invasion extent, and automatic quantification tools for immunohistochemical staining intensity. For example, using tools, such as Mindpeak Breast Ki-67 RoI and Mindpeak ER/PR RoI, which automate the evaluation of Ki-67 and estrogen and progesterone receptors in breast cancer, interobserver agreement among pathologists increases (3). Additionally, AI has been developed to optimize the pathology workflow by triaging case priorities, categorizing cases based on priority, and checking specimen quality and identification (4), (5), (6).
ii) Predicting patient prognosis and supporting treatment decisions
After years of research, several morphological features of histopathological tissues, such as tumor grade and tissue subtypes, have been established and proven useful as indicators for predicting patient prognosis. Similarly, various clinical and genetic data, including treatment effects, responsiveness, resistance obtained from electronic medical records, and cancer gene panel test results, are crucial for predicting patient prognosis. The development of straightforward and accurate prognostic indicators has been reported using AI to effectively correlate and integrate these data with numerous histopathological findings obtained from pathology images, which have not yet been established as prognostic indicators. For example, Shi et al. developed a tumor microenvironment (TME) signature by automatically quantifying TME from pathological images of colorectal cancer, aiding in patient prognosis stratification (7). Reportedly, integrating individual histopathological features and, in some cases, clinical and genetic information, into a single classification system accurately reflects the nature and behavior of tumors. Pathology AI applied to prognosis prediction is expected to contribute to appropriate treatment selection and stratification of patients, promoting precision and personalized medicine (1), (2), (8).
iii) Integrating data with genomic information
The research and development of AI associating the phenotypes of histopathological tissue morphology with genomic profiles is attracting considerable attention. For example, Jaume et al. developed biologically and histopathologically interpretable AI that integrates genomic information and WSI using large datasets, such as the TCGA (9) with WSI and bulk transcriptome data (10). This research is essential for understanding the biological mechanisms underlying cancer and selecting targeted therapies. Furthermore, studies estimating gene mutations and protein expression levels from pathology images to reduce the delay in patient treatment initiation because of the weeks-long time required for clinicians to obtain genetic and immunological test results have been conducted (11), (12). Such research is progressing concerning developing AI that integrates multimodal information and presents it in an interpretable form for pathologists and clinicians.
2. Introduction for Foundation Models
(a) Development of foundation models
Large-scale AI models called foundation models (FMs) have emerged (13) with 1) advancing social networking services, 2) the global spread of COVID-19, which increased the amount of digital data, 3) improved computational efficiency owing to hardware advancements, and 4) development of new AI architectures such as neural networks and transformers. FMs are applicable to a wider range of tasks than traditional AI models. A human-centered AI institute at Stanford defined FM as “a foundation model is any model that is trained on broad data (generally using self-supervision at scale) that can be adapted (e.g., fine-tuned) to a wide range of downstream tasks”(14). Initially, large-scale language models trained on vast text data collected from the web solved various language-related tasks (information retrieval, text generation, sentiment analysis, chatbots, etc.) with high accuracy (15), (16). Subsequently, diverse FMs have been developed using various data modalities, such as images, audio, and point cloud data (17). According to the “AI Foundation Models: Update paper”(18) by the UK government’s Competition and Markets Authority updated in April 2024, giant AI companies like Meta, Google, and OpenAI are investing millions of dollars to acquire developers. In healthcare, the number of papers related to medical foundation models has exponentially increased from 2018 to February 2024, reflecting the growing expectations and interest of healthcare professionals in applying FMs in clinical practice. While there were only a few papers in 2018, >120 papers were published in 2023 alone (19).
(b) Foundation and nonfoundation models
The differences between FMs and non-FMs (deep learning models) are summarized in Table 1 (13). Generally, the more complex the model, the more diverse relationships it can capture (expressiveness), and the more data, the better the model performance (scalability) (20). FMs possess superior expressiveness and scalability based on large models, training data, and parallelizable training methods. A common feature of FMs and non-FMs is the need to obtain and utilize the embeddings generated by the model while making inferences from input data. Embeddings represent the features of the input data as short vectors (or small tensors), and the quality of these embeddings remarkably affects the model accuracy during downstream tasks.
Table 1.
Comparison between Foundation Models and Nonfoundation Models (Adapted from Table 1 in Reference [13]1).
| Characteristics | Foundation model | Nonfoundation model (deep learning model) |
|---|---|---|
| Model architecture | (Mainly) Transformer | Convolutional neural network |
| Model size | Vary large | Medium to large |
| Model applicable task | Many | Single |
| Performance on adapted tasks | State-of-the-art (SOTA) | High to SOTA |
| Performance on untrained tasks | Medium to high | Low |
| Data amount for model training | Very large | Medium to large |
| Using labeled data for model training? | No | Yes |
1Reference [13] was published under the CC BY 4.0 license.
(c) Applications of foundation models in healthcare
FMs in healthcare can be broadly categorized into four types based on the data modality they handle (19), (21):
i) Language Foundation Models for Natural Language Processing (NLP) in Healthcare
Examples of usage: medical report generation, education for medical students/residents, patient self-diagnosis, and mental health support.
ii) Vision Foundation Models (VFMs) for Image Data
Examples of usage: image diagnosis and similar case retrieval, prognosis prediction for specific diseases, and surgical assistance through detection of critical structures and lesions.
iii) Bioinformatics Foundation Models for Omics Data Such as DNA, RNA, and Protein Data
Examples of usage: sequence, interaction, structural, and functional analyses, protein sequence generation, drug response and sensitivity prediction, disease risk prediction, and drug perturbation effect prediction.
iv) Multimodal Foundation Models (MFMs) Integrating Multiple Modalities Such as Language, Image, and Bioinformatics
Examples of usage: comprehensive diagnostic support based on multiple data modalities, medical image report generation, cross-search between text descriptions and chemical structures, promotion of research through dialog with models harboring biological and medical knowledge, and advice on diagnosis and treatment based on patient inquiries and images.
3. Introduction for Histopathology Foundation Models
(a) Need for foundation models in pathology
Representative tasks in computational pathology include patient prognosis prediction, biomarker prediction, cancer and tissue subtype classification, cancer grading, diagnostic prediction, and immunohistochemical staining intensity scoring (Section 1(b)). The development and clinical application of pathology FMs are desired for two reasons:
i) Annotation Cost: Before promoting FM development, the approach to handling individual tasks involved creating supervised learning models based on data annotated by pathologists, which required substantial time and practical effort from pathologists for each disease, organ type, and task type (22). The average salary of a pathologist is $149 h−1 (https://www.salary.com/research/salary/alternate/pathologist-hourly-wages), and assuming 5 min per slide, the annotation cost per pathology slide is approximately $12. The cost of annotation and increasing working hours can be a burden for clinical pathologists (22), (23). Furthermore, the quality of annotation determines the trained model performance, requiring pathologists to conduct high-quality annotation, thereby adding to their burden and responsibility and becoming a bottleneck in the model development of individual tasks. Model development utilizing vast amounts of pathology image data with minimal or simple annotations per case is crucial.
ii) Lack of Public Datasets: At present, approximately 100 publicly available pathology image datasets (24) are accessible to model developers, most of which include hundreds to tens of thousands of WSIs. However, the diseases and organ types included in each dataset are limited, and the data quality varies. TCGA―the largest public dataset―including pathology images, contains tens of thousands of WSIs; however, it is limited to 32 cancer types, making it challenging to cover the diverse diseases encountered in clinical practice.
(b) Examples of pathology foundation models
As of June 2024, more than 10 FMs specifically for pathology images were reported. The origin and scale of datasets, tissue types included, and learning methods vary for each FM. Many models are publicly available and usable for downstream tasks, although some are only available for limited use or are not publicly released (Table 2) (10), (22), (23), (25), (26), (27), (28), (29), (30), (31), (32), (33).
Table 2.
List of Pathology Foundation Models with Published Papers, Including Preprints, between October 2022 and June 2024.
| Foundation model name | CTransPath | (Lunit)* | PLIP | Virchow | UNI | CONCH | PRISM | Prov-GigaPath | TANGLE | RudolphV |
|---|---|---|---|---|---|---|---|---|---|---|
| Publication date | 2022 | 2022 | 2023 | 2024 | 2024 | 2024 | 2024 | 2024 | 2024 | 2024 |
| Journal name | Medical Image Analysis | arxiv | Nature Medicine | Nature Medicine | Nature Medicine | Nature Medicine | arxiv | Nature | CVPR2024 | arxiv |
| Pretraining dataset name | TCGA | TCGA | OpenPath | MSKCC | Mass-100K | Educational sources | MSKCC) | Dataset from Providence | TCGA | Dataset from over 15 different laboratories across the EU and US |
| PAIP | TULIP | Mass-1K | PubMed Central Open Access Dataset | TG-GATEs | TCGA | |||||
| Mass-22K | ||||||||||
| Number of GPUs/type used for training | 48/NVIDIA V100 GPUs | 64/NVIDIA V100 GPUs | Not specified | -/NVIDIA A100 GPUs | 32/NVIDIA A100 GPUs | 8/NVIDIA A100 GPUs | 16/NVIDIA V100 GPUs | 16 nodes × 4/NVIDIA A100 GPUs | 8/NVIDIA A100 GPUs | 16/NVIDIA A100 GPUs |
| Embedding level (patch/slide) | Patch | Patch | Patch | Patch | Patch | Patch | Slide | Slide | Slide | Patch |
| Number of WSIs | 29,763 (TCGA) | 20,994 (TCGA) | Not specified | 1,488,550 | 100,426 | Not specified | 587,196 | 171,189 | 2,074 (TCGA) | 133,998 |
| 2,457 (PAIP) | 15,672 (TULIP) | 6,597 (TG-GATEs) | ||||||||
| Number of patch images (M) | 15 | 32.6 | 0 | 2,000 | 100 | 1 | Not specified | 1,300 | 15 | 1,200 |
| Number of patients | Not specified | Not specified | Not specified | 119,629 | Not specified | Not specified | 195,344 | >30,000 | >1,864 | 34,103 |
| More than 10 types of organs in the dataset? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes |
| Staining types (H&E/H&E + others) | Not specified | H&E | H&E + others | H&E | H&E | H&E + others | H&E | H&E + others | Not specified | H&E + others |
| FFPE/frozen | FFPE, frozen | Not specified | Not specified | FFPE | FFPE | Not specified | Not specified | Not specified | Not specified | FFPE, Frozen |
| VFM/MFM | VFM | VFM | MFM | VFM | VFM | MFM | MFM | MFM | MFM | VFM |
| Model publicly available? | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes |
| Reference number | 33 | 25 | 28 | 27 | 28 | 30 | 31 | 22 | 10 | 32 |
Because of space constraints, not all models are included in this table. For a complete list, please refer to Supplementary Table 1.
*When the model name is not specified in the original paper, the first author’s name is shown.
Herein, we overviewed these pathological FMs by focusing on the following: a) training datasets, modalities, and embedding levels; b) examples of downstream task usage; c) status of public availability; and d) performance comparison between models.
a) Training datasets, modalities, and embedding levels
i) Dataset Origin and Scale: Each model is pretrained on a public dataset (e.g., TCGA, PubMed Central Open Access Dataset [https://ncbi.nlm.nih.gov/pmc/tools/openftlist/], TG-GATEs) (34) or a proprietary clinical pathological image dataset collected from single or multiple institutions). For example, the Virchow (27) model utilizes a dataset of 1,488,550 WSIs from 119,629 patients stored at Memorial Sloan Kettering Cancer Center and is currently the largest dataset of human pathological images.
ii) Organ and Tissue Types: The organs and tissue types, in addition to their ratios, included in datasets are influenced by various factors, such as whether the dataset includes biopsy/surgical specimen WSIs, specific public datasets, or case accumulation trends of the data collection medical facilities. Although identifying consistent trends is difficult, most datasets used to train FMs include images from >10 different organs. Some models, such as Virchow, UNI (29), TANGLE (10), and RudolphV (32), are explicitly trained on datasets that include normal tissues; however, it is not specified whether other models include only tumor datasets or also normal tissue images. To our knowledge, no experiment has examined the extent to which including normal tissues parallel to tumor tissues in FM training contributes to improved model performance in downstream tasks. Chen et al. (29) demonstrated that training on a combination of lab-derived datasets and a public dataset, composed of only normal tissue images, resulted in better performance than that on lab-derived datasets alone for the 43-class cancer-type classification task. However, this improvement cannot be conclusively attributed to the mere increase in image count or the inclusion of normal tissue images.
iii) H&E vs. H&E + Other Stains: Although many pathological FMs are trained using datasets with solely hematoxylin and eosin (H&E)-stained images, some models like CONCH and RudolphV include immunostained and/or specially stained images. CONCH conducted comparative experiments by evaluating models trained with only H&E-stained images compared with models trained with other stain types included. The results indicated that models incorporating various stains performed better on most tasks, including tumor subtyping and grading, image-to-text retrieval, and text-to-image retrieval (8/13). However, in some classification tasks, models trained on only H&E-stained images either outperformed or showed marginally lower performance than models with additional stains. This indicates that incorporating diverse stains does not necessarily yield the best results.
iv) VFM/MFM: Many pathological FMs are VFMs; meanwhile, models like PLIP (28), CONCH, PRISM (31), Prov-GigaPath (22), and TANGLE (10) are MFMs. They handle pathological images and text (e.g., clinical pathology reports, social media, educational sources, and PubMed Central Open Access Dataset captions) and bulk transcriptome data in the case of TANGLE. These MFMs acquire equivalence between paired image and other modality data, enabling model application to tasks such as image-to-text retrieval, text-to-image retrieval, report generation, and gene expression analysis within images.
v) Embedding Levels from Models: WSIs are high-resolution virtual slide images of entire-stained glass slide specimens that often exceed several gigabytes in data size. WSIs have a pyramid structure with multiple layers of images at different magnifications, thereby allowing a comprehensive observation of the entire specimen slide at any zoom level. Memory constraints make it challenging to handle WSIs as whole images during model training; thus, they are typically divided into small patches (tiles) of a few hundred pixels. Therefore, some models output embeddings for each patch, whereas others provide WSI-level embeddings. Models like PRISM, Prov-GigaPath, and TANGLE output WSI-level embeddings, whereas others output patch-level embeddings. Aggregating patch-level embeddings to obtain WSI-level embeddings is possible, and the efficacy of each embedding level for different tasks remains debatable. Prov-GigaPath demonstrated statistically significant improvements over previous patch-level embedding models across 16 tasks, including classification and image-to-text search. However, https://github.com/mahmoodlab/UNI indicates that UNI and CONCH outperformed Prov-GigaPath in four of five classification tasks, including tumor grade classification and immunohistochemical protein expression intensity scoring.
b) Examples of downstream tasks
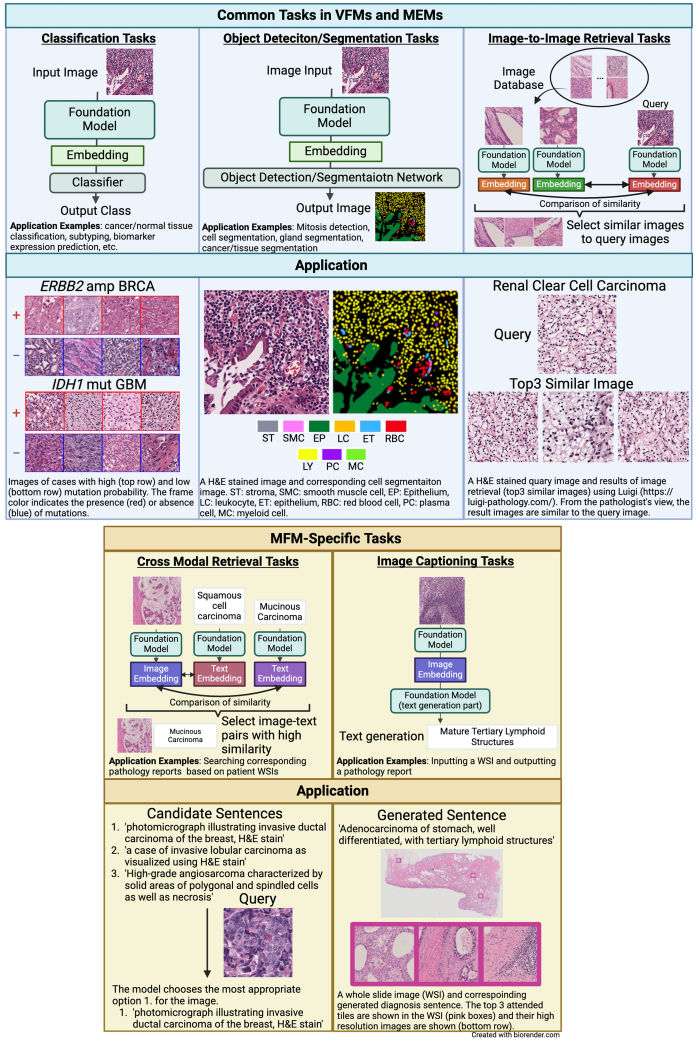
FMs are applicable to various downstream tasks, with appropriate accuracy verification conducted posttraining. The tasks handled by VFMs/MEMs in healthcare are described in Section 2(c), in which we focus on pathological images and list contents by task category (Figure 1). The tasks mentioned herein are based on those listed in FM papers (10), (22), (23), (25), (26), (27), (28), (29), (30), (31), (32), (33).
Figure 1.
Task-specific schematic of the pathology foundation models (FMs).
The figure illustrates five tasks in which pathology FM are applied, including the application example for each task. These include common tasks, such as classification, object detection/segmentation, and image-to-image retrieval, and specific tasks such as cross-modal retrieval and image captioning. The figure highlights the versatility and potential of the pathology FMs in various diagnostic and research contexts.
Common Tasks: Predominantly, classification tasks are applicable to VFMs and MFMs, with fewer object detection, segmentation, and search tasks. VFMs such as Lunit (25), UNI, RandolphV, and CTransPath (33) have been evaluated for these minor tasks. Reportedly, the SegPath dataset, which we previously published, is used as a benchmark for segmentation tasks. The SegPath dataset is a large-scale dataset comprising >1,500 pairs of H&E- and immunohistochemically-stained images (35).
i) Classification Tasks: This task involves predicting which category a given image datum (patch or WSI) belongs to. Examples include disease detection, cancer detection, cancer/normal tissue classification, subtyping, disease diagnosis, rare cancer diagnosis, patient prognosis prediction, cancer recurrence prediction, cancer metastasis prediction, biomarker expression prediction (microsatellite instability, driver mutation, and tumor mutation burden), treatment outcome prediction, and immunohistochemical image protein expression intensity scoring.
ii) Object Detection/Segmentation Tasks: This task involves detecting specific structures in images or predicting the category of each pixel. Examples include mitosis detection, cell segmentation in H&E and immunohistochemical images, gland segmentation, cancer segmentation, and tissue segmentation.
iii) Image-to-Image Retrieval Tasks: This task searches for images with high similarity to a query image.
MFM-specific Tasks:
i) Cross-Modal Retrieval Tasks: This is done using an image or text query to search for corresponding text or image pairs.
Examples include retrieving corresponding pathology reports from a database based on patient WSIs.
ii) Image Captioning Tasks: This task generates summary text for an image.
Examples of such inputs include inputting a WSI and outputting a pathology report.
c) Public availability status
Most FMs are open access, allowing users to apply them to downstream tasks. However, some models (e.g., CONCH) have limited task availability compared to their original papers or are not open access (e.g., Virchow, PRISM, and RudolphV). The usage instructions for available models are typically described on their respective GitHub (a service for sharing and managing program source code online) pages referenced in the original papers.
d) Performance comparison between models
Zeng et al. (36) conducted comparative accuracy experiments for CTransPath, UNI, Virchow, and Prov-GigaPath across 20 tasks in disease detection, biomarker prediction, and treatment outcome prediction. In disease detection tasks, all four models achieved comparable performances. However, in the biomarker and treatment outcome prediction tasks, UNI and Prov-GigaPath consistently demonstrated performance equal to or exceeding that of the other models. For biomarker prediction in the lung tissue, UNI and Prov-GigaPath outperformed the other models. The authors attributed these results to the higher representation of the lung tissue in the pretraining datasets of UNI and Prov-GigaPath, suggesting that the proportion of relevant tissue in the training dataset could enhance the representational power of the model for that tissue type.
4. Issues and Future Directions of Foundation Models
Thus far, we have outlined the development of digital pathology and FMs, their applications in healthcare, the introduction of pathological FMs, and specific examples of their usage. This section focuses on the future implications of FM-related research and discusses potential issues users might face while employing FMs in clinical AI applications.
(a) Hardware requirements
Many FMs require specific hardware because of their immense complexity and scale. Although these models demonstrate remarkable performance, their training and inference processes often require substantial computational resources. For example, running Prov-GigaPath requires high-end Graphics Processing Units (GPUs), such as NVIDIA A100 (37). Hardware investment required for these models can be challenging for resource-constrained medical institutions. Therefore, balancing model performance with hardware feasibility is critical while implementing these models clinically. Future research should optimize these models to operate on less resource-intensive hardware, thereby making them accessible to several medical facilities.
(b) Transition to multimodal AI assistants and generalist medical AI
At present, VFMs are the most developed pathological FMs. However, various MFMs have been developed, focusing on combining language and visual modalities to produce pathology diagnostic reports from image inputs or molecular biology and visual modalities to identify notable genes in model-driven pathology diagnoses. These MFMs are mainstream in pathology and across healthcare (19). The current self-supervised learning methods are not universally generalizable across all modalities and often require tailoring per specific modalities (21). This limitation means that integrating more modalities into FMs in real-world clinical settings is not feasible. In 2023, Lu et al. (38) developed a dialog-based pathological AI assistant capable of prompt engineering, which improved task accuracy by interactively providing appropriate questions and instructions, was built on previously developed pathological MFMs by their group and supported diagnostic assistance by describing histological findings and suggesting additional tests through interactions with pathologists. Recently, the concept of generalist medical artificial intelligence (GMAI) (39) has been proposed, utilizing vast and diverse data types―including images, electronic health records, test results, genomics, graphs, and medical texts―to perform various tasks across different medical specialties. The development of AI assistants integrating FMs from domains beyond pathology will possibly drive comprehensive medical AI research and development toward realizing GMAI.
(c) Hallucinations and interpretability
Models generate embeddings through probabilistic processes instead of truly understanding the logical meaning of data (40). Consequently, FMs might generate nonexistent information (fabricated citations, information not inferable from input data, etc.) during generative tasks such as pathology report generation (hallucinations) (41), (42). Users must recognize these potential hallucinations. Furthermore, the inference processes of large-scale FMs with hundreds of millions to trillions of parameters are incredibly complex and are often described as the “black box” nature of AI. Unlike traditional medical devices that are typically transparent and logical, this characteristic makes it difficult for people to trust the algorithm results (43). Thus, the field of “explainable AI (44)” is advancing, developing techniques to enhance human understanding of how AI models reach their outputs.
(d) Potential for domain shift and information updates
Pathological specimens from Japan and overseas may show remarkable color and texture differences, even with the same staining. Because all publicly available pathological FMs were developed using datasets from overseas facilities, they might not generalize to Japanese specimens (WSIs) because of color and texture distribution (domain shift (45)). Various methods have been developed to efficiently standardize color tones and perform color augmentation. Previously, we created a large-scale pathological image dataset, PLISM (46), which includes tissues stained with H&E in various color tones that facilitate color augmentation. Moreover, there are potential issues pertaining to handling and processing of tissue samples, and the patient specimen data used for training FMs may have inherent biases concerning gender, race, age, and sampling methods. If the sample appearance, such as color tone, or the social and sampling method aspects of the to-be predicted data significantly differ from the training data, the analysis and prediction accuracy of FMs may decrease. Further, the lack of standardized evaluation metrics and benchmarks complicates the validation process, thereby making it difficult for healthcare professionals to accurately assess the performance of these FMs. Additionally, the data used to train these large models, including PubMed and textbook knowledge, can quickly become outdated because of the rapid advancements in medicine, necessitating regular updates to prevent inaccuracies. However, retraining these large-scale models with new data is costly. Researchers aim to improve these model architectures using retrieval-augmented generation (RAG) (15), which allows models to reference external databases while generating responses, consequently enhancing accuracy and explainability.
(e) Concerns about the clinical implementation of foundation models
Despite the rapid development of FMs, their validation in real clinical settings remains insufficient, preventing their clinical application. Similar to other AI models, FMs are regulated as medical devices by the US Food and Drug Administration under a uniform software category developed for specific-use cases (47). The World Health Organization released AI ethics and governance guidance for MFMs in 2024, including recommendations in development and deployment for governments and developers (48). The guidance outlines more than 40 considerable recommendations for governments, technology companies, and healthcare providers to ensure the appropriate use of MFMs for promoting and protecting the population health. However, the current regulatory environment is inadequate for the clinical safety and effective deployment of FMs. Only a few approved AI models have been tested in randomized controlled trials and none have involved FMs (49). The lack of transparency in trial reports and evaluated use cases is a considerable issue. The Standard Protocol Items: Recommendations for Interventional Trials―AI and consolidated standards of reporting trials―AI guidelines (50), which were assessed by an international multistakeholder group from specific fields, such as healthcare, statistics, computer science, law, and ethics, in a two-stage Delphi survey and agreed upon in a 2-day consensus meeting, have been formulated to improve standardization and transparency in clinical trials involving AI; however, most published randomized controlled trials that use AI technology have not strictly followed these established reporting standards (51). Users must be aware of the insufficient algorithm validation of FMs in real clinical settings. At present, there are no pathological FMs approved by the FDA for commercial use, and utilizing pathological FMs remains confined to academic research (52).
5. Conclusion
Advances in digital pathology have expanded the utilization of WSIs, leading to active research and development of related AI technologies. Particularly, the emergence of FMs has broadened the scope of AI applications in medicine, allowing the development of AI models capable of handling various tasks via training on diverse data, such as images, texts, and omics information. In the future, AI-based medical practice using FM-based AI assistants and GMAI will potentially be realized in pathology and real clinical settings, promoting efforts toward precision and personalized medicine. Although medical AI is an extremely useful tool in practice, properly understanding the effectiveness, considerations, and potential issues of using AI centered on FMs to benefit patients is crucial for medical professionals, who are the primary users of FMs.
Article Information
Conflicts of Interest
None
Sources of Funding
This work was supported by the AMED Practical Research for Innovative Cancer Control grant number JP 24ck0106873 and JP 24ck0106904 to S.I., JSPS KAKENHI Grant-in-Aid for Scientific Research (S) grant number 22H04990 to S.I., and JSPS KAKENHI Grant-in-Aid for Scientific Research (B) grant number 21H03836 to D.K.
Author Contributions
M.O. contributed to the search of previous publications and wrote the manuscript draft. D.K. and S.I. reviewed the manuscript critically.
Approval by Institutional Review Board (IRB)
Not applicable.
Supplement
List of pathology foundation models with published papers, including preprints, between October 2022 and June 2024.
*When the model name is not specified in the original paper, the first author’s name is shown.
References
- 1.Rakha EA, Toss M, Shiino S, et al. Current and future applications of artificial intelligence in pathology: a clinical perspective. J Clin Pathol. 2021;74(7):409-14. [DOI] [PubMed] [Google Scholar]
- 2.Bera K, Schalper KA, Rimm DL, et al. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16(11):703-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abele N, Tiemann K, Krech T, et al. Noninferiority of artificial intelligence-assisted analysis of Ki-67 and estrogen/progesterone receptor in breast cancer routine diagnostics. Mod Pathol. 2023;36(3):100033. [DOI] [PubMed] [Google Scholar]
- 4.Serag A, Ion-Margineanu A, Qureshi H, et al. Translational AI and deep learning in diagnostic pathology. Front Med [Internet]. 2019 [cited 2022 May 28]; 6:185. Available from: https://www.frontiersin.org/article/10.3389/fmed.2019.00185 [DOI] [PMC free article] [PubMed]
- 5.Ahmad Z, Rahim S, Zubair M, et al. Artificial intelligence (AI) in medicine, current applications and future role with special emphasis on its potential and promise in pathology: present and future impact, obstacles including costs and acceptance among pathologists, practical and philosophical considerations. A comprehensive review. Diagn Pathol. 2021;16(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong R, Fenyö D. Deep learning and its applications in computational pathology. BioMedInformatics. 2022;2(1):159-68. [Google Scholar]
- 7.Shi L, Zhang Y, Wang H. Prognostic prediction based on histopathologic features of tumor microenvironment in colorectal cancer. Front Med [Internet]. 2023 Apr [cited 2024 Jul 17];10. Available from: https://www.frontiersin.org/journals/medicine/articles/10.3389/fmed.2023.1154077/full [DOI] [PMC free article] [PubMed]
- 8.Srinidhi CL, Ciga O, Martel AL. Deep neural network models for computational histopathology: a survey. Med Image Anal. 2021;67:101813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaume G, Oldenburg L, Vaidya A, et al. Transcriptomics-guided slide representation learning in computational pathology [Internet]. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 2024, pp. 9632-44 [cited 2024 Jul 5]. Available from: http://arxiv.org/abs/2405.11618
- 11.Fujii S, Kotani D, Hattori M, et al. Rapid screening using pathomorphologic interpretation to detect BRAFV600E mutation and microsatellite instability in colorectal cancer. Clin Cancer Res. 2022;28(12):2623-32. [DOI] [PubMed] [Google Scholar]
- 12.Redlich JP, Feuerhake F, Weis J, et al. Applications of artificial intelligence in the analysis of histopathology images of gliomas: a review. Npj Imaging. 2024;2(1):1-16. [Google Scholar]
- 13.Schneider J. Foundation models in brief: a historical, socio-technical focus. arXiv preprint arXiv:2212.08967 [Internet]. 2022 [cited 2024 Jul 7]. Available from: https://arxiv.org/abs/2212.08967v1
- 14.Bommasani R, Hudson DA, Adeli E, et al. On the opportunities and risks of foundation models. arXiv preprint arXiv:2108.07258 [Internet]. 2022 [cited 2024 Jul 5]. Available from: http://arxiv.org/abs/2108.07258
- 15.Lewis P, Perez E, Piktus A, et al. Retrieval-augmented generation for knowledge-intensive nlp tasks. Adv Neural Inf Process Syst [Internet]. 2021 [cited 2024 Jul 5]. Available from: http://arxiv.org/abs/2005.11401
- 16.Izacard G, Lewis P, Lomeli M, et al. Atlas: few-shot learning with retrieval augmented language models. J Mach Learn Res. 2024;24(1):251:11912-54. [Google Scholar]
- 17. uncbiag/Awesome-Foundation-Models [Internet]. uncbiag; 2024 [cited 2024 Jul 5]. Available from: https://github.com/uncbiag/Awesome-Foundation-Models
- 18. CMA. AI Foundation Models technical update report [Internet]. [cited 2024 Jul 5]. Available from: https://assets.publishing.service.gov.uk/media/661e5a4c7469198185bd3d62/AI_Foundation_Models_technical_update_report.pdf
- 19.He Y, Huang F, Jiang X, et al. Foundation model for advancing healthcare: challenges, opportunities, and future directions. arXiv preprint arXiv:2404.03264 [Internet]. 2024 [cited 2024 Jul 7]. Available from: https://arxiv.org/abs/2404.03264v1 [DOI] [PubMed]
- 20.Kaplan J, McCandlish S, Henighan T, et al. Scaling laws for neural language models arXiv preprint arXiv:2001.08361 [Internet]. 2020 [cited 2024 Jul 7]. Available from: http://arxiv.org/abs/2001.08361
- 21.Azad B, Azad R, Eskandari S, et al. Foundational models in medical imaging: a comprehensive survey and future vision. arXiv preprint arXiv:2310.18689 [Internet]. 2023 [cited 2024 Jul 9]. Available from: http://arxiv.org/abs/2310.18689
- 22.Xu H, Usuyama N, Bagga J, et al. A whole-slide foundation model for digital pathology from real-world data. Nature. 2024;630(8015):181-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azizi S, Culp L, Freyberg J, et al. Robust and data-efficient generalization of self-supervised machine learning for diagnostic imaging. Nat Biomed Eng. 2023;7(6):756-79. [DOI] [PubMed] [Google Scholar]
- 24.Stettler M (Duc). maduc7/Histopathology-datasets [Internet]. 2024 [cited 2024 Jul 8]. Available from: https://github.com/maduc7/Histopathology-Datasets
- 25.Kang M, Song H, Park S, et al. Benchmarking self-supervised learning on diverse pathology datasets. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 2023, pp. 3344-54 [Internet]. [cited 2024 Jul 8]. Available from: http://arxiv.org/abs/2212.04690
- 26.Filiot A, Ghermi R, Olivier A, et al. Scaling self-supervised learning for histopathology with masked image modeling [Internet]. medRxiv. 2023 [cited 2024 Jul 8]. Available from: https://www.medrxiv.org/content/10.1101/2023.07.21.23292757v1
- 27.Vorontsov E, Bozkurt A, Casson A, et al. Virchow: a million-slide digital pathology foundation model. arXiv preprint arXiv:2309.07778 [Internet]. 2024 [cited 2024 Jul 8]. Available from: http://arxiv.org/abs/2309.07778
- 28.Huang Z, Bianchi F, Yuksekgonul M, et al. A visual-language foundation model for pathology image analysis using medical Twitter. Nat Med. 2023;29(9):2307-16. [DOI] [PubMed] [Google Scholar]
- 29.Chen RJ, Ding T, Lu MY, et al. Towards a general-purpose foundation model for computational pathology. Nat Med. 2024;30(3):850-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu MY, Chen B, Williamson DFK, et al. A visual-language foundation model for computational pathology. Nat Med. 2024;30(3):863-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaikovski G, Casson A, Severson K, et al. PRISM: A multi-modal generative foundation model for slide-level histopathology. arXiv preprint arXiv:2405.10254 [Internet]. 2024 [cited 2024 Jul 8]. Available from: http://arxiv.org/abs/2405.10254
- 32.Dippel J, Feulner B, Winterhoff T, et al. RudolfV: a foundation model by pathologists for pathologists. arXiv preprint arXiv:2401.04079 [Internet]. 2024 [cited 2024 Jul 8]. Available from: http://arxiv.org/abs/2401.04079
- 33.Wang X, Yang S, Zhang J, et al. Transformer-based unsupervised contrastive learning for histopathological image classification. Med Image Anal. 2022;81:102559. [DOI] [PubMed] [Google Scholar]
- 34.Igarashi Y, Nakatsu N, Yamashita T, et al. Open TG-GATEs: a large-scale toxicogenomics database. Nucleic Acids Res. 2015;43(D1):D921-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komura D, Onoyama T, Shinbo K, et al. Restaining-based annotation for cancer histology segmentation to overcome annotation-related limitations among pathologists. Patterns (N Y). 2023;4(2):100688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campanella G, Chen S, Verma R, et al. A clinical benchmark of public self-supervised pathology foundation models. arXiv preprint arXiv:2407.06508 [Internet]. 2024 [cited 2024 Jul 8]. Available from: http://arxiv.org/abs/2407.06508
- 37.prov-gigapath/prov-gigapath [Internet]. prov-gigapath. 2024 [cited 2024 Jul 9]. Available from: https://github.com/prov-gigapath/prov-gigapath
- 38.Lu MY, Chen B, Williamson DFK, et al. A multimodal generative AI copilot for human pathology. Nature. 2024:1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moor M, Banerjee O, Abad ZSH, et al. Foundation models for generalist medical artificial intelligence. Nature. 2023;616(7956):259-65. [DOI] [PubMed] [Google Scholar]
- 40.Bender EM, Gebru T, McMillan-Major A, et al. On the dangers of stochastic parrots: can language models be too big? In: Proceedings of the 2021 ACM Conference on Fairness, Accountability, and Transparency, New York, NY: Association for Computing Machinery. p. 610-23 [Internet]. 2021 [cited 2024 Jul 9]. Available from: https://doi.org/10.1145/3442188.3445922 [Google Scholar]
- 41.Li Y, Du Y, Zhou K, et al. Evaluating object hallucination in large vision-language models. arXiv preprint arXiv:2305.10355 [Internet]. 2023 [cited 2024 Jul 9]. Available from: http://arxiv.org/abs/2305.10355
- 42.Pal A, Umapathi LK, Sankarasubbu M. Med-HALT: medical domain hallucination test for large language models. arXiv preprint arXiv:2307.15343 [Internet]. 2023 [cited 2024 Jul 9]. Available from: http://arxiv.org/abs/2307.15343
- 43.Azad R, Aghdam EK, Rauland A, et al. Medical image segmentation review: the success of U-Net. 2022 [cited 2024 Jul 9]. Available from: https://arxiv.org/abs/2211.14830 [DOI] [PubMed]
- 44.Arrieta AB, Díaz-Rodríguez N, Del Ser J, et al. Explainable artificial intelligence (XAI): concepts, taxonomies, opportunities and challenges toward responsible AI. Inf Fusion. 2020 [cited 2024 Jul 9];58:82-115. Available from: http://arxiv.org/abs/1910.10045
- 45.Stacke K, Eilertsen G, Unger J, et al. Measuring domain shift for deep learning in histopathology. IEEE J Biomed Health Inform. 2021;25(2):325-36. [DOI] [PubMed] [Google Scholar]
- 46.Ochi M, Komura D, Onoyama T, et al. Registered multi-device/staining histology image dataset for domain-agnostic machine learning models. Sci Data. 2024;11(1):330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meskó B, Topol EJ. The imperative for regulatory oversight of large language models (or generative AI) in healthcare. npj Digit Med. 2023;6(1):120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.WHO. Ethics and governance of artificial intelligence for health: guidance on large multi-modal models [Internet]. Geneva: WHO; 2024 [cited 2024 Jul 9]. Available from: https://www.who.int/publications/i/item/9789240084759
- 49.Lam TYT, Cheung MFK, Munro YL, et al. Randomized controlled trials of artificial intelligence in clinical practice: systematic review. J Med Internet Res. 2022;24(8):e37188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cruz Rivera S, Liu X, Chan AW, et al. Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension. Nat Med. 2020;26(9):1351-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plana D, Shung DL, Grimshaw AA, et al. Randomized clinical trials of machine learning interventions in health care: a systematic review. JAMA Netw Open. 2022;5(9):e2233946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Health C for D and R. Artificial intelligence and machine learning (AI/ML)-enabled medical devices. FDA [Internet]. 2024 [cited 2024 Jul 9]. Available from: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of pathology foundation models with published papers, including preprints, between October 2022 and June 2024.
*When the model name is not specified in the original paper, the first author’s name is shown.