Abstract
Colletotrichum trifolii is a fungal pathogen responsible for anthracnose disease of alfalfa. Previously, a serine/threonine protein kinase gene from this fungus (TB3), which is a functional homolog of the Neurospora crassa COT1 kinase, has been isolated in our laboratory and appears to be associated with hyphal elongation and branching. In this report we show that light treatment rapidly induces TB3 expression and hyphal branching frequency. Western analysis showed TB3 localization in both the cytoplasm and nucleus, but not in membranes. Moreover, indirect immunofluorescence indicated that TB3 levels were most abundant in the nucleus. To further evaluate the subcellular distribution of TB3, a TB3::GFP fusion construct was inserted into C. trifolii. Results indicated that the cellular location of TB3 changed during fungal growth and development. Consistent with previous observations, TB3 was localized in both the cytoplasm and the nucleus but was preferentially localized in the nucleus during extended hyphal growth. The amino terminus of TB3 contains two relatively long polyglutamine repeats. Yeast-based assays showed that these polyglutamine tracts can activate transcription. These results suggest that TB3 may be positioned in a signaling cascade regulating proper hyphal growth and development by functioning as a transcription factor.
A distinctive feature of filamentous fungi is their mode of growth and development via vegetative propagagules known as hyphae. Hyphae grow in a polarized manner and account, in part, for the foraging success of fungi either as saprophytes or pathogens. Coordinated control of hyphal growth and development in filamentous fungi is achieved by activating intracellular signaling cascades in response to environmental stimuli, including nutrient availability, light, and plant host. In addition to these environmental cues that influence hyphal growth, genetic factors have also been identified. In Neurospora crassa, the serine/threonine protein kinase, COT1, has been shown to be required for hyphal elongation (29). Mutations in cot-1 are temperature sensitive; colonial growth occurs at or above 32°C, while normal radial growth is observed at or below 25°C (17). Moreover, disruption of cot-1 results in severe growth defects as disruptants grow little, if at all. cot-1 expression is also photoinducible, and light illumination increases apical hyphal cell branching frequency (16). Immunoblotting with COT1 antibody detects the COT1 kinase in the cytoplasm, membrane, and nucleus (13).
Colletotrichum trifolii is the causal agent of alfalfa anthracnose (1). Pathogenicity of C. trifolii depends on proper growth and appressorium differentiation, which are believed to be regulated through the activation of intracellular signal transduction pathways in response to host stimuli (9). However, our understanding of the genes and biochemical pathways which govern hyphal growth and morphogenesis are not entirely clear. Previously, we cloned and characterized a serine/threonine protein kinase gene, tb3 (3), and the deduced amino acid sequence was similar to the COT1 sequence (70.4% identity). The carboxy-terminal domains of TB3 and COT1 are highly conserved but the amino-terminal regions diverge, and in TB3 a particularly distinct region containing homopolymeric glutamine repeats is present which is not found in COT1. Even with the structural divergence between the predicted cot-1 and tb3 gene products, tb3 complemented the cot-1 mutant of N. crassa, demonstrating the functional conservation of this kinase between a pathogen and a saprophytic fungus (3). This conservation between the TB3 and COT1 kinases suggests that the mechanisms which regulate hyphal growth and branching might be similar in these fungi, despite the fact that they have distinctly different lifestyles (pathogenic and saprophytic, respectively). In addition to N. crassa cot-1, several other TB3 kinase homologs have been identified. The Ustilago maydis ukc1 gene plays an important role in morphogenesis, pathogenesis, and pigmentation of this fungus. Disruption of the ukc1 gene alters cell morphology, blocks the formation of aerial hyphae, and promotes the development of pigmented cells (10). The TB3/COT1 family extends to higher eukaryotes as well. In the fission yeast Schizosaccharomyces pombe, the orb6-encoded protein kinase is required to maintain cell polarity during interphase (26). The Drosophila melanogaster tumor suppressor gene warts controls the amount and direction of cell proliferation and is required for normal morphogenesis (14). Mutations in the human TB3 homolog (DMPK) result in myotonic dystrophy (20). Thus, it is evident that appropriate expression of conserved TB3-like kinases is required for normal cell growth and differentiation across broad taxonomic distances.
In this report we show that, like COT1, TB3 expression is also light inducible. Furthermore, we examined localization of TB3 at different developmental stages in C. trifolii. Unlike COT1, TB3 can localize to the nucleus during hyphal growth. Additionally, in studies employing reporter gene expression in yeast, we show that TB3 kinase, and more specifically the polyglutamine-rich region, is capable of activating transcription.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The wild-type strain of C. trifolii race 1 (7) was used throughout this study. C. trifolii cultures were routinely grown at 25°C on YPSS agar plates (28). For isolation of protoplasts, DNA, RNA, and proteins, mycelia were grown in stationary liquid YPSS for 3 to 7 days. Conidia, germinating conidia, appressoria, and mycelia were collected as described elsewhere (27). Spores and sporulating mycelia were harvested from shake cultures grown for 4 to 5 days. By plating 10 ml of a spore suspension (105 to 106 spores/ml) in sterile glass petri dishes, germinating conidia were obtained in about 3 h and appressoria were formed after 6 h. Escherichia coli strain DH5α (GIBCO BRL) was used for the plasmid DNA transformation. E. coli strains were cultured in Luria-Bertani or 2xYT medium with 100 μg of carbenicillin per ml when required (21).
Nucleic acid techniques, PCR amplification, and DNA sequencing.
Standard techniques were employed in cloning and generation of recombinant plasmids (21). DNA was amplified by Pfu polymerase essentially as described by the manufacturer (Stratagene). DNA was sequenced by the dideoxy chain termination method (22). For Northern blotting, total RNA was isolated from conidia, germinating conidia, mycelia, and appressoria in TRIzol reagent (GIBCO-BRL) according to the manufacturer's instructions. Aliquots (30 μg) of RNA were loaded on 1% formaldehyde denaturing gels in 1× morpholinepropanesulfonic acid buffer and transferred to a charged nylon membrane (21). RNA blots were hybridized in 7% sodium dodecyl sulfate (SDS), 0.5 M Na2HPO4 (pH 7.2), and 2 mM EDTA. The filters were hybridized at 65°C overnight and washed as follows: once for 10 min at room temperature (RT) with low-stringency washing solution (40 mM Na2HPO4 [pH 7.2], 0.5% bovine serum albumin [BSA], 1 mM EDTA, 5% SDS) followed by 10 min at RT and two 10-min washes at 65°C with high-stringency washing solution (40 mM Na2HPO4 [pH 7.2], 1 mM EDTA, 1% SDS). A 25S ribosomal DNA fragment from Colletotrichum gloeosporioides (23) was used as a probe to ensure equivalent loading of RNA.
Western analysis.
C. trifolii mycelia were harvested and immediately frozen in liquid nitrogen, ground to a fine powder with a mortar and pestle, and suspended in extraction buffer (5 mM EDTA, 50 mM Tris-HCl [pH 7.5], 10 mM EGTA, 0.5% Triton, 0.3% β-mercaptoethanol, and fungal protease inhibitor cocktail [PIC; Sigma]). After 30 min of incubation on ice, samples were centrifuged for 15 min at 3,000 to 5,000 × g and the supernatant was placed in a new tube. Protein concentrations were determined by the method of Bradford (2). Protein samples (35 μg/lane) were electrophoretically separated and transferred to nitrocellulose filters. The TB3 antibodies used for immunoblotting were prepared against a 17-amino-acid peptide from an internal TB3 segment with 100% identity to COT1. Signals were detected by the ECL system (Amersham).
Light induction.
Approximately 107 conidia/ml were inoculated into 75 ml of YPSS medium, and cultures were grown at RT for 12 h with agitation (200 rpm) and further incubated for 2 to 3 days without shaking to promote vegetative growth. Mycelia were harvested by filtration, and the resulting mycelial pads were cut in half and either briefly illuminated with saturating white light (energy fluent rate ≥ 1.7 W/m2 in the blue region) or kept in the dark as a control. Light and dark exposures were terminated by quick-freezing the mycelial pads in liquid nitrogen. Total RNA was extracted as previously described and used for Northern analysis.
To evaluate the effect of light on hyphal growth and development, sterile glass microscope slides were overlaid with a thin (approximately 2-mm) layer of molten YPSS agar. A 20-μl aliquot of a spore suspension containing 0.5 × 106 spores per ml was placed at one end of each slide and then streaked across the agar to ensure an even distribution of spores. The inoculated slides were then placed in 150-mm-diameter plastic petri dishes which were then placed in a plastic box containing a water-saturated sponge, and the box was covered with plastic wrap to ensure a humid environment. The cultures were incubated in a lighted incubator. For the dark control, petri dishes were covered with aluminum foil. A coverslip was placed over the resulting colonies, and following selected time points the cultures were examined using a Zeiss microscope with differential interference contrast (DIC) optics. Images were captured via a charge-coupled device camera and processed using Image-Pro Plus 3.0 for Windows (Media Cybernetics).
Construction of TB3::GFP fusions and yeast expression vectors.
To examine the subcellular distribution of TB3, TB3 cDNA including 460 bp of upstream sequence was translationally fused to the green fluorescent protein (GFP) gene (24). A 2.6-kb TB3 DNA fragment was amplified from the fungal cDNA by PCR with the following specific primer pairs that generate EcoRI and BamHI sites at the 5′ ends, respectively: primer TB3-F, 5′-CGGAATTCATTAGTGCCAGCGGGTTG; primer TB3-R, 5′-CGGGATCCACGGAAGTTGTTGTCG. The resultant PCR fragment spans the 5′ end of the TB3 promoter and the 3′ end of the TB3 open reading frame (ORF). pCT74 containing GFP (SGFP-TYG) was used, and a 1.4-kb DNA fragment was generated by PCR using the following additional primers containing a BamHI and XbaI site at the 5′ termini, respectively: primer SGFP-F, 5′-CGGGATCCAATCCCATGGTGA; primer SGFP-R, 5′-GCTCTAGATTCTCATGTTTGAC). For construction of the TB3::GFP fusion, the 2.6-kb DNA fragment from EcoRI and BamHI cleavage was cloned into the EcoRI/BamHI site of vector pBluescript SK(+) to give plasmid pTB3-1. The 1.4-kb BamHI/XbaI DNA fragment was ligated into the BamHI/XbaI site of pTB3-1 to create pTB3-2, which encoded a TB3::GFP fusion consisting of 2.6 kb of the TB3 upstream sequence and full-length ORF fused in frame with the engineered GFP reporter gene. The correct translational fusion was confirmed by DNA sequence analysis. The hygromycin B resistance gene cassette was excised from pCT74 and ligated into the SalI site of pTB3-2 to give plasmid pTB3-3. pTB3-3 containing the TB3::GFP fusion and the hygromycin resistance gene for selection was transformed into protoplasts of C. trifolii as described previously (27).
To analyze the transcriptional activation ability of TB3 in yeast, two yeast expression vectors were constructed as follows. pAS2-1 (Clontech) is a yeast expression vector containing the GAL4 DNA binding domain (GAL4BD) under transcriptional control of the alcohol dehydrogenase 1 promoter (ADH1) and TRP1 as the selectable marker. pAS2-1 was digested with EcoRI and BamHI. The full-length C. trifolii TB3 ORF cDNA fragment was amplified from a previously constructed plasmid (pB42AD-TB3FL) with a pair of primers, FLTB3-L (5′-CGGAATTCATGGATAACAATAATAACCG) and FLTB3-R (5′-CGGGATCCTCAACGGAAGTTGTTGTCGA). In addition, the TB3 polyglutamine tract cDNA fragment was amplified from pTB3 (3) by primers FLTB3-L (5′-CGGAATTCATGGATAACAATAATAACCG) and GLUTB3-R (5′-CGGGAT CCGTTGAAGTATCCAGAAGGTG). Fragments containing the EcoRI site at the 5′ end and the BamHI site at the 3′ end were ligated to similarly digested pAS2-1 to create pGAL4BD-TB3FL and pGAL4BD-TB3GLU, respectively.
The yeast strain Y190 (Clontech) is auxotrophic for TRP1 and LEU2 and contains the GAL1 upstream activating sequence (UAS GAL1) fused to the HIS3 reporter gene. Three tandem copies of the GAL4 consensus sequence were fused to the lacZ reporter gene. Yeast cells were transformed by pBD-TB3FL and pBD-TB3GLU and cultured on SD/Gal/Raf/-Trp. The empty vector (pAS2-1) served as a negative control, and the plasmid (pVA3-1) containing the mouse p53 transcription factor was used as a positive control. Transformants grown on SD/Gal/Raf/-Trp were tested for β-galactosidase activity by both colony filter and o-nitrophenyl-β-d-galactopyranoside (ONPG) assays, as described by the manufacturer (Clontech).
Immunoblotting.
The subcellular fractions of C. trifolii were prepared as described by Gorovits et al. (13). Briefly, C. trifolii mycelial samples were grown with shaking for 2 to 3 days and then frozen in liquid nitrogen, pulverized, and homogenized in protein extraction buffer. After a 20-min low-speed centrifugation (4,000 × g, 4°C), the soluble cytosolic fraction was initially separated from the particulate crude extract. The pellet (nuclei plus membranes) was treated with protein extraction buffer amended with 110 mM KCl for 30 min at 4°C. The particulate fraction was clarified by ultracentrifugation at 100,000 × g (1 h, 4°C). After ultracentrifugation, the soluble supernatant (nuclear fraction, verified by 4′,6′-diamidino-2-phenylindole [DAPI] staining) was removed from the pellet (membrane fraction). The membrane fraction was washed with extraction buffer and resuspended in the same buffer with 110 mM KCl. Glycerol (10%, vol/vol) stocks were prepared from the different fractions and maintained at −70°C prior to Western blotting.
Microscopy.
To determine TB3 localization, C. trifolii mycelia were prepared for indirect immunofluorescence. Conidia (107 to 108/ml) in YPSS medium were inoculated onto coverslips and grown overnight at RT. Coverslips were brought to 4% formaldehyde and 0.25% glutaraldehyde (final concentrations) for 5 min and then resuspended in 2 ml of fresh 4% formaldehyde, 0.25% glutaraldehyde, 50 mM K+-piperazine-N,N′-bis(2-ethanesulfonic acid) (K+-PIPES) (pH 6.9), 25 mM EGTA, 1% dimethyl sulfoxide, and 5 mM MgSO4 and incubated for 90 min at RT with gentle agitation. The cells were washed three times in 50 mM K+-PIPES (pH 6.9), 1 mM EGTA, and 1 mM MgSO4 (PEM). The washed cells were resuspended in 2 ml of PEM supplemented with 1 M sorbitol, 1% BSA (PEMSB), and fungal PIC. The fixed mycelia were digested with lysing enzyme (Sigma) solution as described by Kohn et al. (15). The cells were incubated for 70 min at RT with gentle agitation, washed three times with PEMSB, and resuspended in 0.5 ml of PEMS with PIC. One hundred microliters of permeabilization solution (50 mM K+-PIPES [pH 6.9], 1 mM EGTA, 10% dimethyl sulfoxide, 1 mM MgSO4, 0.2% Nonidet P-40 [NP-40]) was added to the cells and cells were incubated for 10 min. The solution was removed by aspiration and cells were washed once with PEM and then twice with TBST (50 mM Tris-HCl [pH 8.0], 200 mM NaCl, 0.1% Tween 20). To reduce background fluorescence, cells were incubated for 30 min in 0.25 M triethanolamine (pH 7.5), 1 mM EGTA, 1 mM MgSO4, and then washed twice for 2 min each with TBST. Rabbit polyclonal anti-TB3 primary antibodies (1:50 dilution) in TBST plus 1% BSA and 0.04% NP-40 with PIC (incubation buffer) were added and incubated for 1 h in a moist chamber. Cells were then washed five times with TBST for 3 min each and then incubated with the goat anti-rabbit immunoglobulin G secondary antibody conjugated to Texas Red (1:20 dilution; Molecular Probes) in incubation buffer for 1 h in the dark. Following incubation with the secondary antibody, the coverslips containing the treated cells were washed as before and covered with a mounting medium (90% glycerol, 10% phosphate-buffered saline [pH 9.0], and 1 mg of β-phenylenediamine/ml). Finally, coverslips were sealed with clear nail polish as described previously (19). To delineate nuclei, cells were treated with DAPI for 10 min in the dark. Controls included omitting both antibodies, either primary or secondary antibody alone. Slides were viewed with a Zeiss Axioskop microscope using immunofluorescent Texas Red- and DAPI-specific filters, respectively.
Phosphatase assay.
Hyphae were harvested by inoculation of 500 ml of YPSS in a 1-liter flask with a ≈15-mm3 agar-mycelium plug of C. trifolii obtained from the advancing margin of a YPSS culture. This liquid culture was incubated at RT, with constant shaking at 200 rpm, for 2 days and then incubated for an additional 3 to 6 days without shaking. The culture was harvested by filtration and lyophilized. The dried samples were ground to a powder in liquid nitrogen and suspended in 300 μl of extraction buffer (5 mM EDTA, 50 mM Tris-HCl [pH 7.5], 10 mM EGTA, 0.5% Triton, 0.3% β-mercaptoethanol, and fungal PIC). After 30 min of incubation on ice, samples were centrifuged for 15 min at 3,000 to 5,000 × g, and the supernatant was placed in a new tube. Protein concentrations were determined by the method of Bradford (2). An aliquot of the cell extracts was incubated for 1 h at 30°C in 100 μl of 50 mM Tris, 0.1 mM EDTA, 5 mM dithiothreitol, 0.01% Brij 35, pH 7.5, in the presence or absence of 400 U of lambda protein phosphatase (New England Biolabs). Samples were separated by gel electrophoresis and either stained with Coomassie blue or detected by Western blotting with TB3 antibody.
RESULTS
Light increases hyphal branching frequency and TB3 expression in C. trifolii.
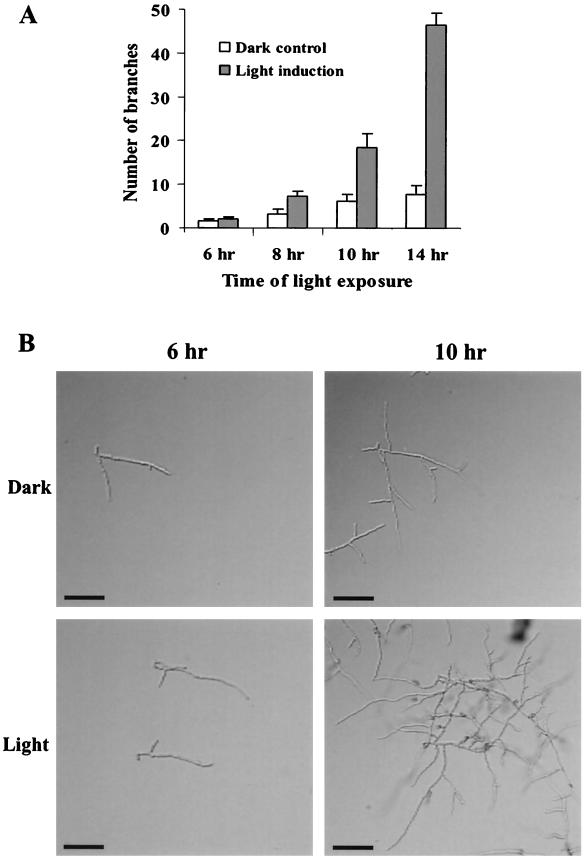
Light has been shown to be an important signal regulating developmental programs in filamentous fungi (16). To evaluate effects of light on C. trifolii hyphal growth and development, spores were inoculated onto sterile glass microscope slides containing a thin layer of YPSS agar and grown in the presence or absence of constant light. A time course was conducted with various intervals of light treatment, and the number of branches from the original germ tube was recorded 6 h after germination (Fig. 1A). Figure 1B shows hyphal branching following 6- and 10-h illumination periods. Results indicated that illumination significantly increased apical hyphal branching frequency.
FIG. 1.
Effect of light on hyphal morphology in C. trifolii. (A) Spores were inoculated onto sterile glass microscope slides with a thin layer of YPSS agar and grown in constant light or in complete darkness (control). Following the indicated time intervals, the number of branches from the original germ tube was recorded 6 h after germination. Results represent the means (± standard deviations) of three separate experiments. (B) Hyphal branching following 6- and 10-h illumination periods. Cultures were observed by using DIC. Bars, 20 μm.
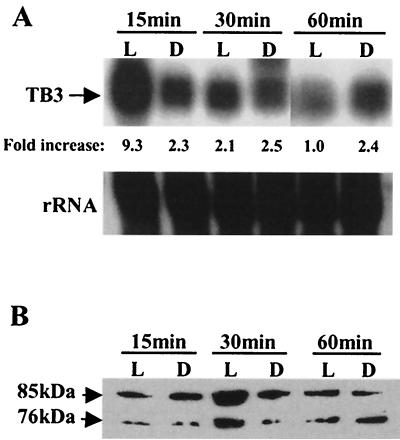
We then examined the effect of light on tb3 expression. Mycelia were grown in YPSS and exposed to light for different time intervals. RNA and protein were extracted from mycelial samples, and the expression of tb3 was monitored by Northern and Western blotting (Fig. 2A and B, respectively). As for N. crassa cot-1, tb3 expression was also light inducible. Following 15 min of exposure to white light, significantly high levels of tb3 transcription were observed. tb3 transcription levels lowered after 30 min of illumination and further decreased as the illumination period increased. Consistent with the changes in mRNA levels, TB3 protein levels peaked after 30 min of light exposure. TB3 expression remained constant under dark conditions. These results show a correlation between light, tb3 expression, and hyphal branching. In these experiments, two immunoreactive forms of the TB3 protein (≈76 and ≈85 kDa) were detected (Fig. 2B). The ≈76-kDa protein corresponds to the predicted amino acid sequence, based on the tb3 gene sequence.
FIG. 2.
Light regulation of TB3 expression in C. trifolii. Mycelia were grown overnight in the dark in YPSS medium and harvested by filtration. The mycelial pads were cut in half and either illuminated (L lanes) for 15, 30, or 60 min or kept in the dark (D lanes). Light and dark treatments were terminated by quick-freezing the mycelial pads in liquid nitrogen. (A) Northern blots of total RNA (30 μg/lane) prepared from C. trifolii mycelia after light exposure, with TB3 as a probe. The 25S ribosomal DNA fragment from C. gloeosporioides was used to show equivalent RNA loading. (B) Western blots of lysates (35 μg of total protein per lane) prepared from the same treated mycelia as in panel A. TB3 protein was detected with TB3 antibody.
Developmental expression of TB3.
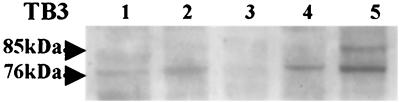
Previous results (3) showed that tb3 mRNA was most highly expressed 1 h after conidial germination under inducing conditions (hard surface and nutrient-rich media). To further characterize expression patterns of TB3, TB3 steady-state protein levels were compared during various fungal developmental stages. Basal levels of TB3 were observed in conidia and germinating conidia (Fig. 3, lanes 1 and 2). TB3 expression was not detected in appressoria (Fig. 3, lane 3). The highest levels of TB3 expression were observed during hyphal growth (Fig. 3, lanes 4 and 5). These data suggest that TB3 is required during proliferative growth and is unnecessary or down regulated during isotropic or nonpolar growth. Interestingly, the two immunoreactive forms of TB3 were detected by TB3 antibody in still mycelial culture, but not in shake culture (Fig. 3, lanes 4 and 5).
FIG. 3.
Developmental Western analysis of C. trifolii TB3. Equivalent concentrations of total fungal proteins were used, as determined by Bradford assays (2). Lane 1, conidia; lane 2, germinating conidia; lane 3, mature appressoria; lane 4, mycelial shake culture; lane 5, mycelial still culture.
Treatment of the TB3 protein with phosphatase.
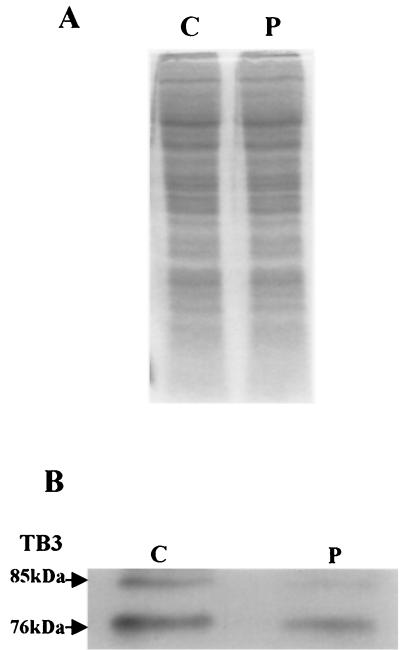
As mentioned, two forms of TB3 protein (of ≈76 and ≈85 kDa) were detected in fungal hyphae undergoing growth in still culture (Fig. 2B and 3, lane 5). To examine whether these different migrating forms of TB3 are due to different phosphorylation states, aliquots of protein extracts were incubated with lambda phosphatase and analyzed by Western blotting. Phosphatase treatment eliminated the ≈85-kDa band, suggesting that posttranslational modification of TB3 by phosphorylation occurs and may be responsible for the difference in migration of the two protein bands (Fig. 4).
FIG. 4.
Phosphatase treatment of the TB3 protein. C. trifolii extracts were prepared as described in Materials and Methods and divided in half, with one portion treated with lambda phosphatase. Samples were separated by gel electrophoresis and either stained with Coomassie blue (A) or treated with TB3 antibody (B). C, no phosphatase; P, phosphatase added.
Localization of TB3: immunofluorescence microscopy.
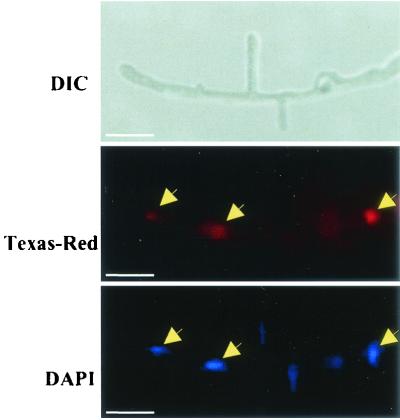
TB3 localization was examined by indirect immunofluorescence microscopy (18). C. trifolii hyphae were fixed and treated with purified TB3 antibody and secondary antibody conjugated to Texas Red. These samples were also treated with the DNA stain DAPI. In this assay, TB3 appears red (Texas Red), while nuclei appear blue. In C. trifolii hyphae, antibodies prepared against TB3 polypeptide revealed nuclear association in preparations from C. trifolii hyphae (Fig. 5, middle and bottom panels)
FIG. 5.
Immunofluorescence microscopy of TB3. Hyphae were fixed and dual stained with TB3 antibody coupled to anti-rabbit immunoglobulin G conjugated to Texas Red. Hyphae were observed by using DIC and by fluorescence microscopy (Texas Red). Nuclei were visualized with DAPI. Arrows indicate the nuclear localization of the TB3 proteins. The lack of equivalent Texas Red staining is presumably due to antibody inaccessibility to these nuclei. Bars, 10 μm.
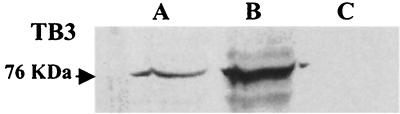
Localization of TB3: immunoblotting.
Three different subcellular preparations including cytoplasmic, membrane, and nuclear fractions were obtained from C. trifolii mycelia. Western analysis (Fig. 6) indicated that TB3 was present in both the nuclear and cytoplasmic fractions (lanes A and B, respectively), but it was undetectable in membrane fractions (lane C).
FIG. 6.
TB3 distribution in subcellular fractions of C. trifolii. Nuclear (A), cytoplasmic (B), and membrane (C) fractions were prepared from hyphae grown for 2 to 3 days with constant shaking, and total proteins were hybridized with TB3 antibodies by Western analysis.
Localization of TB3: reporter gene.
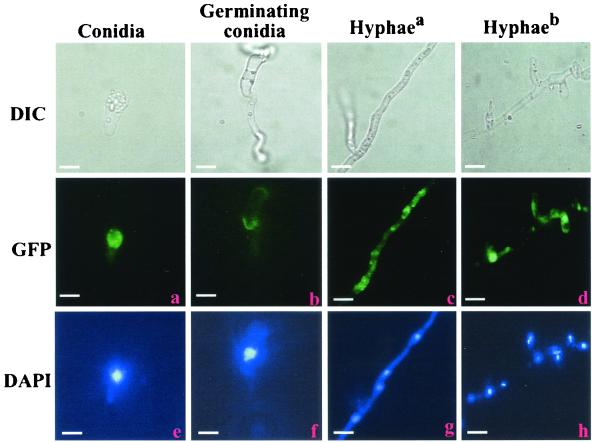
A plasmid vector was constructed in which GFP was fused to full-length tb3 under the control of the tb3 promoter. Following transformation, 25 hygromycin-resistant strains expressing the TB3::GFP fusion were isolated, and 10 independent single-copy insertion transformants (determined by Southern blotting) (data not shown) were examined by fluorescence microscopy. Nuclear DNA was stained with DAPI. Comparison of GFP fluorescence (Fig. 7a to d) with DAPI fluorescence (Fig. 7e to h) showed that TB3 localization varied depending on developmental stage. During conidiation, TB3 was primarily cytoplasmic (Fig. 7a and e). As germ tubes elongated, TB3 proteins were concentrated in the regions close to membranes (Fig. 7b and f). In hyphae grown for 2 to 4 days, TB3 appeared in the cytoplasm with little fluorescence in the nucleus (Fig. 7c and g), but as hyphal growth increased over 1 week's time TB3 proteins were primarily found in the nucleus (Fig. 7d and h).
FIG. 7.
Cellular distribution of TB3::GFP fusion proteins. TB3-GFP fusion proteins were expressed in wild-type C. trifolii, and cells were observed as described in the legend for Fig. 5. Hyphaea, hyphae grown for 2 to 3 days; Hyphaeb, hyphae grown for 1 week. Bars, 10 μm.
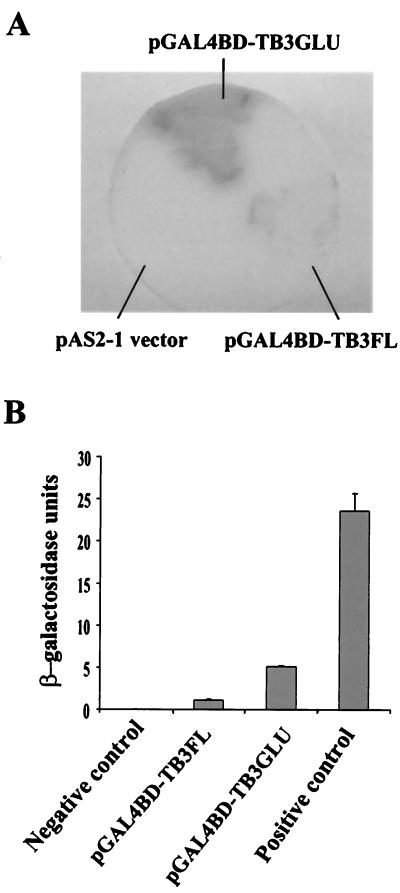
Transcriptional activity of TB3 protein in yeast.
The N-terminal regulatory domain of TB3 contains two relatively long stretches of glutamine residues (32 of 43 residues); this is a primary and perhaps important distinction between the C. trifolii TB3 and N. crassa COT1 kinases (3). Polyglutamine tracts are associated with mediation of protein-protein interactions, often involving transcriptional activation (12, 25). Coupled with the observation that TB3 was localized to the nucleus, we were interested in determining whether TB3 and more specifically the polyglutamine-rich region can activate transcription. TB3 transcriptional activation was therefore evaluated in yeast. The full-length TB3 ORF and the TB3 polyglutamine tracts were fused to the GAL4 DNA binding domain (GAL4BD-TB3GLU and GAL4BD-TB3FL), respectively, and transformed into yeast Y190. The transformants grown on SD/Gal/Raf/-Trp had β-galactosidase activity (Fig. 8). The β-galactosidase activity in the truncated TB3GLU transformants was greater than that in the full-length TB3FL transformants. Thus, full-length TB3 as well as TB3 polyglutamine tracts could activate transcription in yeast, but the glutamine tracts alone were more effective in activating transcription.
FIG. 8.
(A) β-Galactosidase activity analyses by yeast colony filter assay. Yeast were transformed by pAS2-1, pGAL4BD-TB3FL, or pGAL4BD-TB3GLU and grown on SD/Gal/Raf/-T plates. The blue color is indicative of β-galactosidase activity expressed by yeast. (B) β-Galactosidase activity analyses in a yeast ONPG assay. The data represent the means (± standard deviations) of three separate experiments.
DISCUSSION
All living cells sense and respond to environmental cues. Since mycelia are the primary means by which fungi forage for nutrients, proper hyphal regulation is extremely important. In filamentous fungi, light has been known to play a key role in both conidiation as well as vegetative growth. For example, conidiation of Colletotrichum graminicola is light dependent. Dark-grown cultures of this fungus have a fluffy hyphal colony morphology and are unable to form conidia, whereas illumination of such cultures results in abundant conidia with fewer hyphae (J. Wang and R. M. Hanau, 18th Fungal Genet. Conf., poster 37, 1995). The vegetative phase of N. crassa is, in part, regulated by blue light which, as discussed, stimulates hyphal branching frequency (16). In this paper, we show that expression of the C. trifolii TB3 kinase was induced by illumination and correlated to an increase in hyphal branching. The data presented here provide evidence for a relationship between hyphal growth, expression of TB3, and light. As with N. crassa, light illumination increased apical hyphal branching frequency in C. trifolii, suggesting a role for light in hyphal proliferation. Previous studies have demonstrated that N. crassa COT1 is required for hyphal elongation and branching, and disruption of cot-1 prevents hyphal growth (29). Since C. trifolii tb3 complemented the cot1 mutant to normal hyphal growth (3) and since light influenced hyphal branching frequency and TB3 expression similar to that of COT1, it is reasonable to hypothesize that TB3 may function to regulate hyphal growth in C. trifolii in a manner similar to that of COT1. Disruption of the tb3 gene in C. trifolii is under way.
As shown in Fig. 3, TB3 expression was regulated during morphogenesis. Higher levels of the TB3 proteins were found during hyphal growth but were undetectable during appressorium formation (Fig. 3, lane 3). Appressoria are highly differentiated and develop as cell division ceases. The down regulation of TB3 in appressorium development, coupled with the induction of expression during hyphal growth, suggests TB3 affects proliferative growth.
In N. crassa, two COT1 isoforms (≈73 and ≈67 kDa) are expressed in both wild-type and cot-1 strains grown at permissive temperature (25°C). The lower-molecular-mass isoform was not detected in cot-1 strains grown at the restrictive temperature. Interestingly, following a shift from restrictive to permissive temperatures, the ≈67-kDa COT1 isoform reappeared (13), which suggested that this isoform may be required for or regulate hyphal elongation. Consistent with N. crassa COT1 expression, TB3 antibody also detected two TB3 kinase isoforms (≈76 and ≈85 kDa) readily observed in stationary culture (Fig. 2B and 3, lane 5). Besides the ≈76-kDa peptide, which corresponds to the predicted amino acid sequence, the unique presence of the higher-molecular-mass ≈85-kDa TB3 isoform in stationary culture may be functionally significant. As protein phosphorylation-dephosphorylation is a common posttranslational modification regulating signal transduction pathways (8), we investigated whether this modification was responsible for these TB3 isoforms. Results indicated that the higher-molecular-mass form of TB3 protein (≈85 kDa) disappeared following phosphatase treatment. Under nonshaking growth conditions, C. trifolii grows almost exclusively vegetatively, suggesting that the ≈85-kDa TB3 isoform may be involved in regulation of hyphal growth.
Interestingly, the immunoblotting assay only detected the ≈76-kDa protein, with a higher level in the cytoplasmic fraction than in the nuclear fraction. We speculate that this is due to the early time point of sample harvest and the antibody specificity. TB3 antibody only detects the ≈76-kDa protein in shake culture, with TB3 largely distributed in cytoplasm during early growing periods (2 to 3 days). In the immunoblotting assay, the samples were harvested later in the growing period (see Materials and Methods).
Two stretches of polyglutamine residues are found in the N-terminal regulatory domain of C. trifolii TB3 but not in N. crassa COT1 or fission yeast ORB6 (3, 10, 29). Homopolymeric tracts of glutamine are not uncommon and have been found in a number of mammalian transcription factors (5, 6, 11, 12, 25). Moreover, recent studies have shown that homopolymeric stretches of glutamines are also found in other serine/threonine protein kinases which have a high degree of similarity with C. trifolii TB3. U. maydis ukc1 encodes two homopolymeric glutamine tracts in the N-terminal one-third of the polypeptide (10). Glutamine-rich regions are also found in the polypeptide encoded by the Drosophila tumor suppressor gene warts (14). While it is tempting to speculate that these products function as transcriptional activators, the role of the highly conserved regions has not yet been determined in any of TB3 family members. Here we used a yeast two-hybrid system to examine whether TB3 had the capability for transcriptional activation. β-Galactosidase activity was detected in yeast cells transformed with the TB3GLU-GAL4BD or TB3FL-GAL4BD fusion constructs. We also found that the β-galactosidase activity in the TB3GLU (TB3 polyglutamine repeats) transformants was higher than that in the TB3FL (full-length TB3) transformants, although both activities were considerably less than that of the positive yeast control. These data indicate that TB3FL and TB3GLU stimulate transcription in yeast. The different activities between the full-length and polyglutamine constructs might be due to differences in protein conformation. TB3GLU is composed of only 95 amino acids and contains only two stretches of polyglutamine repeats. It is likely that the TB3GLU-GAL4BD fusion binds more readily to transcription factors and thus is more active. Alternatively, the entire protein might contain elements which negatively modulate the ability for transactivation which are not present in the truncated construct. Thus, based on the transcriptional activation of TB3 in yeast, TB3 nuclear localization may have functional relevance.
Supporting this idea, residues 229 to 232 (KRAR) of the TB3 protein satisfy the requirement of a minimal nuclear localization signal (NLS) by having the tetrameric sequence K(R/K)X(R/K), in which X is K, R, P, V, or A (4). Flanking the putative NLS are potential phosphorylation sites. Consistent with an NLS in the deduced amino acid sequence is the fact that TB3 was found in the nucleus. Indirect immunofluorescence microscopy showed TB3 signals in nuclei, and Western analysis showed the TB3 protein in both the nuclear and cytoplasmic fractions of hyphae. Moreover, C. trifolii transformed with a TB3::GFP fusion construct showed strong nuclear fluorescence in older hyphae. These observations are in contrast to studies in N. crassa where the COT1 protein was primarily associated with membrane fractions and dispersed throughout the cytoplasm but was found only in the cytoplasmic fractions when a cot-1 mutant was grown at the restrictive temperature (13). Our evidence thus suggests that TB3 may be a transcription factor mediating light-dependent response pathways involved in fungal hyphal growth and development.
Acknowledgments
We thank Stephen Memmott and Youngsil Ha for technical suggestions and helpful comments on the manuscript.
This work was supported by BARD grant no. US2814-96.
REFERENCES
- 1.Barnes, D. K., S. A. Ostazeske, J. A. Shillinger, and C. H. Hanson. 1969. Effect of anthracnose (Colletotrichum trifolii) infection on yield, stand and vigor of alfalfa. Crop Sci. 9:344-346. [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Buhr, T. L., S. Oved, G. M. Truesdell, C. Huang, O. Yarden, and M. B. Dickman. 1996. A kinase-encoding gene from Colletotrichum trifolii complements a colonial growth mutant of Neurospora crassa. Mol. Gen. Genet. 251:565-572. [DOI] [PubMed] [Google Scholar]
- 4.Chelsky, D., R. Ralph, and G. Jonak. 1989. Sequence requirements for synthetic peptide-mediated translocation to the nucleus. Mol. Cell. Biol. 9:2487-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Courey, A. J., and R. Tjian. 1988. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell 55:887-898. [DOI] [PubMed] [Google Scholar]
- 6.Cox, G. W., L. S. Taylor, J. D. Willis, G. Melillo, R. L. White III, S. K. Anderson, and J. J. Lin. 1996. Molecular cloning and characterization of a novel mouse macrophage gene that encodes a nuclear protein comprising polyglutamine repeats and interspersing histidines. J. Biol. Chem. 271:25515-25523. [DOI] [PubMed] [Google Scholar]
- 7.Dickman, M. B. 1988. Whole cell transformation of the alfalfa fungal pathogen Colletotrichum trifolii. Curr. Genet. 14:241-246. [Google Scholar]
- 8.Dickman, M. B., and O. Yarden. 1999. Serine/threonine protein kinases and phosphatases in filamentous fungi. Fungal Genet. Biol. 26:99-117. [DOI] [PubMed] [Google Scholar]
- 9.Dickman, M. B., T. L. Buhr, G. M. Truesdell, V. Warwar, and C. Huang. 1995. Molecular signals during the early stages of alfalfa anthracnose. Can. J. Bot. 73(Suppl. 1):1169-1177. [Google Scholar]
- 10.Durrenberger, F., and J. Kronstad. 1999. The ukc1 gene encodes a protein involved in morphogenesis, pathogenicity and pigment formation in Ustilago maydis. Mol. Gen. Genet. 261:281-289. [DOI] [PubMed] [Google Scholar]
- 11.Emili, A., J. Greenblatt, and C. J. Ingles. 1994. Species-specific interaction of the glutamine-rich domain of Sp1 with the TATA-box binding protein. Mol. Cell. Biol. 14:1582-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerber, H. P., K. Seipel, O. Georgiev, M. Hofferer, M. Hug, S. Rusconi, and W. Schaffner. 1994. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science 263:808-811. [DOI] [PubMed] [Google Scholar]
- 13.Gorovits, R., K. A. Sjollema, J. H. Sietsma, and O. Yarden. 2000. Cellular distribution of COT1 kinase in Neurospora crassa. Fungal Genet. Biol. 30:63-70. [DOI] [PubMed] [Google Scholar]
- 14.Justice, R. W., O. Zilian, D. F. Woods, M. Noll, and P. J. Bryant. 1995. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9:534-546. [DOI] [PubMed] [Google Scholar]
- 15.Kohn, L. M., E. Stasovski, I. Carbone, J. Royer, and J. B. Anderson. 1991. Mycelial incompatibility and molecular markers identify genetic variability in field populations of Sclerotinia sclerotiorum. Phytopathology 81:480-485. [Google Scholar]
- 16.Lauter, F. R., U. Marchfelder, V. E. A. Russo, C. T. Yamashiro, E. Yatzkan, and O. Yarden. 1998. Photoregulation of cot-1, a kinase-encoding gene involved in hyphal growth in Neurospora crassa. Fungal Genet. Biol. 23:300-310. [PubMed] [Google Scholar]
- 17.Mitchell, M. B., and H. K. Mitchell. 1954. A partial map of linkage group D in Neurospora crassa. Proc. Natl. Acad. Sci. USA 40:436-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peleg, Y., R. Addison., R. Aramayo, and R. L. Metzenberg. 1996. Translocation of Neurospora crassa transcription factor NUC-1 into the nucleus is induced by phosphorus limitation. Fungal Genet. Biol. 20:185-191. [DOI] [PubMed] [Google Scholar]
- 19.Pringle, J. R., R. A. Preston, A. E. Adams, T. Stearns, D. G. Drubin, B. K. Haarer, and E. W. Jones. 1989. Fluorescence microscopy methods for yeast. Methods Cell Biol. 31:357-435. [DOI] [PubMed] [Google Scholar]
- 20.Sabouri, L. A., M. S. Mahadevan, M. Narang, D. S. Lee, L. C. Surh, and R. G. Korneluk. 1993. Effect of the myotonic dystrophy (DM) mutation on mRNA levels of the DM gene. Nat. Genet. 4:233-238. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. A. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephensen, S. A., J. R. Green, J. M. Manners, and D. J. Maclean. 1997. Cloning and characterization of glutamine synthetase gene from Colletotrichum gloeosporioides and demonstration of elevated expression during pathogenesis on Stylosanthes guianensis. Curr. Genet. 31:447-454. [DOI] [PubMed] [Google Scholar]
- 24.Susana, G., R. Odile, B. Evelyne, B. Spencer, and G. Pierre. 1998. Mitochondria localization of a NADR-dependent isocitrate dehydrogenase isoenzyme by using the green fluorescent protein as a marker. Proc. Natl. Acad. Sci. USA 95:7813-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka, M., W. M. Clouston, and W. Herr. 1994. The oct-2 glutamine-rich and proline-rich activation domains can synergize with each other or duplicate of themselves to activate transcription. Mol. Cell. Biol. 14:6046-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verde, F., D. J. Wiley, and P. Nurse. 1998. Fission yeast orb6, a Ser/Thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc. Natl. Acad. Sci. USA 95:7526-7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang, Z., and M. B. Dickman. 1999. Colletotrichum trifolii mutants disrupted in the catalytic subunit of cAMP-dependent protein kinase are nonpathogenic. Mol. Plant Microbe Interact. 12:430-439. [DOI] [PubMed] [Google Scholar]
- 28.Yang, Z., and M. B. Dickman. 1997. Regulation of cAMP and cAMP-dependent protein kinase during conidial germination and appressorial formation in Colletotrichum trifolii. Physiol. Mol. Plant Pathol. 50:117-127. [Google Scholar]
- 29.Yarden, O., M. Plamann, D. J. Ebbole, and C. Yanofsky. 1992. cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J. 11:2159-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]