Kinetoplast DNA (kDNA) is the most structurally complex mitochondrial DNA in nature. Unique to the single mitochondrion of unicellular flagellates of the order Kinetoplastida, kDNA is best known as a giant network of thousands of catenated circular DNAs (an electron micrograph of a network is shown in Fig. 1A). The kDNA circles are of two types, maxicircles and minicircles. Maxicircles usually range from 20 to 40 kb, depending on the species, and are present in a few dozen identical copies per network. Minicircles, present in several thousand copies per network, are usually nearly identical in size (0.5 to 10 kb, depending on the species) but are heterogeneous in sequence. Maxicircles encode typical mitochondrial gene products (e.g., rRNAs and subunits of respiratory chain complexes) but, remarkably, some of the protein-coding genes are encrypted. To generate functional mRNAs, the cryptic maxicircle transcripts undergo posttranscriptional modification via an intricate RNA editing process that involves insertion and deletion of uridine residues at specific sites in the transcripts. The genetic information for editing is provided by guide RNAs (gRNAs) that are mostly encoded by minicircles, although a few are encoded by maxicircles. Encoding gRNAs is the only known function of minicircles, and some organisms that edit extensively (such as Trypanosoma brucei) possess about 200 different minicircle sequence classes in their network to provide sufficient gRNAs. For reviews on RNA editing, see references 13, 17, and 46.
FIG. 1.
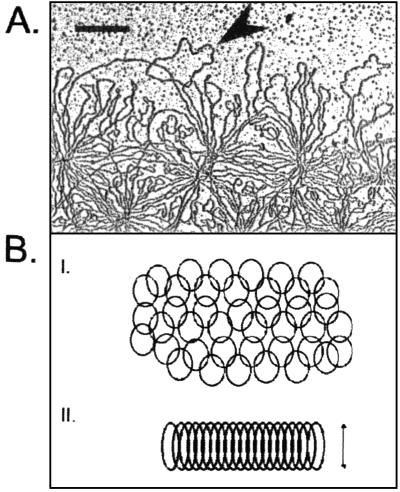
kDNA network structure. (A) Electron micrograph of the periphery of an isolated kDNA network from T. avium. Loops represent interlocked minicircles (the arrowhead indicates a clear example). Bar, 500 nm. (B) Diagrams showing the organization of minicircles. (I) Segment of an isolated network showing interlocked minicircles in a planar array. (II) Section through a condensed network disk in vivo showing stretched-out minicircles. The double-headed arrow indicates the thickness of the disk, which is about half the circumference of a minicircle.
Almost all knowledge of kDNA, involving both its unusual network structure and its novel mechanism of RNA editing, is derived from studies of a small group of familiar organisms of the suborder Trypanosomatina, all of which are parasitic. These organisms have been extensively investigated either because they are pathogenic (e.g., T. brucei, Trypanosoma cruzi, Leishmania spp., and Phytomonas spp.) or because they are models for these pathogens (e.g., Crithidia fasciculata and Leishmania tarentolae). Studies of these organisms have led to the strong impression that all kDNAs are essentially the same, with only minor variations. These many studies have provided a classical view of kDNA structure.
However, there were early indications of variations on this classical theme. Electron microscopy studies during the 1970s revealed that some members of the early-branching suborder Bodonina, which includes free-living as well as parasitic species, had kDNA that in vivo seemed at odds with the classical network structure (6, 7, 52). For example, in some species, the kDNA seemed to be dispersed throughout the mitochondrial matrix, either uniformly or in multiple foci, rather than being condensed in one region, as it is in species containing a network. This situation was clarified in 1986 when molecular studies revealed that the kDNA of Bodo caudatus contained minicircles that are not catenated (20). These early reports inspired us to initiate a survey of kDNA structures from a variety of kinetoplastid organisms. In the last few years, our laboratories and others have found that kDNA actually exists in a wonderful diversity of structures.
In this review, we first summarize briefly what is known about the classical kDNA network structure and its mode of replication. Next, we address recently discovered novel kDNA structures. We then provide a phylogenetic analysis to trace a probable pathway for the evolution of the kDNA network. Finally, we discuss some biological implications of the kDNA network and other kDNA structures.
kDNA: CLASSICAL VIEW
Network structure.
The best known kDNA structure is the network from C. fasciculata. This network contains 5,000 minicircles and about 25 maxicircles. The circles are catenated to form a planar network that has a topology resembling that of chain mail in medieval armor (Fig. 1B). The isolated network, when viewed by electron microscopy, is elliptically shaped and measures about 10 by 15 μm (45). Each minicircle is catenated to about three neighbors, and each linkage is a single interlock (8, 41). Minicircles are covalently closed, except when the network undergoes replication; in addition, unlike circular DNAs in other cell types, they are not supercoiled (41). Minicircles not only contain sequences encoding one or more gRNAs but also have other characteristics (Table 1). One is the sequence-dependent bent helix, found in minicircles of most trypanosomatids (34). Another is the universal minicircle sequence (UMS), a 12-nucleotide motif that is conserved in all trypanosomatids and that is part of the minicircle replication origin (37). The topology of network maxicircles is less well understood, although it is clear that they are all catenated to each other as well as to minicircles (44).
TABLE 1.
Characteristics of minicircles and minicircle-like sequences
| Species | kDNA type | Encodes gRNAs | UMSa | Bent DNAb |
|---|---|---|---|---|
| T. brucei | Network | Yes | GGGGTTGGTGTAc | Yes |
| B. saltans | Pro-kDNA | Yes | GGGGTTGATATA | Yes |
| D. trypaniformis | Poly-kDNA | ? | ? | ? |
| C. helicis | Pan-kDNA | ? | GGGGTTGATGAG | Yes |
| T. borreli | Mega-kDNA | Yes | GGTGTTGATGTA | No |
The UMS is part of the replication origin.
Phased A tracts that cause bending are found in minicircles from most trypanosomatids. T. cruzi is an exception.
This sequence is conserved in all trypanosomatids examined.
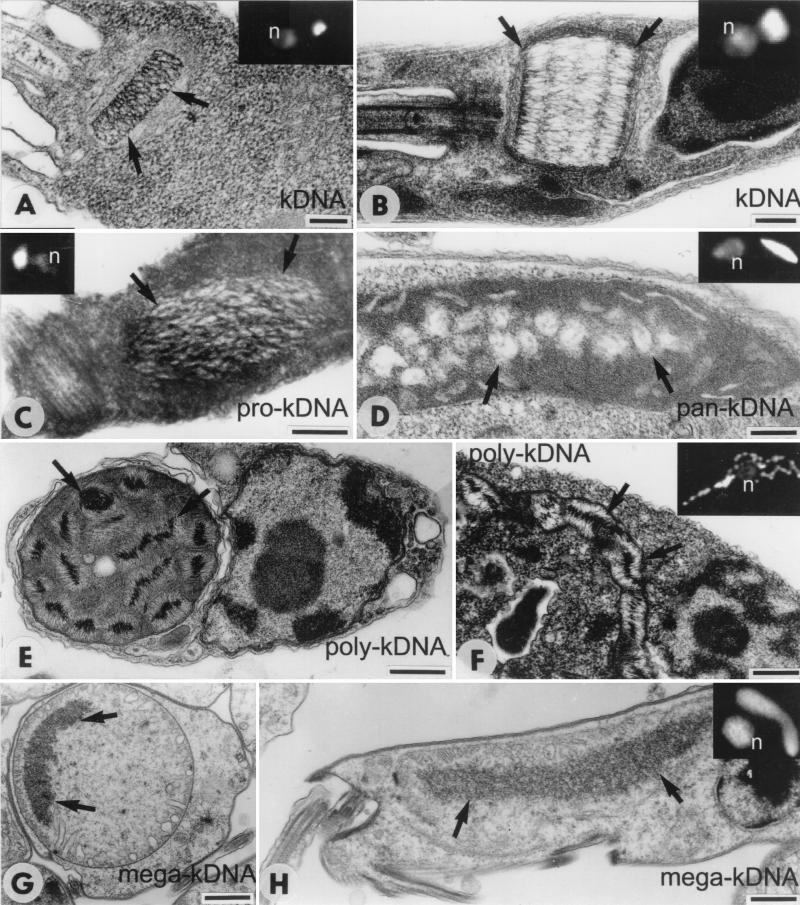
Within the mitochondrial matrix, the C. fasciculata network is condensed into a highly organized disk-shaped structure that measures about 1 μm in diameter and 0.35 μm in thickness. Within the disk, the minicircles are stretched out and aligned side-by-side approximately perpendicular to the planar face of the disk. Therefore, the thickness of the disk is about half the circumference of a minicircle (29, 45). Figure 1B is a diagram of a section through a kDNA disk showing individual minicircles. Figures 2A and B show electron micrographs of thin sections cut perpendicular to a face of the disk. Figure 2A shows the kDNA disk of C. fasciculata (whose minicircles are 2.5 kb), and Fig. 2B shows a comparable image of a much thicker disk from Trypanosoma avium (whose minicircles are 10 kb) (29). In C. fasciculata, there are histone-like proteins, termed kinetoplast-associated proteins, that are involved in organizing the kDNA network in the disk-like structure (28, 57). The kDNA disk is positioned in a fixed region of the mitochondrial matrix, near the basal body of the flagellum (Fig. 2A and B and 2B, inset). The axis of the disk aligns with the axis of the flagellum to which it is physically linked (19, 42; D. Robinson and P. T. Englund, unpublished results).
FIG. 2.
Images of trypanosomatid and bodonid cells showing kDNA. For light microscopy, cells were fixed in 4% paraformaldehyde for 3 min at room temperature, incubated in phosphate-buffered saline containing 0.1 μg of DAPI/ml for 3 min at room temperature, and examined with a Zeiss Axioplan 100 microscope. For electron microscopy, cells were fixed in 2% glutaraldehyde in 0.2 M cacodylate buffer at 4°C overnight, postfixed in 2% osmium tetroxide for 1 h at room temperature, and embedded in Epon-Araldite. Thin sections stained with uranyl acetate and lead citrate were examined in a JEOL 1010 microscope. Arrows in electron micrographs indicate kDNA. Insets show DAPI-stained cells (n, nucleus); kDNA is stained brightly. (A) Longitudinal section through the classical disk-shaped kDNA of C. fasciculata. The disk thickness is about half the minicircle circumference (2.5 kb). (B) Longitudinal section through the kDNA disk of T. avium. The disk appears cylindrical due to the large minicircle size (10 kb), but the organization is similar to that of C. fasciculata. (C) Pro-kDNA bundle of B. saltans in a dilated region of the mitochondrion, close to the basal bodies of the flagella. (D) Pan-kDNA of C. helicis, composed of multiple electron-lucent loci in the mitochondrial lumen. (E and F) Transverse (E) and longitudinal (F) sections of the mitochondrion of D. trypaniformis showing multiple poly-kDNA nucleoids with the DNA fibrils radiating from a dense core. (G and H) Transverse (G) and longitudinal (H) sections of T. borreli in which a dense body of mega-kDNA is spread throughout the mitochondrial lumen. Bars, 200 nm in panels A to F and 1 μm in panels G and H. Cells in insets are all at the same scale.
Replication of the kDNA network.
kDNA synthesis requires a complex replication machine composed of multiple proteins situated in a variety of fixed positions surrounding the kDNA disk. One crucial consequence of the replication process, and a likely reason for its complexity, is to ensure that each daughter cell receives a complete repertoire of minicircles so that essential gRNA species will be available for RNA editing. Although the system is not perfectly precise, as it allows drift in the minicircle copy number (48), it is adequate for survival of the cell population (see below for a more extensive discussion of this issue). For further information on kDNA replication, see reference 25.
Network replication initiates, near the beginning of the nuclear S phase, with the topoisomerase-catalyzed release of covalently closed minicircles from the network. Minicircles are released vectorially into the kinetoflagellar zone (KFZ), a region between the kDNA disk and the mitochondrial membrane nearest the flagellar basal body (10). Within the KFZ, minicircles encounter key proteins that are localized specifically in this region. These proteins include the minicircle origin recognition protein (1), primase (27), and two DNA polymerases (M. Klingbeil, S. Motyka, and P. T. Englund, submitted for publication). These proteins, and probably others, presumably assemble on the minicircle replication origin, allowing replication to initiate. Minicircles, either in the form of advanced replication intermediates or segregated minicircle progeny, then migrate from the KFZ to the antipodal sites, two loci that flank the kDNA disk. The antipodal sites contain a distinct set of replication enzymes, and within them some minicircle processing reactions are thought to occur. These include the removal of RNA primers by SSE1, an enzyme with RNase H activity (12); repair by DNA polymerase β of some but not all of the many minicircle gaps that have been introduced during replication (50); and reattachment of the still-gapped progeny by topoisomerase II (36) to the network periphery (55).
Although gapped progeny minicircles are attached initially to the network at two peripheral loci, adjacent to the antipodal sites, they subsequently become distributed around the entire network periphery (38, 47). This remarkable fact strongly suggests that there is relative movement between the kDNA disk and the antipodal sites, and a simple explanation would be that the kDNA disk actually spins during replication (38). There appears to be a spinning kinetoplast in C. fasciculata, T. cruzi, L. tarentolae, and Phytomonas serpens but, surprisingly, not in T. brucei (18). In the last parasite, gapped minicircles accumulate adjacent to the antipodal sites; therefore, its kinetoplast appears to remain stationary during the replication process (14). When all minicircles have replicated, the network has increased in size from 5,000 minicircles, all covalently closed, to 10,000 minicircles, all containing gaps. At this time, the gaps are repaired and the network splits in two. The mechanism of the latter process is not understood, but it is probably mediated by a topoisomerase that unlinks minicircles along the cleavage line. The segregation of progeny kDNA networks into daughter cells is thought to be mediated by their connection with flagellar basal bodies (42).
There are other molecular transactions involving minicircles and maxicircles. During mating of T. brucei and T. cruzi, not only exchange of nuclear genes (21, 33) but also exchange of intact minicircles (16) and maxicircles (33, 51) occur. Therefore, hybrid networks in the progeny contain components derived from the kDNA network of each parent. Although the mechanism of minicircle and maxicircle exchange is unknown, it must involve fusion of the mitochondria from the two parental cells.
VARIATIONS IN kDNA STRUCTURE
Here we review the recently discovered diversity in kDNA structure. We discuss kDNA structures known as pro-kDNA, poly-kDNA, pan-kDNA, and mega-kDNA. This nomenclature has been used earlier (54), except for pro-kDNA and mega-kDNA, which are new terms introduced in this review.
Pro-kDNA.
Electron microscopy of thin sections of Bodo saltans (a late-diverging free-living bodonid isolated from a lake) revealed a single bundle-like structure in the mitochondrial matrix that superficially resembles a kDNA disk (Fig. 2, compare panel C with panels A and B). As with a kDNA network, the pro-kDNA bundle is situated near the basal body of the flagellum, although there is no information as to whether there are molecular connections between the two. 4′,6′-Diamidino-2-phenylindole (DAPI) staining (Fig. 2C, inset) as well as in situ hybridization with a minicircle probe confirmed that this structure contains kDNA (I. Gažiová and J. Lukeš, submitted for publication). Molecular analysis of pro-kDNA revealed that it is composed not of networks but of individual 1.4-kb minicircles, with only a few very small catenanes. As in kDNA networks, these minicircles are mostly covalently closed and, significantly, are topologically relaxed (2). It is not known whether they develop gaps, as do kDNA network minicircles, after they have undergone replication. Each minicircle encodes two gRNAs and, like classical kDNA minicircles, contains sequences that cause DNA bending (A tracts phased every 10 bp) (2). The minicircles also contain a short sequence, within a 350-bp conserved region, that resembles the UMS replication origin (Table 1). The B. saltans maxicircle is unusually large (∼70 kb), and a 4-kb fragment that has been sequenced contains typical maxicircle genes. However, the gene order and editing patterns differ from those of trypanosomatids (3).
Other bodonids may have pro-kDNA, as electron microscopy of thin sections has revealed kDNA similar to that of B. saltans in structure and in location within the mitochondrial matrix. Examples include the free-living Bodo designis, Procryptobia (Bodo) sorokini, Rhynchomonas nasuta, and Cephalothamnium cyclopi (7, 15, 54). However, there have been no studies on the molecular nature of their kDNAs.
Poly-kDNA.
Inspection of DAPI-stained cells or electron micrographs of the early-branching bodonids Dimastigella trypaniformis (a commensal of the intestine of a termite) (Fig. 2E and F and 2F, inset), Dimastigella mimosa (a free-living bodonid isolated from a sewage plant), and Cruzella marina (a parasite of the intestine of a sea squirt) revealed a kDNA packaging pattern distinct from that of B. saltans. Instead of being condensed into a single globular bundle (Fig. 2C), the kDNA is distributed among various discrete foci throughout the mitochondrial lumen (Fig. 2E and F and 2F, inset) (5, 53). Molecular studies have shown that poly-kDNA, like pro-kDNA, does not exist in the form of a network. Instead, it consists of monomeric minicircles (1.2 to 2.0 kb, depending on the species), many of which are covalently closed but not supercoiled (one faint band detected by gel electrophoresis of D. trypaniformis kDNA migrated as expected for supercoils but could also be a smaller minicircle) (49). Minicircle dimers, but no larger oligomers, were also found, and they were relatively abundant only in C. marina (A. Zíková and J. Lukeš, unpublished results). No sequence information is available for poly-kDNA minicircles or maxicircles.
Other bodonids apparently have poly-kDNA. Based on Giemsa staining, these include free-living Rhynchobodo spp., Hemistasia phaeocysticola, and ectoparasitic Ichthyobodo (Costia) necatrix (6, 11, 22).
Pan-kDNA.
The kDNA of Cryptobia helicis (a parasite of the receptaculum seminis of snails) fills most of the mitochondrial matrix (Fig. 2D and inset). Like pro-kDNA and poly-kDNA, pan-kDNA does not exist in the form of a network, and almost all of its 4.2-kb minicircles are monomeric. However, one major difference from all the kDNA forms discussed so far is that C. helicis minicircles are not relaxed but are supercoiled (32). Although most minicircles are present as supercoiled monomers, dimers and oligomers are also present. C. helicis minicircles contain typical minicircle motifs, including a UMS-related sequence and a bent helix (Table 1). Maxicircles are ∼43 kb, and the two genes partially sequenced so far encode RNAs that are not edited (32).
Pan-kDNA may also occur in free-living B. caudatus (as judged from the published data) (20) and Cryptobia branchialis, a parasite of fish (7).
Mega-kDNA.
The most unusual kDNA (from the perspective that the network is the conventional structure) is that of the fish parasite Trypanoplasma borreli. This kDNA is distributed fairly uniformly throughout a large region of the mitochondrial matrix (Fig. 2G and H and 2H, inset). However, molecular studies have indicated that it does not contain minicircles at all (35). Instead, minicircle-like sequences are tandemly linked into large molecules (possibly circular) of approximately 200 kb. Each minicircle-size unit (1 kb each, cut once by ScaI) encodes gRNAs that are unusual in having uridine tails on both 5′ and 3′ ends (46, 48, 58). Cloned ScaI fragments contain a UMS-related sequence (35). The gene order and editing patterns of maxicircle genes in this species are significantly different from those of trypanosomatids (30, 35).
In addition to T. borreli, light and electron microscopy images suggest that similarly organized mega-kDNA may occur in other species of Trypanoplasma and in Jarrellia, a parasite of whales (40).
EVOLUTION OF kDNA
The function of kDNA, whether in the form of a network, monomeric circles, or tandemly repetitious minicircle-like sequences, is to encode the substrates for RNA editing. Editing occurs through the transfer of sequence information from the predominantly minicircle-encoded gRNAs to encrypted transcripts encoded by maxicircles. How does this function relate to the unprecedented structure of the kDNA network? How did kDNA networks arise? What are the biological factors driving their evolution? Alternatively, why did kDNA networks arise at all, since organisms containing much simpler kDNA structures are still extant and widespread? We cannot provide clear answers to all of these questions, but from an evolutionary perspective we can provide some insights.
Phylogeny of the Kinetoplastida.
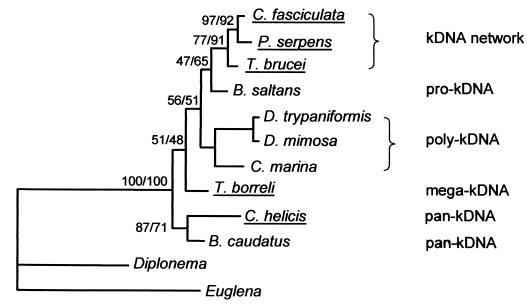
The order Kinetoplastida was originally subdivided into the suborders Bodonina and Trypanosomatina based on morphological characteristics (54). Subsequently, phylogenetic trees constructed from nuclear rRNA genes confirmed the morphology-based subdivisions and the paraphyletic status of the Bodonina. These trees also established the monophyly as well as the derived character of the Trypanosomatina (31, 56). These results have been further supported by comparative analyses of the mitochondrial gene order on maxicircle DNA and the RNA editing patterns of these genes and by phylogenetic analysis of the cytochrome oxidase subunit I and II genes (2). However, the most extended bodonid data set, analyzed with maximum parsimony and maximum likelihood, has failed to resolve the branching order of the early-diverging bodonid species (9). Therefore, until more conserved genes can be analyzed, the precise evolution of the early-branching bodonids cannot be definitively traced; Fig. 3 shows the current phylogenetic tree. However, it is currently believed that C. helicis is among the earliest of the bodonids and that B. saltans is among the last to diverge.
FIG. 3.
Kinetoplastid phylogenetic tree. Majority-consensus maximum-likelihood tree constructed by using a small-subunit rRNA alignment (alignment 10 at http://www.rna.ucla.edu/trypanosome/alignments.html) narrowed to species for which kDNA structural information is available (see the text). Bootstrap analysis was performed with 1,000 replicates. Bootstrap values for maximum likelihood and maximum parsimony are shown (to left and right of slashes, respectively). Parasitic species are underlined. Pro-kDNA contains monomeric relaxed minicircles condensed in a single region of the mitochondrial matrix. Poly-kDNA contains monomeric relaxed minicircles condensed in multiple foci. Pan-kDNA contains monomeric supercoiled minicircles distributed throughout a large region of the mitochondrial matrix. Mega-kDNA contains molecules with tandemly linked minicircle-like sequences. The B. caudatus strain used by Hajduk et al. (20) appears to have pan-kDNA, although electron microscopy data on thin sections is lacking. See the text for further discussion.
Evolution of kDNA structures.
If one superimposes on the phylogenetic tree shown in Fig. 3 the variety of kDNA structures together with their compaction patterns within the mitochondrial matrix, a straightforward and logical pathway for the evolution of kDNA structures can be deduced. We propose that the pan-kDNA of C. helicis is the form most similar to the ancestral state. In pan-kDNA, the size, supercoiling, monomeric status, and distribution of minicircles in the mitochondrial matrix resemble those of plasmids (Fig. 2D and 4). We postulate that the precursor to modern minicircles was derived from a plasmid harbored within the mitochondrion of an ancient flagellate. That C. helicis minicircles contain UMS-related and bent DNA sequences is consistent with the hypothesis that they are an ancestor of the minicircles in kDNA networks.
FIG. 4.
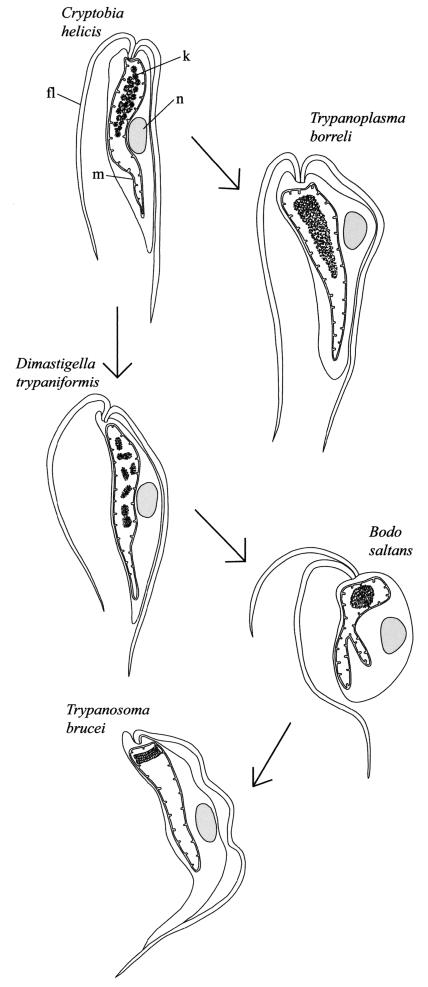
Proposed evolution of kinetoplastids, emphasizing differences in kDNA organization and compaction. kDNA (k) is the structure within the mitochondrial matrix. fl, flagellum; m, mitochondrion; n, nucleus. kDNA in C. helicis is pan-kDNA, that in T. borreli is mega-kDNA, that in D. trypaniformis is poly-kDNA, that in B. saltans is pro-kDNA, and that in T. brucei is a kDNA network.
From Cryptobia we propose a branched pathway, with one branch leading ultimately to the kDNA network of trypanosomatids and the other leading to the mega-kDNA of T. borreli. We focus first on the branch that leads to the network and discuss mega-kDNA below. There are two events that must have occurred on the pathway to the network. One is the loss of supercoiling that occurred with the advent of poly-kDNA in Dimastigella and Cruzella, with relaxed minicircles also being characteristic of pro-kDNA and a kDNA network (Fig. 5). Concurrent with the loss of supercoiling was the gradual compaction of monomeric minicircles (shown in the diagrams in Fig. 4). In poly-kDNA, minicircles are bundled in multiple foci throughout the mitochondrial matrix, and in pro-kDNA, they are compacted in a single focus. In a kDNA network, at the end of this pathway, minicircles are even more tightly condensed into a disk. The compaction of kDNA minicircles could resemble that of bacterial plasmids, some of which aggregate in multiple foci within a bacterial cell (39).
FIG. 5.
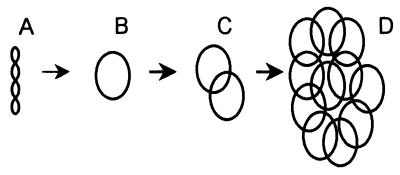
Proposed stages in the evolution of a kDNA network. (A) Supercoiled noncatenated minicircles present in early-branching bodonids. (B) Relaxed noncatenated minicircles also found in early-branching bodonids. (C) Small catenanes of relaxed minicircles, relatively abundant in some late-branching bodonids. (D) kDNA network, present only in late-emerging trypanosomatids.
Compaction and absence of supercoiling are major factors that allow the formation of a network. As demonstrated by in vitro reactions, plasmid DNA aggregated by spermidine in the presence of topoisomerase will form massive networks if the DNA is relaxed but only small catenated oligomers if it is supercoiled (26). The reason that relaxed circles more readily form a network is that they take up more space and can more easily penetrate each other. According to this logic, it is inevitable that relaxed, compacted DNA circles in the presence of topoisomerase will form a network.
However, conundrums remain. For example, there is a price to pay for abandoning supercoiling. The tension from superhelicity promotes unwinding of the DNA helix, which in turn drives fundamental processes such as replication, transcription, and recombination (23). Therefore, the kinetoplastid protozoa must have gained something to compensate for the loss of minicircle supercoiling. One possible gain could be the ability to form a network, but this does not appear to be the case because cells with poly-kDNA and pro-kDNA do not catenate their minicircles even though they are relaxed and compacted (even more compact in the case of pro-kDNA). One could argue that monomeric minicircles are not catenated because there is no topoisomerase associated with the minicircle bundles. However, that explanation appears to be incorrect. It was recently revealed by immunofluorescence that B. saltans topoisomerase II, about 48% identical in sequence to mitochondrial topoisomerase II from C. fasciculata and T. brucei, localizes predominantly with the pro-kDNA bundles within the mitochondrion of the cell (Gažiová and Lukeš, submitted). Therefore, even though these minicircles are compacted, relaxed, and associated with topoisomerase, they do not form a network, as they would in vitro. We do not know the reason for this behavior.
Evolution of mega-kDNA.
As already mentioned, there was an early branch point in the evolution of kDNA structures (Fig. 4). One branch, discussed above, led to the kDNA network, while the other led to the mega-kDNA of T. borreli. Mega-kDNA could have arisen easily by recombination of pan-kDNA minicircles such as those in C. helicis, forming concatemers of tandemly linked minicircle-like sequences, each encoding a gRNA. Such recombination has resulted in a structure completely different from that of a kDNA network, although as we discuss below, it could serve some of the same functions.
Minicircle segregation dilemma.
The repertoire of minicircles and maxicircles has not been thoroughly characterized in any kinetoplastid protozoan with noncatenated circles. However, it likely consists of multiple identical maxicircle copies and numerous different minicircle species (each encoding one or more essential gRNAs) present in unique copy numbers. When the maxicircles and minicircles undergo replication, their copy numbers should double. The problem is how the progeny segregate into the two daughter cells. It is difficult to imagine a molecular mechanism that would provide precise segregation of every copy of these multiple noncatenated species into the two daughter cells. In contrast, if the maxicircles and minicircles segregated randomly, then there would be a rapid loss of essential circles (as predicted by mathematical modeling) (43) and the cells would soon die. If the cells do not have a molecular mechanism for proper segregation of their kDNA components, it is likely that they compensate for minicircle loss by undergoing frequent genetic exchange. This process, involving the exchange of intact DNA circles, could homogenize the minicircle content and maintain cell viability, as has been suggested for T. brucei (43). It is likely that the evolution of mega-kDNA and kDNA networks was driven by a need to improve on the mechanism of minicircle segregation.
Mega-kDNA and the minicircle segregation dilemma.
Mega-kDNA in T. borreli contains tandemly linked minicircle-like sequences. We speculate that the mega-kDNA repertoire consists of multiple maxicircle copies and multiple molecules containing minicircle-like sequences. Although segregation of the progeny of these molecules could still be random, the ratio of minicircle-like sequences would remain fixed because they are tandemly linked. gRNA-encoding sequences could be lost only by recombination or if a daughter cell lost all copies of the tandemly repetitious molecules.
Evolution of the kDNA network.
A major driving force for the formation of a network was likely to provide a solution to the minicircle segregation dilemma, an idea that was already expressed explicitly (4). Organization into a network may allow essential minicircles to be present in low copy numbers without placing them at risk for the rapid loss that would occur if minicircle segregation occurred randomly. We do not yet know how a network facilitates minicircle segregation, but we have suggested two models (for a further discussion and earlier references, see reference 25). One model is based on the finding that minicircle replication initiates and proceeds in the KFZ (1, 10). If replication is also completed in this zone (as of yet there is no evidence that that is the case) and if the progeny minicircles segregate there, one could postulate a mechanism that delivers one sister minicircle to each antipodal site. The sister minicircles would then be reattached on opposite sides of the network, ultimately destined for different daughter cells when the double-size network splits in two. A second model for minicircle segregation could function if minicircle replication is not completed until the replicating molecule arrives at the antipodal site. In this scenario, the two progeny minicircles could attach to the network at neighboring positions, making it likely that they would be distributed into the same daughter network. However, with C. fasciculata kDNA, we found that the progeny minicircles do not attach to the network simultaneously; multiply gapped progeny are attached after a delay (24). Therefore, rotation of the kinetoplast during replication could result in the attachment of sister minicircles at opposite sides of the network, favoring their distribution to different daughter cells at the time of network division (38). A problem, of course, is T. brucei, whose network does not rotate (see above). Solutions to this problem could be that its minicircles segregate by the first model or that minicircle exchange occurs during mating to homogenize and sustain the minicircle repertoire (43). More experiments are needed to clarify the molecular mechanism by which the network structure facilitates minicircle segregation.
Acknowledgments
This work was supported by grants from the Grant Agency of the Czech Academy of Sciences (A6022903) and the Ministry of Education of the Czech Republic (MSMT-123100003) (to J.L.) and from the National Institutes of Health (GM27608) (to P.T.E.).
We thank members of our laboratories and Dave Barry, Alastair Simpson, and Barbara Sollner-Webb for stimulating discussions and helpful comments. We also thank Guy Brugerolle for kindly providing the electron micrographs of D. trypaniformis, Jana Fišáková for artistic talent, and Libor Grubhoffer for continuous support.
REFERENCES
- 1.Abu-Elneel, K., D. R. Robinson, M. E. Drew, P. T. Englund, and J. Shlomai. 2001. Intramitochondrial localization of universal minicircle sequence-binding protein, a trypanosomatid protein that binds kinetoplast minicircle replication origins. J. Cell Biol. 153:725-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blom, D., A. de Haan, J. van den Burg, M. van den Berg, P. Sloof, M. Jirků, J. Lukeš, and R. Benne. 2000. Mitochondrial minicircles in the free-living bodonid Bodo saltans contain two gRNA gene cassettes and are not found in large networks. RNA 6:121-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blom, D., A. de Haan, M. van den Berg, P. Sloof, M. Jirkù, J. Lukeš, and R. Benne. 1998. RNA editing in the free-living bodonid Bodo saltans. Nucleic Acids Res. 26:1205-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst, P. 1991. Why kinetoplast DNA networks? Trends Genet. 7:139-141. [DOI] [PubMed] [Google Scholar]
- 5.Breunig, A., G. Brugerolle, K. Vickerman, H. Hertel, and H. König. 1993. Isolation and ultrastructural features of a new strain of Dimastigella trypaniformis Sandon 1928 (Bodonina, Kinetoplastida) and comparison with a previously isolated strain. Eur. J. Protistol. 29:416-424. [DOI] [PubMed] [Google Scholar]
- 6.Brugerolle, G., and J.-P. Mignot. 1979. Distribution et organisation de l'ADN dans le complexe kinétoplaste-mitochondrie chez un Bodonidé, protozoaire kinétoplastidé; variation au cours du cycle cellulaire. Biol. Cell 35:111-114. [Google Scholar]
- 7.Brugerolle, G., J. Lom, E. Nohýnková, and L. Joyon. 1979. Comparaison et évolution des structures cellulaires chez plusieurs espèces de Bodonidés et Cryptobiidés appartenant aux genres Bodo, Cryptobia et Trypanoplasma (Kinetoplastida, Mastigophora). Protistologica 15:197-221. [Google Scholar]
- 8.Chen, J., C. A. Rauch, J. H. White, P. T. Englund, and N. R. Cozzarelli. 1995. The topology of the kinetoplast DNA network. Cell 80:61-69. [DOI] [PubMed] [Google Scholar]
- 9.Doležel, D., M. Jirků, D. A. Maslov, and J. Lukeš. 2000. Phylogeny of the bodonid flagellates (Kinetoplastida) based on small subunit rRNA gene sequences. Int. J. Syst. E vol. Microbiol. 50:1943-1951. [DOI] [PubMed] [Google Scholar]
- 10.Drew, M. E., and P. T. Englund. 2001. Intramitochondrial location and dynamics of Crithidia fasciculata kinetoplast minicircle replication intermediates. J. Cell Biol. 153:735-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elbrächter, M., E. Schnepf, and I. Balzer. 1996. Hemistasia phaeocysticola (Scherffel) comb. nov.; rederscription of a free-living, marine, phagotrophic kinetoplastid flagellate. Arch. Protistenkd. 147:125-136. [Google Scholar]
- 12.Engel, M. L., and D. S. Ray. 1999. The kinetoplast structure-specific endonuclease I is related to the 5′ exo/endonuclease domain of bacterial DNA polymerase I and colocalizes with the kinetoplast topoisomerase II and DNA polymerase β during replication. Proc. Natl. Acad. Sci. USA 96:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estévez, A. M., and L. Simpson. 1999. Uridine insertion/deletion RNA editing in trypanosome mitochondria—a review. Gene 240:247-260. [DOI] [PubMed] [Google Scholar]
- 14.Ferguson, M. F., A. F. Torri, D. Pérez-Morga, D. C. Ward, and P. T. Englund. 1994. Kinetoplast DNA replication: mechanistic differences between Trypanosoma brucei and Crithidia fasciculata. J. Cell Biol. 126:631-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frolov, A. O., S. A. Karpov, and A. P. Mylnikov. 2001. The ultrastructure of Procryptobia sorokini (Zhukov) comb.nov. and rootlet homology in kinetoplastids. Protistology 2:85-95. [Google Scholar]
- 16.Gibson, W. C., and L. Garside. 1990. Kinetoplast DNA minicircles are inherited from both parents in genetic hybrids of Trypanosoma brucei. Mol. Biochem. Parasitol. 42:45-53. [DOI] [PubMed] [Google Scholar]
- 17.Gott, J. M., and R. B. Emeson. 2000. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 34:499-531. [DOI] [PubMed] [Google Scholar]
- 18.Guilbride, D. L., and P. T. Englund. 1998. The replication mechanism of kinetoplast DNA networks in several trypanosomatid species. J. Cell Sci. 111:675-679. [DOI] [PubMed] [Google Scholar]
- 19.Gull, K. 1999. The cytoskeleton of trypanosomatid parasites. Annu. Rev. Microbiol. 53:629-665. [DOI] [PubMed] [Google Scholar]
- 20.Hajduk, S. L., A. M. Siqueira, and K. Vickerman. 1986. Kinetoplast DNA of Bodo caudatus: a noncatenated structure. Mol. Cell. Biol. 6:4372-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenni, L., S. Marti, J. Schweizer, B. Betschart, R. W. F. LePage, J. M. Wells, A. Tait, P. Paindavoine, E. Pays, and M. Steinert. 1986. Hybrid formation between African trypanosomes during cyclical transmission. Nature 322:173-175. [DOI] [PubMed] [Google Scholar]
- 22.Joyon, L., and J. Lom. 1969. Étude cytologique, systématique et pathologique d'Ichthyobodo necator (Henneguy, 1883) Pinto, 1928 (Zooflagelle). J. Protozool. 6:703-719. [Google Scholar]
- 23.Kanaar, R., and N. R. Cozzarelli. 1992. Roles of supercoiled DNA structure in DNA transactions. Curr. Opin. Struct. Biol. 2:369-379. [Google Scholar]
- 24.Kitchin, P. A., V. A. Klein, and P. T. Englund. 1985. Intermediates in the replication of kinetoplast DNA minicircles. J. Biol. Chem. 260:3844-3851. [PubMed] [Google Scholar]
- 25.Klingbeil, M. M., M. E. Drew, Y. Liu, J. C. Morris, S. A. Motyka, T. T. Saxowsky, Z. Wang, and P. T. Englund. 2001. Unlocking the secrets of trypanosome kinetoplast DNA network replication. Protist 152:255-262. [DOI] [PubMed] [Google Scholar]
- 26.Krasnow, M. A., and N. R. Cozzarelli. 1982. Catenation of DNA rings by topoisomerases. Mechanism of control by spermidine. J. Biol. Chem. 257:2687-2693. [PubMed] [Google Scholar]
- 27.Li, C., and P. T. Englund. 1997. A mitochondrial DNA primase from the trypanosomatid Crithidia fasciculata. J. Biol. Chem. 272:20787-20792. [DOI] [PubMed] [Google Scholar]
- 28.Lukeš, J., J. C. Hines, C. J. Evans, N. K. Avliyakulov, V. P. Prabhu, J. Chen, and D. S. Ray. 2001. Disruption of the Crithidia fasciculata KAP1 gene results in structural rearrangement of the kinetoplast disc. Mol. Biochem. Parasitol. 117:179-186. [DOI] [PubMed] [Google Scholar]
- 29.Lukeš, J., and J. Votýpka. 2000. Trypanosoma avium: novel features of the kinetoplast structure. Exp. Parasitol. 96:178-181. [DOI] [PubMed] [Google Scholar]
- 30.Lukeš, J., G.-J. Arts, J. van den Burg, A. de Haan, F. Opperdoes, P. Sloof, and R. Benne. 1994. Novel pattern of editing regions in mitochondrial transcripts of the cryptobiid Trypanoplasma borreli. EMBO J. 13:5086-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukeš, J., M. Jirků, D. Doležel, I. Král'ová, L. Hollar, and D. A. Maslov. 1997. Analysis of ribosomal RNA genes suggests that trypanosomes are monophyletic. J. Mol. Evol. 44:521-527. [DOI] [PubMed] [Google Scholar]
- 32.Lukeš, J., M. Jirků, N. Avliyakulov, and O. Benada. 1998. Pankinetoplast DNA structure in a primitive bodonid flagellate, Cryptobia helicis. EMBO J. 17:838-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machado, C. A., and F. J. Ayala. 2001. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc. Natl. Acad. Sci. USA 98:7396-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marini, J. C., S. D. Levene, D. M. Crothers, and P. T. Englund. 1982. Bent helical structure in kinetoplast DNA. Proc. Natl. Acad. Sci. USA 79:7664-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maslov, D. A., and L. Simpson. 1994. RNA editing and mitochondrial genomic organization in the cryptobiid kinetoplastid protozoan Trypanoplasma borreli. Mol. Cell. Biol. 14:8174-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melendy, T., C. Sheline, and D. S. Ray. 1988. Localization of type II DNA topoisomerase to two sites at the periphery of the kinetoplast DNA of Crithidia fasciculata. Cell 55:1083-1088. [DOI] [PubMed] [Google Scholar]
- 37.Ntambi, J. M., T. A. Shapiro, K. A. Ryan, and P. T. Englund. 1986. Ribonucleotides associated with a gap in newly replicated kinetoplast DNA minicircles from Trypanosoma equiperdum. J. Biol. Chem. 261:11890-11895. [PubMed] [Google Scholar]
- 38.Pérez-Morga, D., and P. T. Englund. 1993. The attachment of minicircles to kinetoplast DNA networks during replication. Cell 74:703-711. [DOI] [PubMed] [Google Scholar]
- 39.Pogliano, J., T. Q. Ho, Z. Zhong, and D. R. Helinski. 2001. Multicopy plasmids are clustered and localized in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poynton, S. L., B. R. Whitaker, and A. B. Heinrich. 2001. A novel trypanoplasm-like flagellate Jarrellia atramenti n. g., n. sp. (Kinetoplastida: Bodonidae) and ciliates from the blowhole of a stranded pygmy sperm whale Kogia breviceps (Physeteridae): morphology, life cycle and potential pathogenicity. Dis. Aquat. Organ. 44:191-201. [DOI] [PubMed] [Google Scholar]
- 41.Rauch, C. A., D. Pérez-Morga, N. R. Cozzarelli, and P. T. Englund. 1993. The absence of supercoiling in kinetoplast DNA minicircles. EMBO J. 12:403-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson, D. R., and K. Gull. 1991. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature 352:731-733. [DOI] [PubMed] [Google Scholar]
- 43.Savill, N. J., and P. G. Higgs. 1999. A theoretical study of random segregation of minicircles in trypanosomatids. Proc. R. Soc. London Ser. B 266:611-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shapiro, T. A. 1993. Kinetoplast DNA maxicircles: networks within networks. Proc. Natl. Acad. Sci. USA 90:7809-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shapiro, T. A., and P. T. Englund. 1995. The structure and replication of kinetoplast DNA. Annu. Rev. Microbiol. 49:117-143. [DOI] [PubMed] [Google Scholar]
- 46.Simpson, L. 1999. RNA editing—an evolutionary perspective, p. 585-608. In F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Simpson, L., and A. M. Simpson. 1976. Pulse-labeling of kinetoplast DNA: localization of 2 sites of synthesis within the networks and kinetics of labeling of closed minicircles. J. Protozool. 23:583-587. [DOI] [PubMed] [Google Scholar]
- 48.Simpson, L., O. H. Thiemann, N. J. Savill, J. D. Alfonzo, and D. A. Maslov. 2000. Evolution of RNA editing in trypanosome mitochondria. Proc. Natl. Acad. Sci. USA 97:6986-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Štolba, P., M. Jirků, and J. Lukeš. 2001. Polykinetoplast DNA structure in Dimastigella trypaniformis and Dimastigella mimosa (Kinetoplastida). Mol. Biochem. Parasitol. 113:323-326. [DOI] [PubMed] [Google Scholar]
- 50.Torri, A. F., T. A. Kunkel, and P. T. Englund. 1994. A β-like DNA polymerase from the mitochondrion of the trypanosomatid Crithidia fasciculata. J. Biol. Chem. 269:8165-8171. [PubMed] [Google Scholar]
- 51.Turner, C. M. R., J. Sternberg, N. Buchanan, and A. Tait. 1995. Trypanosoma brucei: inheritance of kinetoplast DNA maxicircles in a genetic cross and their segregation during vegetative growth. Exp. Parasitol. 80:234-241. [DOI] [PubMed] [Google Scholar]
- 52.Vickerman, K. 1977. DNA throughout the single mitochondrion of a kinetoplastid flagellate: observations on the ultrastructure of Cryptobia vaginalis. J. Protozool. 24:221-233. [Google Scholar]
- 53.Vickerman, K. 1978. The free-living trypanoplasms: description of three species of the genus Procryptobia n.g., and redescription of Dimastigella trypaniformis Sandon, with notes on their relevance to the microscopic diagnosis of disease in man and animals. Trans. Am. Microsc. Soc. 97:485-502. [PubMed] [Google Scholar]
- 54.Vickerman, K. 1990. Phylum Zoomastigina; class Kinetoplastida, p. 215-238. In L. Margulis, J. O. Corliss, M. Melkonian, and D. J. Chapman (ed.), Handbook of Protoctista. Jones & Bartlett Publishers, Boston, Mass.
- 55.Wang, Z., and P. T. Englund. 2001. RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. EMBO J. 20:4674-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wright, A.-D. G., S. Li, S. Feng, D. S. Martin, and D. H. Lynn. 1999. Phylogenetic position of the kinetoplastids, Cryptobia bullocki, Cryptobia catostomi, and Cryptobia salmositica and monophyly of the genus Trypanosoma inferred from small subunit ribosomal RNA sequences. Mol. Biochem. Parasitol. 99:69-76. [DOI] [PubMed] [Google Scholar]
- 57.Xu, C. W., J. C. Hines, M. L. Engel, D. G. Russell, and D. S. Ray. 1996. Nucleus-encoded histone H1-like proteins are associated with kinetoplast DNA in the trypanosomatid Crithidia fasciculata. Mol. Cell. Biol. 16:564-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasuhira, S., and L. Simpson. 1996. Guide RNAs and guide RNA genes in the cryptobiid kinetoplastid protozoan, Trypanoplasma borreli. RNA 2:1153-1160. [PMC free article] [PubMed] [Google Scholar]