Abstract
Background
Split-hand/foot malformation (SHFM) is a congenital disability characterized by the absence or hypoplasia of the central ray of the hands and/or feet. This study reports a causative variant in the TP63 gene in a Chinese family exhibiting limb anomalies associated with SHFM4.
Methods
Enrolled in this study was a Chinese family with limb anomalies without any other clinical features. Karyotype analysis and chromosomal microarray analysis (CMA) were conducted to identify chromosomal abnormalities. Whole exome sequencing (WES) was utilized to investigate sequence variants, while RNA sequencing assessed differentially expressed genes, with findings confirmed through quantitative PCR (qPCR).
Results
Karyotype analysis and CMA revealed no chromosomal abnormalities in the family. Subsequently, WES identified a rare heterozygous variant of NM_003722.5: c.956G > A (p.Arg319His) in the TP63 gene in the proband, which was inherited from her father who also presented with limb deformities. However, both of the sister and grandfather of the proband had the same variant but exhibited normal limb morphology. RNA sequencing results demonstrated an increased expression level of TP63 and its downstream genes (PERP, CDH3, and DLX5) compared with the controls, indicating an enrichment of cell adhesion molecules the differentially expressed genes in the patient. However, significant differences were noted only for the CDH3 and DLX5 genes in qPCR analysis (p<0.05).
Conclusion
This study identifies, for the first time, the TP63 gene variant c.956G > A (p.Arg319His) as a causative factor for SHFM4 in Chinese individuals with incomplete penetrance. In addition, we hypothesize that the p.Arg319His variant functions as a gain-of-function variant, leading to the upregulation of cell adhesion target genes. Such upregulation then disrupts the p63-Dlx signaling pathway and causes AER stratification failure.
Keywords: Split-hand/foot malformation, Whole exome sequencing, RNA sequencing, TP63, Pathogenesis
Introduction
Split-hand/foot malformation (SHFM) is a congenital limb malformation characterized by the absence of central rays in the autopod, presenting with syndactyly, median clefts of the hands and feet, and aplasia and/or hypoplasia of the phalanges, metacarpals, and metatarsals. Variants in several genes, including TP63, DLX5, DLX6, FGF8, FGFR1, WNT10B, and BHLHA9, have been implicated as causative factors for SHFM [1–5]. According to the classification proposed by Umair and Hayat, isolated SHFM is categorized as Type 1, which encompasses isolated hand and/or foot malformations (SHFM Types 1 to 6). Conversely, Type 2 is associated with deficiencies in the long bones (SHFLD Types 1 to 3) [6, 7]. The apical ectodermal ridge (AER) is a critical transient signaling center for proximodistal growth and distal limb development. Previous studies have suggested that gene variants associated with SHFM may lead to dysregulation of FGF8 within AER cells located in the central regions of the hands and feet. This dysregulation may further disrupt the Wnt-Bmp-Fgf signaling pathways in the AER, contributing to the development of SHFM [1, 8].
The tumor protein p63 (TP63) gene contains 15 exons located at chromosome 3q27, which plays an important role in regulating epithelial, limb and craniofacial development [9]. A previous study [10] noted that p63−/− mouse model showed reduced FGF8 expression in the AER, subsequently leading to split hand/foot malformation. Pathogenic variants within the TP63 gene are classified into six TP63-related disorders, including ankyloblepharon-ectodermal defects-cleft lip/palate (AEC) syndrome, acro-dermo-ungual-lacrimal-tooth (ADULT) syndrome, ectrodactyly, ectodermal dysplasia, cleft lip/palate syndrome 3 (EEC3), limb-mammary syndrome (LMS), split-hand/foot malformation type 4 (SHFM4), isolated cleft lip/cleft palate [11].
In this report, we present a family with SHFM4 from the Quanzhou region of Southeast China. Whole exome sequencing (WES) identified a missense c.956G > A (p.Arg319His) variant of TP63 gene. This variant, first reported in Chinese individuals, demonstrates incomplete penetrance. Our findings offer valuable insights for genetic counseling in clinical practice.
Materials and methods
Subjects
In this study, a family with SHFM from Fujian province, Southeast China was recruited. The proband, along with his immediate family members, was recruited for further genetic analysis. Following the acquisition of informed consent, we conducted karyotype analysis, chromosomal microarray analysis, and whole exome sequencing. Approval for the study was granted by the Institutional Ethics Committee of Quanzhou Women’s and Children’s Hospital (Approval No. 2020No.31).
Karyotype analysis
Approximately 10 ml amniotic fluid in the fetus and 2 ml peripheral blood in each family member were collected for karyotype analysis. According to our previous reported protocol [12], the cultured peripheral blood lymphocytes cells and amniotic fluid cells were harvested using the Sinochrome ChromprepII automatic chromosome harvesting system (Shanghai Lechen Biotechnology Co., Ltd.).
Chromosomal microarray analysis
A total of 10 mL of amniotic fluid was obtained from the fetus, along with 3 to 5 mL of peripheral blood from each family member, for the purpose of chromosomal microarray analysis and whole exome sequencing. The extraction of genomic DNA from both the amniotic fluid and peripheral blood samples was performed using the QIAamp DNA Blood Kit (QIAGEN, Germany).
According to the protocol described previously [13], chromosomal microarray analysis was carried out using single-nucleotide polymorphism based Affymetrix Cytoscan 750 K chip (Life Technologies, American) to detect single-nucleotide polymorphism and copy number variants (CNVs). Data analysis was performed utilizing the Genotyping Console and Chromosome Analysis Suite Software. The pathogenicity of the identified CNVs was assessed using several databases, including the Database of Genomic Variants (DGV), Online Mendelian Inheritance in Man (OMIM), DECIPHER and PubMed. Subsequently, the CNVs were classified as pathogenic, likely pathogenic, variants of uncertain significance, likely benign, or benign, in accordance with the standards and guidelines set forth by the American College of Medical Genetics (ACMG) and the Clinical Genome Resource (ClinGen) [14].
Whole exome sequencing
The genomics DNA of the fetus and the other family members were further subjected to WES analysis. The Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA) was used for whole exome sequencing. The procedure involved several steps, including DNA quantification, DNA shearing, library preparation of targeted regions, and quantification of the sequencing libraries. Data analysis was conducted in accordance with the methodologies outlined in our previous study [15]. This analysis included variant calling, annotation and variant screening. To identify minor allele frequencies for all known variants, we utilized several established databases, including dbSNP, the 1000 Genomes Project, the Exome Aggregation Consortium, and the Exome Variant Server. Variants were selected based on a comprehensive evaluation that integrated clinical information from the proband with the aforementioned annotated data, with particular emphasis placed on de novo variants, compound heterozygotes, homozygotes, and hemizygotes. Detected candidate variants were classified as pathogenic, likely pathogenic, variants of uncertain significance, likely benign, or benign according to the ACMG guidelines [16].
RNA sequencing
The total RNA was extracted from the patient (n = 1) and three age matched controls (n = 3, without any birth defects), using fetal aborted tissue according to the instruction manual of the Trizol reagent (Life technologies, California, USA). The concentration and purity of the extracted RNA were measured using NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE). RNA integrity was assessed using the RNA Nano 6000 assay kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). A total of 1 µg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using the Hieff NGS Ulitma Dual-mode mRNA Library Prep Kit for Illumina (Yeasen Biotechnology (Shanghai) Co., Ltd.) following manufacture’s recommendations, and index codes were added to attribute sequences to each sample. The libraries were sequenced on an Illumina NovaSeq platform to generate150 bp paired-end reads, in accordance with the manufacturer’s instructions.
The raw data, in fastq format, underwent initial processing using custom perl scripts. Gene expression levels were then estimated using the fragments per kilobase of transcript per million fragments mapped (FPKM) method. In the case of samples with biological replicates, differential expression analysis between two conditions/groups was performed using DESeq2. DESeq2 employs statistical algorithms based on the negative binomial distribution to determine differential expression in digital gene expression data. To account for multiple testing, the resulting P-values were adjusted using the Benjamini and Hochberg’s approach to control the false discovery rate. Genes with an adjusted P-value < 0.01 & Fold Change ≥ 2 found by DESeq2 were classified as differentially expressed. For samples without biological replicates, differential expression analysis between two samples was conducted using edgeR. The FDR < 0.01 & Fold Change ≥ 2 was set as the threshold for significantly differential expression.
Results
Subject information
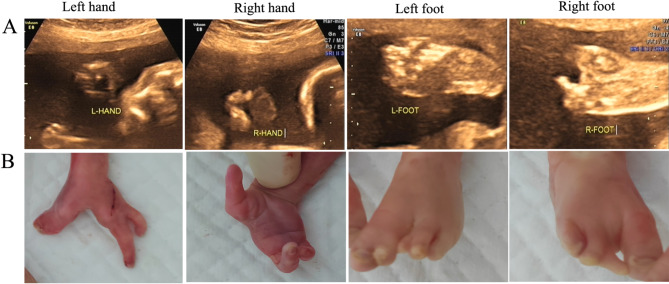
A family from Quanzhou region Southeast China with SHFM was recruited in this study (Fig. 1). The proband (IV2) is a female who was born weighing 2900 g and measuring 50 cm in height. At the time of the study, she is 4 years old and has reached normal developmental milestones, albeit with limb deformities that include a widening gap between the index and middle fingers of her right hand, as well as syndactyly and polydactyly in her right foot (Fig. 2). Surgical intervention was performed when she was 2 years old. As demonstrated in Fig. 1, the proband’s father (III2), and other family members listed in this pedigree (II3, IV7) also exhibit limb deformities. The proband’s father (III2) presents with widening gaps between his index and middle fingers on both hands, in addition to bipedal syndactyly and ectrodactyly. Moreover, individuals II3 and IV7 display syndactyly, although no specific observations have been made regarding the nature of their conditions. In contrast, the proband’s sister, who is 5 years old, has achieved normal developmental milestones and exhibits no significant limb deformities. In the family’s third pregnancy, at a gestational age of 25+ 2 weeks, prenatal ultrasound examination revealed that the fetus displayed various limb deformities, including clefts in both hands and the left foot, as well as syndactyly in the left hand and both feet. Ectrodactyly was also noted in both hands and the right foot (Fig. 3). The family chose to terminate the pregnancy, leading to the delivery of a male infant who exhibited limb deformities consistent with the findings from the prenatal ultrasound examination (Fig. 3).
Fig. 1.
Pedigree Information of the Enrolled Family. The arrow indicates the proband in the family. In this pedigree, II3, III2, IV2, IV3 and IV7 exhibited limb abnormalities
Fig. 2.

Limb Anomalies Observed in the Proband (IV2). A–B: In the proband, a noticeable widening of the gap between the index and middle fingers of the right hand was observed, accompanied by normal phalanges (E). Additionally, as shown in F, the right foot exhibited syndactyly involving the first and second toes, along with polydactyly of the third toe. Surgical intervention was performed on her right foot, with relevant details provided in C and D
Fig. 3.
Ultrasound Abnormalities Observed in the Fetus. A: The ultrasound examination of the fetus revealed the presence of split and ectrodactyly in both hands, as well as syndactyly in the left hand. Additionally, similar split and syndactyly were noted in both feet, with ectrodactyly occurring solely in the right foot. B: The limb anomalies observed in the fetus following induction were consistent with the previously obtained ultrasound findings
Karyotype and chromosomal microarray analysis results
Karyotype analysis and SNP array analysis revealed no evident chromosomal abnormalities or CNVs in the proband, as well as in the fetus, the proband’s parents, and her sister.
Whole exome sequencing detection results
WES technology was employed to investigate the genetic etiology of limb deformities observed in this family. The results of the trio-WES analysis identified a rare heterozygous variant of NM_003722.5: c.956G > A (p.Arg319His) in the TP63 gene of the proband, which was inherited from her father who also had limb deformities. The presence of this variant was further confirmed through Sanger sequencing (Fig. 4). Notably, the fetus also harbored the NM_003722.5: c.956G > A (p.Arg319His) variant in the TP63 gene, which was associated with more severe limb deformities. Interestingly, the proband’s older sister (IV1) and grandfather (II1) also possessed the same variant but displayed normal physical features. Conversely, the proband’s uncle (III5) and cousin (IV6) did not have the variant and exhibited normal characteristics. The detected variant was absent from population databases such as gnomAD and the 1000 genomes project, indicating its potential pathogenicity. Computational analyses suggested that this variant is likely to impact protein structure and function (MetaSVM_score:0.955; GERP++_RS:5.61). Moreover, no additional pathogenic variants associated with related clinical phenotypes were detected within this family. A high degree of conservation was observed for the TP63 variant across different species (Fig. 5). The computer-aided software homology modeling results indicated that the hydrogen bond connection was changed following the variant, which could potentially destabilize the protein (Fig. 5). According to the ACMG guidelines, the c.956G > A (p.Arg319His) variant in the TP63 gene was interpreted as a likely pathogenic variant (PM1, PS4_Supporting, PM2_Supporting, PP1, PP3).
Fig. 4.
Results of Whole Exome Sequencing and Sanger Sequencing in the Enrolled Family. A: Whole exome sequencing identified a c.956G > A (p.Arg319His) variant in the TP63 gene of the proband, which was verified by Sanger sequencing (B). Parental Sanger sequencing results indicated the variant in TP63 gene was inherited from the father (D), but it was absent in the proband’s mother (C). In addition, the c.956G > A (p.Arg319His) variant in the TP63 gene was also detected in the fetus (E) and the proband’s older sister (F)
Fig. 5.
Bioinformatics Analysis of the Detected Variant in the TP63 Gene. A: The c.956G > A (p.Arg319His) variant is situated within the P53 DNA-binding domain. B: Bioinformatic predictions regarding the subcellular localization of the TP63 gene suggest that it is primarily found in the cell nucleus. C: A high level of conservation was observed for the detected variant of the TP63 across different species. D, E, F: Prior to the variant, Arg319 formed hydrogen bonds with Tyr231, Glu239, and Met316. However, following the variant, His319 interacts through hydrogen bonds with His230 and Met316, potentially affecting protein stability
RNA sequencing detection results
In order to further reveal the potential pathogenic mechanism of SHFM4 associated with rare TP63 variants, we collected fetal skeletal muscle abortion tissues for RNA sequencing. The RNA sequencing results revealed that 1,251 genes exhibited significant differential expression when compared to the control group. Notably, we observed a marked upregulation of TP63 expression in the patient carrying the TP63 variants. In addition, in accordance with existing literature on the molecular mechanisms underlying TP63 deficiency and its role in SHFM4, several downstream genes of TP63—including PERP, CDH3, and DLX5—were also found to be upregulated (Table 1). Subsequent quantitative PCR (qPCR) analysis demonstrated significant differences in the expression levels of CDH3 and DLX5 when compared to controls; however, no significant differences were observed for TP63 and PERP (Fig. 6).
Table 1.
The dysregulated genes associated with TP63 variants
| Genes | mRNA expression | p value | Log2FC | Regulated | |||
|---|---|---|---|---|---|---|---|
| Control 1 | Control 2 | Control 3 | Patient | ||||
| TP63 | 5.967392 | 6.007119 | 7.322112 | 48.424246 | 1.25E-10 | 2.982539165 | up |
| PERP | 0.880717 | 0.166465 | 2.325963 | 474.396118 | 2.41E-11 | 8.369835839 | up |
| CDH3 | 0.250423 | 0.196128 | 0.615285 | 18.383955 | 7.91E-05 | 4.849359762 | up |
| DLX5 | 0.796084 | 0.275651 | 1.363152 | 59.041502 | 2.27E-08 | 5.813319306 | up |
Fig. 6.
GO and KEGG Analysis of Differentially Expressed Genes. A: The volcano plot illustrates that a total of 1,251 genes exhibited significant expression differences when compared to the control group. B, C, D: The results of the GO analysis highlight the differential expression of genes across three categories: molecular function, cellular component, and biological process, respectively. E: The KEGG analysis reveals the enriched signaling pathways associated with the differentially expressed genes. F: Quantitative PCR (qPCR) results indicate that the expression levels of CDH3 and DLX5 were significantly upregulated compared to the controls, corroborating the findings from RNA sequencing (p < 0.05). Conversely, no significant differences were observed for the TP63 and PERP genes (p > 0.05). Asterisks indicate statistical significance: ***p < 0.001; **p < 0.05
Further analyses of gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were conducted on the differentially expressed genes. As delineated in Fig. 6, the GO analysis results of the differentially expressed genes were consistent with those of RNA samples obtained from skeletal muscle, which are associated with molecular function (structural constituent of muscle, muscle alpha-actinin binding and actinin binding), biological process (muscle organ development, muscle contraction, skeletal muscle tissue development and mvofibril assembly) and cellular component (myofibril, sarcomere, obsolete contractile fiber part, M band, I band, troponin complex, contractile fiber and A band). In addition, KEGG analysis results indicated that a significant number of differentially expressed genes are enriched in various signaling pathways, including the PI3K-Akt signaling pathway, cAMP signaling pathway, MAPK signaling pathway, and pathways related to cell adhesion molecules, among others.
Discussion
The increasing application of chromosome microarray analysis in prenatal diagnosis has facilitated clear genetic diagnosis for a growing number of fetuses. Nevertheless, approximately half of these cases remain undiagnosed. The WES technology demonstrates significant advantages in identifying sequence variants and has been proposed for application in prenatal genetic etiology diagnosis, particularly for fetuses exhibiting ultrasonic structural abnormalities. In the present study, WES was performed on a family with limb anomalies, leading to the first reported identification of a missense variant, c.956G > A (p.Arg319His), in the TP63 gene, associated with SHFM4 and exhibiting variable penetrance.
The TP63 gene, which can be detected in a variety of human and mouse tissues, including proliferating basal cells of epithelial layers in the epidermis, cervix, urothelium, and prostate [17], plays an important role in regulating epithelial, limb and craniofacial development. Variants within the TP63 gene are linked to six distinct disorders [11]. Although the phenotypes of these diseases overlap to a large extent, they all have their own characteristics [18, 19]. The full-length p63 protein consists of five functional domains: a transactivation domain, a DNA-binding domain, an oligomerization domain, a sterile-alpha motif (SAM) domain, and a transactivation inhibitory (TI) domain. Generally, the pattern of variants within the TP63 gene results in related but clinically distinguishable syndromes, establishing a specific correlation between TP63 alleles and these syndromes [20]. Causative variants located in the DNA-binding domain of the TP63 gene are associated with EEC3 syndrome, while the majority of variants situated in the SAM and TI domains lead to the development of AEC syndrome [18, 21]. Moreover, variants in ADULT syndrome, limb-mammary syndrome, and SHFM4 are distinctive to these respective syndromes [20]. Previous research has identified four variants in the TP6 gene in patients with SHFM4 and EEC3, all of which reside within the DNA-binding domain. Among these, two SHFM4-associated variants appear to be crucial for maintaining the structural integrity of the DNA-binding domain, whereas the TP63 variants leading to EEC3 are situated at amino acid residues that interact directly with DNA [22]. Furthermore, a previous study [23] indicated that the proteins resulting from EEC and AEC variants exhibited reduced transcriptional activity. Conversely, the proteins resulting from SHFM variants demonstrated transcriptional activity on several skin-specific gene promoters. This finding suggests an underlying regulatory feedback mechanism for TP63 that connects transcriptional activity to the modulation of protein homeostasis.
Despite that, in recent years, some studies revealed that the same variants in the TP63 gene can result in distinct disorders. For instance, research conducted by Ianakiev et al. [22] identified a c.673 C > T variant in the TP63 gene in a patient with SHFM who exhibited isolated split hands and feet. However, the same c.673 C > T variant identified in a fetus from China was associated with EEC syndrome [24]. Although SHFM4 and EEC syndrome share similar limb abnormalities, EEC syndrome is defined by additional clinical features, including cleft lip and palate, lacrimal duct atresia, hypospadias, hypopigmentation, and anomalies of the teeth, hair, and nails [25]. Moreover, the present study identifies a c.956G > A (p.Arg319His) variant in the TP63 gene, which was first reported by van Bokhoven H et al. [26] in a patient with EEC syndrome. A subsequent study [27] identified the same variants in a patient with ectrodactyly and syndactyly in both feet, along with tooth shape abnormalities, finally diagnosed as ELA (EEC/LM/ADULT) syndrome. However, both our findings and previous reports [28] indicate that patients with the c.956G > A variant may solely exhibit isolated limb abnormalities characteristic of SHFM4. The underlying genetic mechanisms accounting for divergent clinical phenotypes arising from the same variants remain unclear, potentially implicating epigenetic factors or post-transcriptional modifications.
EEC syndrome is characterized by reduced penetrance. Variants in the TP63 gene, specifically c.955 C > T (p.Arg319Cys), have been associated with EEC syndrome and SHFM, demonstrating this reduced penetrance [22, 29]. Additionally, a previous study [27] has identified the c.956G > A (p.Arg319His) variant in the TP63 gene, which is also associated with EEC syndrome and shows a similar pattern of reduced penetrance. However, it is important to note that there have been no reports suggesting that patients carrying the p.Arg319His variant leading to SHFM4 exhibit incomplete penetrance. In the present study, we present the first identification of the p.Arg319His variant in the TP63 gene within a Chinese family, demonstrating its association with SHFM4 and highlighting its incomplete penetrance.
AER is the transitory major signaling center for proximodistal growth and distal limb development [30]. Variants in genes associated with split-hand/split-foot malformation (SHFM) have been implicated in the dysregulation of FGF8 within the central region of the AER. This dysregulation can lead to the misexpression of several downstream target genes, resulting in the failure of AER stratification and, ultimately, the development of SHFM [1]. Previous studies indicated that TP63 plays a crucial role in forming AER and controlling AER functions via transcriptional regulation of AER-restricted target genes including DLX5, FGF8, SP6, SP8, and MSX1 [1]. The AER stratification failure caused by the interference of the p63-Dlx signaling pathway may be due to the dysregulation of TP63 target genes related to cell adhesion, such as PERP and CDH3 [1]. In this study, the patient with TP63 variants exhibited an increased level of TP63 mRNA. In addition, TP63 target genes related to cell adhesion, specifically PERP and CDH3, were found to be dysregulated, indicating further interference with the p63-Dlx signaling pathway that contributes to AER stratification failure. Results from KEGG analysis further corroborated an enrichment of cell adhesion molecules in the patient’s samples. As illustrated in Table 1; Fig. 6, the expression levels of TP63 and PERP genes were significantly upregulated; however, qPCR validation did not demonstrate a significant increase, potentially due to limitations in the controls employed. It is well known that FGF8 signaling is essential for AER induction and maintenance, and it is a direct target of TP63. Additionally, a previous study [10] reported that p63−/− mouse model (loss-of-function) exhibited decreased level of FGF8 in the AER, leading to the manifestation of split hand/foot malformation. In contrast, the variant of c.956G > A (p.Arg319His) observed in this study showed no significant difference in the expression level of FGF8 compared with the controls. Based on our findings, we propose that the p.Arg319His variant in TP63 gene may up-regulate the cell adhesion target genes of PERP and CDH3, disrupting the p63-Dlx signaling pathway, which ultimately results in the failure of AER stratification. This mechanism may represent a significant pathogenic pathway underlying the observed phenotypes in this study.
A previous study indicated that SHFM variants proteins exhibit transcriptional activity on several skin-specific gene promoters. Furthermore, the DNA-binding and sterile alpha motif (SAM) domain variants demonstrate extended half-lives and accumulate in the skin of patients with EEC and AEC syndromes in vitro [23]. Notably, the present study revealed an upregulation of TP63 expression in a patient with a variant located in the DNA-binding domain, suggesting a potential gain-of-function mechanism—although this mechanism was not conclusively determined in the current investigation. This gain of function may be associated with the observed extended half-lives of the variant proteins. Nevertheless, additional research is required to further elucidate the molecular mechanisms by which the TP63 variant contributes to SHFM4.
In summary, our study is the first to identify the c.956G > A (p.Arg319His) variant in the TP63 gene associated with SHFM4 in Chinese individuals, emphasizing the phenomenon of incomplete penetrance. Additionally, we hypothesize that p.Arg319His in the TP63 gene may result in gain-of-function effects, leading to the upregulation of cell adhesion target genes. This dysregulation has been shown to disrupt the p63-Dlx signaling pathway, ultimately contributing to the failure of AER stratification and the subsequent manifestation of SHFM4.
Acknowledgements
We extend our sincere gratitude to Huaqiao University, Fujian Provincial Science and Technology Project and the Quanzhou Science and Technology Bureau for their financial support of this research. We also wish to thank the patient who participated in the study for their invaluable contribution.
Author contributions
JZ conceived the study and authored the manuscript. HZ, YC, and YL conducted karyotype analysis, recruited participants, and provided clinical consultations. SL, MH, and JZ executed whole exome sequencing (WES), RNA sequencing analysis, and subsequent data analysis. CC and JZ undertook the revision and refinement of the manuscript. All authors have reviewed and approved the final version of the article.
Funding
This research was sponsored by Huaqiao University Joint of Hospital and University Innovation Project (2023YX001), Science and Technology Innovation Joint Fund project of Fujian Province (2023Y9255) and Quanzhou City Science and Technology Program of China (2023NS068).
Data availability
The raw sequence data reported in this study have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: https://ngdc.cncb.ac.cn/gsa-human, accession number: HRA009232).
Declarations
Ethics approval and consent to participate
Ethics approval and consent to participate: The Institutional Ethics Committee of Quanzhou Women’s and Children’s Hospital granted approval for the commencement of the study (2020No.31). All patients provided written informed consent and consented to the use of relevant data and information for scientific research.
Consent for publication
All subjects participating in this study provided written informed consent for publication of their own and their children’s genetic data and relevant information.
Competing interests
The authors declared no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jianlong Zhuang, Email: 415913261@qq.com.
Chunnuan Chen, Email: chenchunnuan1983@aliyun.com.
References
- 1.Kantaputra PN, Carlson BM. Genetic regulatory pathways of split-hand/foot malformation. Clin Genet. 2019;95(1):132–9. [DOI] [PubMed] [Google Scholar]
- 2.Ullah A, Hammid A, Umair M, Ahmad W. A novel heterozygous intragenic sequence variant in DLX6 probably underlies first case of autosomal Dominant Split-Hand/Foot malformation type 1. Mol Syndromol. 2017;8(2):79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villanueva C, Jacobson-Dickman E, Xu C, et al. Congenital hypogonadotropic hypogonadism with split hand/foot malformation: a clinical entity with a high frequency of FGFR1 mutations. Genet Med. 2015;17(8):651–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malik S, Percin FE, Bornholdt D, et al. Mutations affecting the BHLHA9 DNA-binding domain cause MSSD, mesoaxial synostotic syndactyly with phalangeal reduction, Malik-Percin type. Am J Hum Genet. 2014;95(6):649–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conte D, Garaffo G, Lo Iacono N, et al. The apical ectodermal ridge of the mouse model of ectrodactyly Dlx5;Dlx6-/- shows altered stratification and cell polarity, which are restored by exogenous Wnt5a ligand. Hum Mol Genet. 2016;25(4):740–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Umair M, Hayat A. Nonsyndromic Split-Hand/Foot Malformation: recent classification. Mol Syndromol. 2020;10(5):243–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukowska-Olech E, Sowińska-Seidler A, Wierzba J, Jamsheer A. SHFLD3 phenotypes caused by 17p13.3 triplication/ duplication encompassing fingerin (BHLHA9) invariably. Orphanet J Rare Dis. 2022;17(1):325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petit F, Sears KE, Ahituv N. Limb development: a paradigm of gene regulation. Nat Rev Genet. 2017;18(4):245–58. [DOI] [PubMed] [Google Scholar]
- 9.Yang A, Schweitzer R, Sun D, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398(6729):714–8. [DOI] [PubMed] [Google Scholar]
- 10.Restelli M, Lopardo T, Lo Iacono N, et al. DLX5, FGF8 and the Pin1 isomerase control ∆Np63α protein stability during limb development: a regulatory loop at the basis of the SHFM and EEC congenital malformations. Hum Mol Genet. 2014;23(14):3830–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sutton VR, van Bokhoven H. TP63-Related disorders. In: Adam MP, Everman DB, Mirzaa GM editors GeneReviews®. Seattle (WA): University of Washington, Seattle; June 8, 2010. [PubMed]
- 12.Zhuang J, Wang Y, Zeng S, Lv C, Lin Y, Jiang Y. A prenatal diagnosis and genetics study of five pedigrees in the Chinese population with Xp22.31 microduplication. Mol Cytogenet. 2019;12:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhuang J, Zhang N, Fu W, et al. Cytogenetic and molecular analysis of distal 4q duplication with distinctive phenotype using single-nucleotide polymorphism array. Mol Cytogenet. 2021;14(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riggs ER, Andersen EF, Cherry AM et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen) [published correction appears in Genet Med. 2021;23(11):2230]. Genet Med. 2020;22(2):245–257. [DOI] [PMC free article] [PubMed]
- 15.Zhuang J, Chen C, Chen Y, et al. Identification of a rare variant of c.1777G > A (p.G593S) in the COL1A1 gene as the etiology of recurrent Osteogenesis Imperfecta by whole-exome sequencing. Front Pediatr. 2022;10:816090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang A, Kaghad M, Wang Y, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2(3):305–16. [DOI] [PubMed] [Google Scholar]
- 18.Sawardekar SS, Zaenglein AL. Ankyloblepharon-ectodermal dysplasia-clefting syndrome: a novel p63 mutation associated with generalized neonatal erosions. Pediatr Dermatol. 2011;28(3):313–7. [DOI] [PubMed] [Google Scholar]
- 19.Niculescu L, Wagner M, Westphal DS, et al. A case of Ankyloblepharon-ectodermal defects-cleft Lip/Palate-syndrome with Choanal Atresia and skin erosions: phenotypic variability of TP63-related disorders. Acta Derm Venereol. 2019;99(1):111–2. [DOI] [PubMed] [Google Scholar]
- 20.van Bokhoven H, Brunner HG. Splitting p63 [published correction appears in Am J Hum Genet. 2003;72(3):779]. Am J Hum Genet. 2002;71(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soares E, Zhou H. Master regulatory role of p63 in epidermal development and disease. Cell Mol Life Sci. 2018;75(7):1179–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ianakiev P, Kilpatrick MW, Toudjarska I, Basel D, Beighton P, Tsipouras P. Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am J Hum Genet. 2000;67(1):59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Browne G, Cipollone R, Lena AM, et al. Differential altered stability and transcriptional activity of ∆Np63 mutants in distinct ectodermal dysplasias. J Cell Sci. 2011;124(Pt 13):2200–7. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Cheng L, Weng X, et al. [Identification of a Tp63 gene variant in an abortus with Ectrodactyly, ectodermal dysplasia, cleft lip/palate syndrome by whole-exome sequencing]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2020;37(2):139–41. [DOI] [PubMed] [Google Scholar]
- 25.Sutton VR, van Bokhoven H. TP63-Related disorders. In: Adam MP, Everman DB, Mirzaa GM editors GeneReviews®. Seattle (WA): University of Washington, Seattle; June 8, 2010. [PubMed]
- 26.van Bokhoven H, Hamel BC, Bamshad M, et al. p63 gene mutations in eec syndrome, limb-mammary syndrome, and isolated split hand-split foot malformation suggest a genotype-phenotype correlation. Am J Hum Genet. 2001;69(3):481–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otsuki Y, Ueda K, Nuri T, Satoh C, Maekawa R, Yoshiura KI. EEC-LM-ADULT syndrome caused by R319H mutation in TP63 with ectrodactyly, syndactyly, and teeth anomaly: a case report. Med (Baltim). 2020;99(44):e22816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bilal M, Hayat A, Umair M, et al. Sequence variants in the WNT10B and TP63 genes underlying isolated Split-Hand/Split-Foot Malformation. Genet Test Mol Biomarkers. 2020;24(9):600–7. [DOI] [PubMed] [Google Scholar]
- 29.Yang Y, Huang LY, Han J, Li DZ. Prenatal diagnosis of Ectrodactyly-Ectodermal dysplasia-cleft (EEC) syndrome in a Chinese woman with a TP63 mutation. Eur J Obstet Gynecol Reprod Biol. 2017;213:146–7. [DOI] [PubMed] [Google Scholar]
- 30.Lo Iacono N, Mantero S, Chiarelli A, et al. Regulation of Dlx5 and Dlx6 gene expression by p63 is involved in EEC and SHFM congenital limb defects. Development. 2008;135(7):1377–88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw sequence data reported in this study have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: https://ngdc.cncb.ac.cn/gsa-human, accession number: HRA009232).