Abstract
Sgt1p is a highly conserved eucaryotic protein that is required for both SCF (Skp1p/Cdc53p-Cullin-F-box)-mediated ubiquitination and kinetochore function in yeast. We show here that Sgt1p is also involved in the cyclic AMP (cAMP) pathway in Saccharomyces cerevisiae. SGT1 is an allele-specific suppressor of cdc35-1, a thermosensitive mutation in the leucine-rich repeat domain of the adenylyl cyclase Cyr1p/Cdc35p. We demonstrate that Sgt1p and Cyr1p/Cdc35p physically interact and that the activity of the cAMP pathway is affected in an sgt1 conditional mutant. Sequence analysis suggests that Sgt1p has features of a cochaperone. Thus, Sgt1p is a novel activator of adenylyl cyclase in S. cerevisiae and may function in the assembly or the conformational activation of specific multiprotein complexes.
The targeted proteolysis of cell cycle regulators is one of the major mechanisms controlling cell proliferation (13, 37). The SCF (Skp1p/Cdc53p-Cullin-F-box) complexes direct proteins such as Cdk inhibitors and G1 cyclins to the proteasome through ubiquitination (15). The SCF ubiquitin ligase catalytic core (Cdc53p, Cdc34p, and Rbx1p) is linked by Skp1p to F-box protein subunits that recognize the SCF substrates. Skp1p is also a subunit of the CBF3 (centromere binding factor 3) kinetochore complex together with Ndc10p, Cep3p, and the F-box protein Ctf13p (14, 66). Skp1p is essential for the activities of both complexes and is necessary for both entry into S phase and entry into anaphase (1, 14).
SGT1 (suppressor of the G2 allele of SKP1) was first described in yeast as a suppressor of skp1-4, a temperature-sensitive mutation of SKP1 that arrests cell division preferentially in G2/M at the restrictive temperature because of kinetochore defects (38). SGT1 is an essential gene that encodes a highly conserved eucaryotic protein, as shown by the ability of the human homolog of Sgt1p to restore the viability of the budding yeast null mutant. A study of sgt1-3 and sgt1-5 thermosensitive mutants showed that Sgt1p, like Skp1p, is needed for both entry into S phase and kinetochore function (38). Sgt1p physically associates with Skp1p and is required for both SCF and CBF3 activities, but its exact function in these processes is not known.
We show here that Sgt1p also contributes to the activity of the cyclic AMP (cAMP) pathway and physically interacts with the yeast adenylyl cyclase Cyr1p/Cdc35p. Adenylyl cyclases catalyze the synthesis of cAMP from ATP. All eucaryotic and some bacterial adenylyl cyclases contain structurally similar catalytic domains that are functional in a homo- or heterodimeric configuration (31). They can be classified in four groups. Class I cyclases, found in metazoans, contain 12 transmembrane segments and two cytosolic catalytic domains. Class II enzymes, found in Dictyostelium and protozoa, have an extracellular domain, a single transmembrane segment, and a cytosolic catalytic domain. Class III cyclases are peripherally associated plasma membrane proteins with one cytosolic catalytic domain and are found in yeasts and fungi. Class IV cyclases are soluble proteins found in bacteria and mammalian testes. They are part of signal transduction pathways in both procaryotes and eucaryotes that regulate a wide range of biological phenomena, including nutrient and stress responses (17); the regulation of cell growth, division, and differentiation (25, 40); hormonal responses (30); circadian rhythms; and long-term memory (48). In eucaryotes, adenylyl cyclases are regulated by GTP binding proteins (30). In budding yeast, a Gα subunit-type protein called Gpa2p and the GTP binding Ras proteins are both implicated in adenylyl cyclase activation in response to nutrient-rich conditions (71). Ras interacts with a leucine-rich repeat (LRR) region of yeast adenylyl cyclase and with a cyclase-associated protein called Cap/Srv2p (20, 62, 67).
The major effector of cAMP in eucaryotes is the cAMP-dependent protein kinase, or protein kinase A (PKA) (5). In the absence of cAMP, the catalytic subunit of PKA is found in an inactive complex with the regulatory subunit. The binding of cAMP by the regulatory subunit leads to dissociation of the complex and activation of the catalytic subunit (69). In budding yeast, the adenylyl cyclase pathway is notably involved in cell growth control and stress responses (71), but it also regulates the cell cycle by modulating G1 cyclin expression (2, 29, 72) and the activities of the anaphase-promoting complex/cyclosome and the SCF pathway (32). In terms of the stress response, the activation of adenylyl cyclase and PKA antagonizes the action of the Msn2p and Msn4p transcription factors, which activate the transcription of stress-responsive genes (7, 44, 59, 65). The nuclear accumulation of Msn2p and Msn4p is inversely correlated with the activity of the adenylyl cyclase/PKA pathway (27). Glycogen accumulation is also inversely correlated with the activity of the adenylyl cylase/PKA pathway (22). Our results thus implicate Sgt1p in the activities of pathways controlling cell growth and stress responses in addition to its previously implicated role in pathways controlling cell division.
MATERIALS AND METHODS
Strains and plasmids.
Table 1 lists the genotypes of the yeast strains used in this study. Table 2 lists the plasmids used in this study. The PDE2 deletion was made by digesting pJMΔpde2 with SacI-XbaI, transforming yeast cells with the products of digestion, and selecting for G418 resistance. SGT1 and CYR1 were epitope tagged by using the PCR cassettes pFA6a-13myc-TRP1 and pFA6a-HIS3-pGAL1-3HA as described previously (41). We constructed pFA6a-KanMX6-ANB1-UB-R-LacI-4HA by exchanging the BglII-PacI fragment of pFA6a-kanMX6-pGAL1-3HA for the ANB1-UB-R-lacI-4HA region of ZM168 (49). This cassette contains a ubiquitin-arginine-lacI-4HA peptide under the control of the ANB1 promoter (49).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| CMY389 | MATacyr1-2 leu2Δura3-52 trp1Δ1 ade2-101 lys2-801 | This study |
| CMY391 | MATacdc35-10 ura3-52 trp1Δ1 leu2Δ ade2-101 lys2-801 | This study |
| CMY282 | MATacdc35-1 sgt1-S371N ura3-52 trp1Δ1 his7 lys2-801 (A364a) | This study |
| CMY248 | MATasgt1-S371N ura3-52 his7 (A364a) | Lee Hartwell |
| CDY33 | MATaSGT1-13myc::TRP1 pGAL-3HA-CYR1::HIS3 ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2-801 ade2-101 | This study |
| CDY34 | MATaSGT1-13myc::TRP1 ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2-801 ade2-101 | This study |
| CDY35 | MATapGAL-3HA-CYR1::HIS3 ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2-801 ade2-101 | This study |
| YPH499 | MATaura3-52 lys2-801 ade2-10 trp1-Δ63 his3-Δ200 leu2-Δ1 | 64 |
| YKK57 | sgt1-5::LEU2 ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2-801 ade2-101 | 38 |
| CDY1 | MATapde2Δ::kanMX ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2-801 ade2-101 | This study |
| CDY3 | pde2Δ::kanMX sgt1-5::LEU2 ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2-801 ade2-101 | This study |
| CDY5 | MATapde2Δ::kanMX cdc35-1 sgt1-S371N ura3-52 trp1Δ1 his7 lys2-801 | This study |
| ZMY60 (CDY27) | MATapACE1-UBR1 pACE1-ROX1 trp1-Δ1 ura3-Δ52 leu2::PET56 ade2-101 | 49 |
| CDY26 | MATaANB1-UB-R-lacI-4HA-SGT1::kan pACE1-UBR1 pACE1-ROX1 trp1-Δ1 ura3-Δ52 leu2::PET56 ade2-101 | This study |
| CDY31 | MATapSTRE-lacZ::URA3 pACE1-UBR1 pACE1-ROX1 trp1-Δ1 ura3-Δ52 leu2::PET56 ade2-101 | This study |
| CDY32 | MATapSTRE-lacZ::URA3 ANB1-UB-R-lacI-4HA-SGT1::kan pACE1-UBR1 pACE1-ROX1 trp1-Δ1 ura3-Δ52 leu2::PET56 ade2-101 | This study |
| CDY116 | MATapGAL-3HA-cdc35-1::TRP1 sgt1-S371N ura3-52 his7 lys2-801 | This study |
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristic(s) | Reference(s) or source |
|---|---|---|
| YCp50-CYR1 | CYR1 CEN URA3 | 35 |
| ZM168 | pBS-ANB1-UB-R-LacI-HA | 49 |
| CDp4 | pFA6a-Kan-ANB1-UB-R-LacI-4HA | This study |
| PMJΔpde (CDp12) | pde2Δ::kanMX | 74 |
| CDp15 | pMM2-STRE-LacZ URA3 | 8, 44 |
| pSGT1 (CMP435) | pRS316(CEN-URA3)-SGT1; 2.3-kb EcoRI fragment | This study |
| pOBD2-SGT1 | pOBD2(CEN-TRP1)-pADH- GAL4DBD-SGT1 | This study |
| YCp50-TPK1 | TPK1 CEN URA3 | This study |
| YCp50-SGT1 | SGT1 CEN URA3 | This study |
Sequencing of the CYR1/CDC35 gene in the A364a strain background.
Complete sequencing of the wild-type CYR1/CDC35 gene in the A364a strain background (CMY282) revealed the following nucleotide differences compared with the published sequence of this gene in the S288C strain background (S288C sequence on the left and A364a sequence on the right): A9G, G911A, T2455C, T2508C, A2605G, G2956A, T3110C, A4305C, G4405A, and A5890G. Sequencing also revealed the following 24-bp insert (positions 1514 to 1537) in the A364a sequence relative to the S288C sequence: ACTCTCGTCATCGTAAGAACCGAC.
Glycogen staining and β-galactosidase assay.
Patches of yeast cells growing on 1% yeast extract-2% Bacto Peptone-2% dextrose (YPD) plates (2% agar) for one of the experiments (see Fig. 5A) or on synthetic complete medium containing 500 μM CuSO4 for another experiment (see Fig. 6E) were stained for glycogen by overlaying the plates with a solution of 0.4% I2-0.2% KI (12). β-Galactosidase activity was quantified as described previously (28).
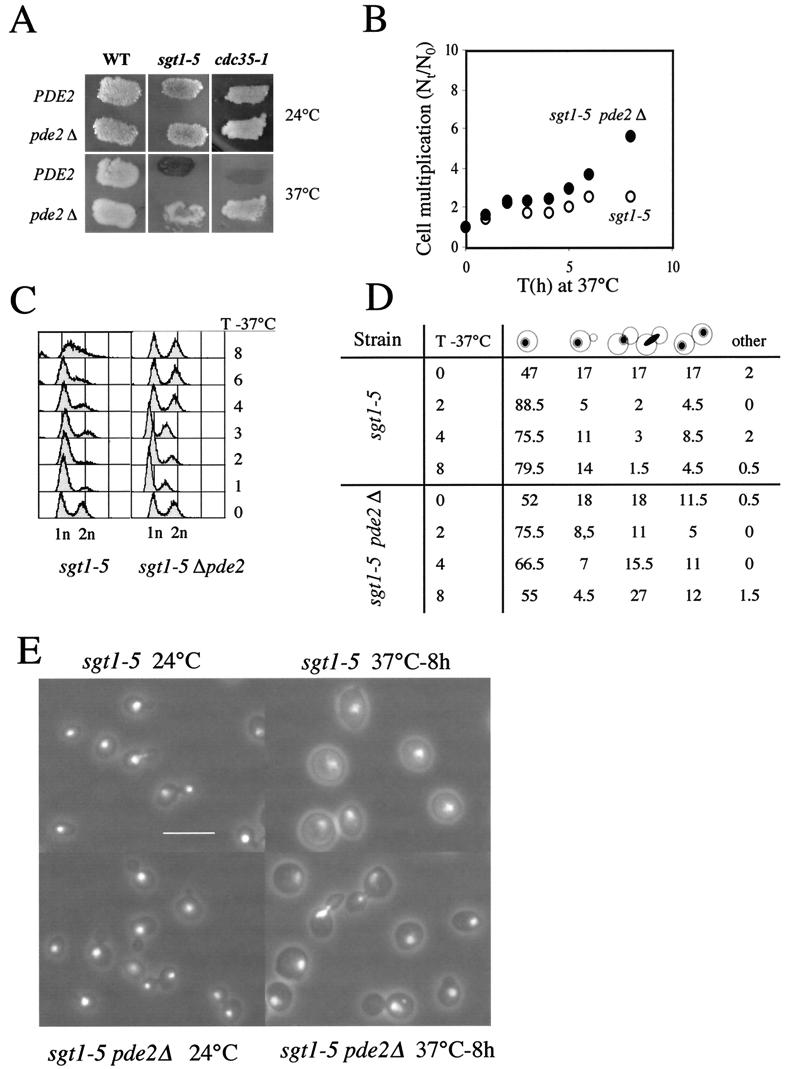
FIG.5.
Elevation of intracellular cAMP levels affects sgt1-5 phenotypes. (A) Wild-type (WT; YPH499), sgt1-5 (YKK57), cdc35-1 (CMY282), pde2Δ (CDY1), sgt1-5 pde2Δ (CDY3), and cdc35-1 pde2Δ (CDY5) strains were stained with iodine to visualize the accumulation of glycogen in patches of yeast colonies replicated on YPD plates at 24 and 37°C. The darkly staining sgt1-5 and cdc35-1 patches of cells incubated at 37°C indicate a high level of glycogen in these cells. (B) Exponentially growing sgt1-5 cells (open circles) and sgt1-5 pde2Δ cells (filled circles) on YPD plates at 24°C were transferred to 37°C, and the cell multiplication factor (number of cells at various times [Nt] divided by the number of cells at time zero [N0]) was determined at the indicated times (T). (C) Fluorescence-activated cell sorting analysis of DNA content. Times (T) are given in hours. 1n, G1-phase DNA content; 2n, G2-phase DNA content. (D) Morphological analysis of sgt1-5 and sgt1-5 pde2Δ cells at 24°C (time zero) and after transfer to 37°C for the indicated times (T, in hours). (E) Fluorescent phase-contrast images of sgt1-5 and sgt1-5 pde2Δ cells stained with propidium iodide to visualize nuclear DNA. Bar, 10 μm.
FIG. 6.
Induced proteolysis of N-degron-Sgt1p leads to arrest of cell growth and activation of the expression of a STRE-LacZ reporter construction. (A) Schematic diagram of the N-degron system (49) used to create a conditional null mutant of Sgt1p by induced proteolysis. (B to E) The addition of 0.5 mM CuSO4 to strain CDY26 (N-degron-Sgt1p) growing on synthetic complete medium at 30°C but not parental strain ZMY60 blocks cell growth (B); induces the proteolysis of N-degron-4HA-Sgt1p after 2, 4, and 6 h of incubation with CuSO4, as determined by immunoblotting with anti-HA antibodies (arrow, position of R-LacI-4HA-Sgt1p) (C); activates the expression of STRE-LacZ (β-gal., β-galactosidase) (D); and triggers glycogen accumulation specifically in strain CDY26 after 8 h of incubation, as shown by iodine staining of patches of cells replicated on agar medium plates (E).
Immunofluorescence microscopy and flow cytometry.
Flow cytometric analysis of yeast strains was performed as described previously (43). Immunofluorescence analysis of yeast cells was performed essentially as described previously (53) by using purified anti-myc mouse monoclonal antibody 9E10 at a final concentration of 7 μg/ml and Alexa 594-labeled goat anti-mouse immunoglobulin G (Molecular Probes) at a final concentration of 20 μg/ml. DNA was stained with 0.5 μg of 4′,6′-diamidino-2-phenylindole (DAPI)/ml. Twenty optical z sections separated by 150 nm were acquired by using a Leica DMRXA fluorescence microscope equipped with a ×100 oil immersion objective mounted on a piezoelectric motor and MetaMorph software (Universal Imaging Inc). Out-of-plane fluorescence was removed from each stack section by using a nearest-neighbor deconvolution algorithm with the default parameters provided by the MetaMorph software package.
Two-hybrid screen.
Gal4 DNA binding domain (DBD)-Sgt1p (expressed from vector pOB2) was used as bait, and interacting protein fragments coded for in the yeast genome were selected in the presence of 25 mM 3-aminotriazole as described previously (21). Clones were then checked for the expression of β-galactosidase from the second reporter gene in the screening strain.
IP.
Yeast strains CDY33, CDY34, and CDY35 were grown in 500 ml of 1% yeast extract-2% Bacto Peptone-2% galactose (YP-Gal) medium and harvested when the optical density at 600 nm reached 0.7. Cells were washed once with immunoprecipitation (IP) buffer (50 mM Tris [pH 7.5], 100 mM NaCl, 10 mM EDTA, 15% glycerol); resuspended in 700 μl of IP buffer containing 1 mM Pfabloc and 2 μg each of aprotinin, leupeptin, and pepstatin/ml; and broken in an Eaton press. Extracts were kept for 30 min on ice after the addition of 2 ml of IP buffer containing 1% Triton X-100. Extracts were centrifuged (JA20 rotor; 10 min, 4°C, 3,000 rpm), and the protein concentration in the supernatant was determined with the Bradford reagent (Bio-Rad). Crude extracts (1 to 4 mg of protein for anti-hemagglutinin [HA] IP or 4 mg for anti-myc IP in a final volume of 300 μl) were precleared with 25 μl of protein A-Sepharose beads that had been washed with IP buffer-1% Triton X-100 (1 h of incubation with rotation at 4°C and then centrifugation for 15 s at 100 × g) and then were incubated with rotation for 1 h at 4°C with 2 μl of 12CA5 (anti-HA) ascites fluid or 9E10 (anti-myc) purified antibody (0.7 mg/ml). Protein A-Sepharose beads were saturated for nonspecific protein binding by incubation for 1 h at 4°C with 4 mg of total protein extract from wild-type strain YPH499. A 25-μl quantity of saturated protein A-Sepharose beads was then incubated for 1 h at 4°C with anti-HA or anti-myc immune complexes. The beads were washed four times with IP buffer-1% Triton X-100, and immunoprecipitated proteins were solubilized by heating in the presence of sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer. Precleared extracts (10 to 40 μg), the IP supernatant (10 to 40 μg), and immunoprecipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, incubated with 12CA5 or 9E10 antibody followed by anti-mouse immunoglobulin G antibody coupled to horseradish peroxidase, and revealed by chemiluminescence detection. For one of the experiments (see Fig. 3D), CDY116 was grown in YP-Gal at 24°C, whereas CDY116(pSGT1) was grown at 24°C on synthetic medium containing Casamino Acids, adenine, tryptophan, and 2% galactose but no uracil in order to maintain selection for plasmid pSGT1. Protein extracts and immunoprecipitates were prepared as described above, except that protein A-Sepharose beads containing covalently coupled 12CA5 anti-HA antibody were used. Immunoprecipitates used for anti-Sgt1p immunoblots were solubilized with sample buffer in the absence of β-mercaptoethanol in order to reduce the liberation of immunoglobulin heavy chains from the protein A-anti-HA antibody beads that would otherwise obscure the Sgt1p signal on the immunoblots.
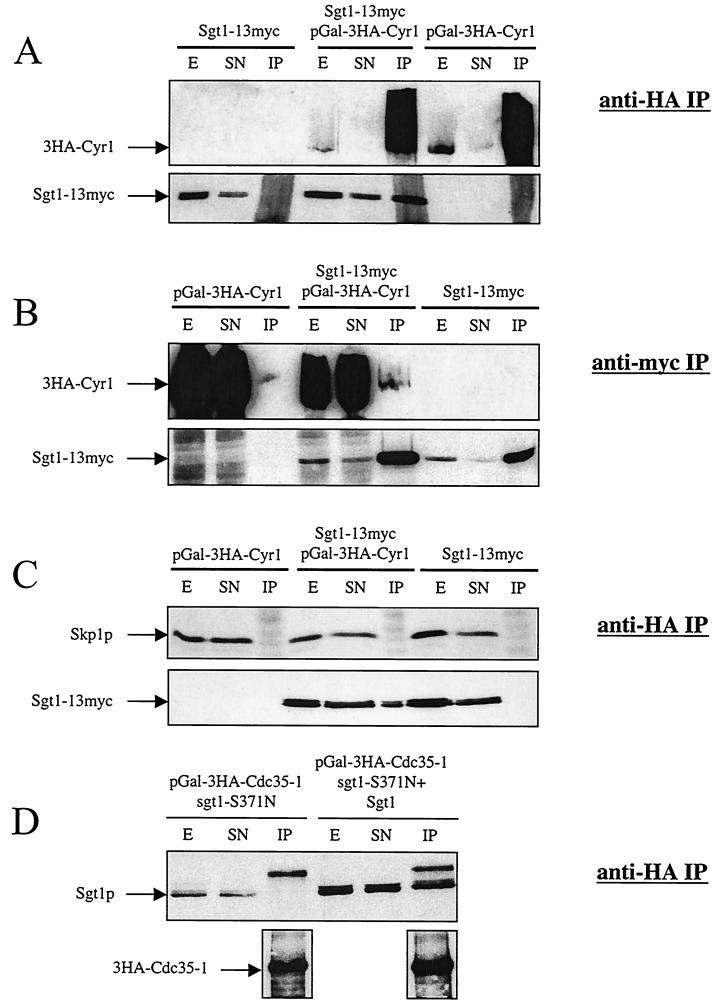
FIG. 3.
Sgt1p, but not Skp1p, coimmunoprecipitates with Cyr1p. (A) 3HA-Cyr1p overexpressed from a galactose-inducible promoter in strains CDY33 and CDY35 was immunoprecipitated with anti-HA antibody 12CA5, and the immunoprecipitates were analyzed by Western blotting to determine the efficiency of 3HA-Cyr1p IP and the fraction of coprecipitated Sgt1-13myc. Ten micrograms of the crude extract (E) and 10 μg of the supernatant (SN) are shown along with the total amount of the immunoprecipitated material (IP) from 1 mg of protein extract. (B) Sgt1-13myc expressed from its endogenous promoter in strains CDY33 and CDY34 was immunoprecipitated with anti-myc antibody 9E10, and the immu-noprecipitates were analyzed by Western blotting to determine the efficiency of Sgt1-13myc IP and the fraction of coprecipitated overexpressed 3HA-Cyr1p. Ten micrograms of the crude extract (E) and 10 μg of the supernatant (SN) are shown along with the total amount of the immunoprecipitated material (IP) from 4 mg of protein extract. (C) 3HA-Cyr1p overexpressed from a galactose-inducible promoter was immunoprecipitated with anti-HA antibody 12CA5, and the immunoprecipitates were analyzed by Western blotting to determine the fraction of coprecipitating Sgt1-13myc and Skp1p. Forty micrograms of the crude extract (E) and 40 μg of the supernatant (SN) are shown along with the total amount of the immunoprecipitated material (IP) from 4 mg of protein extract. Rabbit anti-Skp1p antibodies were kindly provided by Wade Harper. (D) 3HA-Cdc35-1p overexpressed from a galactose-inducible promoter in strain CDY116 with or without pSGT1 was immunoprecipitated from 1 mg of protein extract with protein A-Sepharose beads containing covalently coupled anti-HA antibody 12CA5. The immunoprecipitated material was then analyzed by immunoblotting with rabbit anti-Sgt1p polyclonal antibodies (upper panel) to test for coprecipitation of Sgt1-S371N or wild-type Sgt1p with Cdc35-1p or anti-HA antibodies to determine the level of 3HA-Cdc35-1p immunoprecipitated from each cellular extract (lower panels). In the upper panel, 20 μg each of crude extract (E) and supernatant (SN) was loaded next to the total amount of the immunoprecipitated material (IP).
Sequence-structure threading and analysis.
The sequence analysis of and the structure prediction for the different Sgt1p domains of Saccharomyces cerevisiae were carried out by considering independently the following sequence regions: positions 1 to 133, 168 to 219, and 220 to 395. The compatibility of the individual sequences and sequence alignments with known three-dimensional (3D) protein structures was tested by using the sequence-structure threading methods FUGUE (61) (http://www-cryst.bioc.cam.ac.uk/∼fugue/) and 3D-PSSM (36) (http://www.bmm.icnet.uk/servers/3dpssm/). These algorithms recognize protein folds by using one-dimensional and 3D sequence profiles coupled with secondary structure and solvation potential information. They allow for the detection of remote sequence homology and for the optimization of sequence alignments.
For the region from positions 1 to 133, a preliminary multiple-sequence alignment was calculated by using the Clustal W algorithm (33) with S. cerevisiae Sgt1p-homologous sequences from Schizosaccharomyces pombe, Neurospora crassa, Homo sapiens, Arabidopsis thaliana, and Oryza sativa. The multiple-sequence alignment was analyzed with the threading methods 3D-PSSM and FUGUE, both of which identified with high levels of confidence (over 95 and 99%, respectively) the tetratricopeptide repeat (TPR) domain of protein phosphatase 5 as the most compatible fold (sequence identity, 13%; PDB no. 1A17). The scores for the prediction were an E value of 1.87 × 10−4 and a Z score of 8.12 with the 3D-PSSM and the FUGUE methods, respectively. The 3D-PSSM method also identified the TPR domains of the Hop protein (PDB no. 1ELR and 1ELW) as highly probable folds for Sgt1 (positions 1 to 133), although their sequence identity with Sgt1p was rather low (15 and 16% for TPR1 and TPR2, respectively). The protein phosphatase 5 and Hop TPR domains bind the acidic C terminus of Hsp90 (56, 58). Interestingly, analysis of the S. cerevisiae Sgt1p sequence alone without the use of the multiple-sequence alignment did not allow any confident prediction.
For the second region (positions 168 to 219; CS domain), analysis of the S. cerevisiae Sgt1p sequence alone allowed the 3D-PSSM and FUGUE methods to identify with high levels of confidence (over 95 and 99%, respectively) the fold of the Hsp90 cochaperone p23 as the most probable fold (sequence identity, 18%; PDB no. 1EJF). The scores were an E value of 8.12 × 10−6 and a Z score of 11.05 with the 3D-PSSM and the FUGUE methods, respectively. Similar values were obtained for the sequence of H. sapiens Sgt1 (21% sequence identity with p23) and the sequence of the Siah-interacting protein (SIP) (16% sequence identity with p23) (46).
Next, the sequence alignment was optimized and a structural model was generated by using the Centre de Bioinformatique Structurale server and the incremental threading optimization evaluation method (16, 39) (http://bioserv.infobiosud.univ-montp1.fr/bioserver/). Finally, regarding the 3D structure of the C-terminal region spanning the adenylyl cyclase-interacting domain (AC domain), no prediction could be proposed with confidence. Nevertheless, the Psi-Pred2 method for secondary structure prediction (34) (http://bioinf.cs.ucl.ac.uk/psipred/) detected with a high level of confidence a helix-turn-helix motif in the region that is conserved in both the Sgt1p and the SIP families.
A structural model of the Cyr1p LRR domain was generated to analyze the location of mutated residue L901 in the LRR structure. The structure of the internalin B LRR domain (PDB no. 1D0B) was used as a template because it had the highest sequence identity with Cyr1p in the region of the mutation. The structural model was generated as described above.
RESULTS
SGT1 suppresses the thermosensitivity of a cdc35-1 mutant.
cdc35-1 is a temperature-sensitive allele of the adenylate cyclase gene CYR1/CDC35 that was isolated in the A364a genetic background (54). In genetic crosses with CYR1+/CDC35+ A364a strains, tetrads with 2TS+:2ts− segregation were obtained, as expected. However, in outcrosses with CYR1+/CDC35+ strains in the S288C/YPH background, 3TS+:1ts− segregation was observed, consistent with the presence of an unlinked suppressor locus in the S288C/YPH background. Further crosses showed that this suppressor acted in a dominant fashion. We thus sought to identify it by transforming the cdc35-1 A364a mutant with an S288C genomic DNA library in the YCp50 centromeric plasmid (55) and selecting for temperature-resistant growth. Three different genes were isolated in this manner (Fig. 1A): CYR1, encoding the wild-type adenylyl cyclase; TPK1/SRA3, encoding one of the catalytic subunits of PKA (9); and SGT1, encoding an Skp1p-interacting protein (38).
FIG. 1.
Isolation of SGT1 and structure of the encoded protein. (A) SGT1 is an allele-specific suppressor of cdc35-1. cdc35-1 (CMY282), cdc35-10 (CMY391), and cyr1-2 (CMY389) mutants were transformed with the indicated plasmids at 23°C, with selection for growth on synthetic medium without uracil. Patches of cells were then replica plated and incubated at 23 and 36°C to test for the suppression of thermosensitive growth. Ycp50-CYR1 and Ycp50 are positive and negative controls, respectively, for complementation. (B) Schematic diagram of domains found within the Sgt1p sequence. The TPR and Sgt1 CHORD (CS domain) motifs were previously described (38, 63), and the highly conserved C-terminal sequence may be a domain that interacts with adenylyl cyclase (AC domain) in yeast cells. (C) Sequence and secondary structure prediction of the highly conserved C-terminal domain of Sgt1p from a series of eucaryotic organisms (S.c., S. cerevisiae; S.p., S. pombe; N.c., N. crassa; C.e., Caenorhabditis elegans; O.s., O. sativa, H.s., H. sapiens, D.m., Drosophila melanogaster) showing the positions of the sgt1-S371N (A364a) mutation and one of the two sgt1-5 mutations. The secondary structure prediction (Pred) (H, alpha helix; E, β strand; C, coil) was obtained with the Psi-Pred2 algorithm (34) (http://bioinf.cs.ucl.ac.uk/psipred/). The height of the bars is an estimate of the confidence (Conf) of the prediction. The region of Sgt1p between amino acids (AA) 338 and 365 is predicted with a high level of confidence to adopt a helix-turn-helix structure. Amino acid residues that are identical for at least five of the seven Sgt1p homologs are shown in bold.
These data suggested that SGT1 contributes to adenylyl cyclase activity and that the sgt1 allele found in the A364a strain background is deficient for this function. Sequencing of the sgt1 allele in the A364a background showed that it encoded a protein that was identical to that encoded by the SGT1 allele in the S288C background, except for an S371N substitution. Ser-371 is absolutely conserved in all known Sgt1p sequences and is found in a highly conserved C-terminal region of the protein (Fig. 1B and C). We found no phenotype associated with the sgt1-S371N allele in the A364a background in the presence of a wild-type CDC35/CYR1 gene. Interestingly, one of the two amino acid substitutions found in the sgt1-5 mutant (E364K) affects another highly conserved residue that maps close to the S371N substitution (Fig. 1C), and the sgt1-5 mutant has a thermosensitive G1 arrest phenotype that resembles that of cdc35/cyr1 mutants (38). Thus, the sgt1-5 mutant may also have an adenylyl cyclase deficiency (see below).
The cdc35-1 allele has a point mutation in the LRR domain of Cyr1p.
YCp50-CYR1 and YCp50-TPK1 complemented two other temperature-sensitive alleles for adenylyl cyclase, cyr1-2 and cdc35-10, whereas Ycp50-SGT1 could suppress only the thermosensitivity of cdc35-1 (Fig. 1A) and is thus an allele-specific suppressor. The cyr1-2 mutation creates a UGA stop codon that is thought to lead to low levels of adenylyl cyclase by readthrough translation (50), and the cdc35-10 mutation creates thermolabile adenylyl cyclase activity (6). In contrast, the cdc35-1-encoded adenylyl cyclase activity in membrane preparations was not thermosensistive and did not respond to G protein stimulation in vitro, suggesting that cdc35-1 may affect a Cyr1p regulatory site (10, 68). We sequenced the entire cdc35-1 allele and found that it differs from the wild-type allele in the same genetic background by only a T2702A transversion that leads to an L901H substitution affecting a conserved leucine in the LRR domain of the protein (Fig. 2A and B). A structural model of this region of the Cyr1p LRR domain shows that L901 is likely to be part of the hydrophobic core of an LRR (Fig. 2C). The leucine-to-histidine mutation is predicted to destabilize the structure of the hydrophobic core in this local region. The LRR domain has been implicated in Ras2 activation of Cyr1p (20, 62, 67). We thus tested whether the suppression of cdc35-1 by SGT1 required RAS2. A cdc35-1 SGT1 strain grew at 37°C, whereas a cdc35-1 SGT1 ras2Δ strain showed thermosensitive growth similar to that of a cdc35-1 sgt1-S371N mutant (unpublished data). SGT1 and RAS2 are thus both required to suppress the thermosensitive growth of the cdc35-1 mutant.
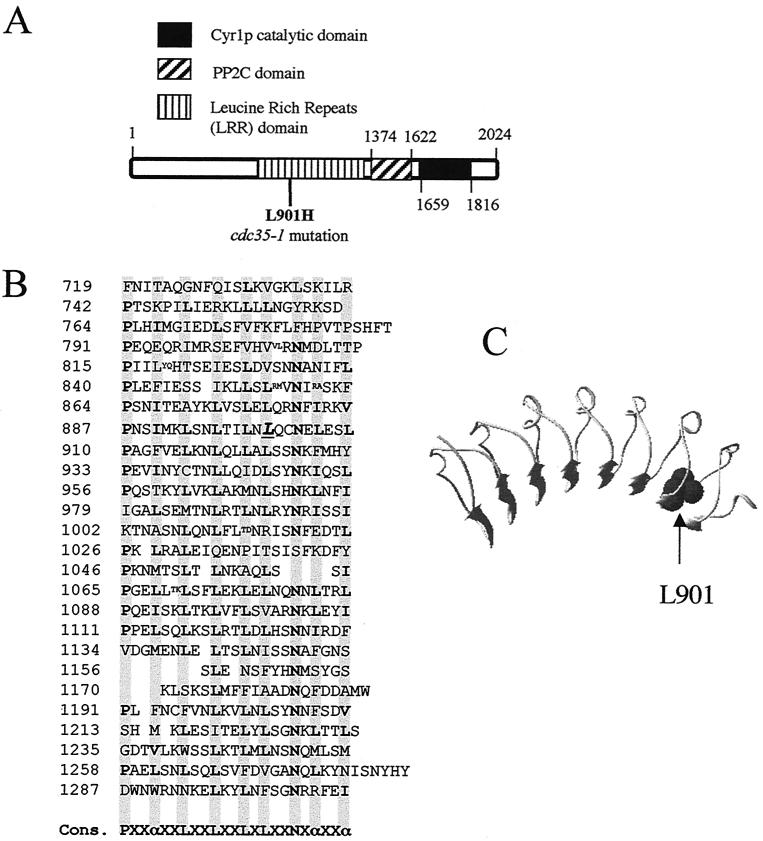
FIG. 2.
The cdc35-1 mutation affects a conserved leucine in the LRR domain of adenylyl cyclase. (A) Schematic diagram of the Cyr1p/Cdc35p sequence showing the position of the L901H substitution within the LRR domain of the cdc35-1 mutant and the positions of the protein phosphatase 2C (PP2C) and adenylyl cyclase catalytic domains, as proposed by the Pfam database (3). (B) Sequence of the Cyr1p LRR domain showing the position of Leu-901 (underlined and in italic type). Note that this sequence corresponds to that of the wild-type CYR1/CDC35 gene in the A364a background, which differs at a number of positions from the published sequence of the gene in the S288C background (see Materials and Methods). Cons., consensus; α, preferred hydrophobic residue. Shaded columns show the positions of preferred amino acids in the leucine-rich repeats. Small letters show sites of amino acid insertions within the repeats. (C) Structural model of the Cyr1p LRR domain in the environment of Leu-901.
Cyr1p and Sgt1p can be coimmunoprecipitated.
The localization of the cdc35-1 mutation to the LRR domain and the allele specificity of cdc35-1 suppression by SGT1 suggested that Sgt1p and Cyr1p may physically interact. This notion was tested by immunoprecipitating tagged versions of each protein and determining whether the putative partner was coprecipitated. Sgt1-13myc was expressed from its normal chromosomal promoter. Attempts to visualize by immunoblotting Cyr1-3HA expressed from its normal chromosomal promoter were unsuccessful, so we overexpressed 3HA-Cyr1p from a galactose-inducible promoter. Under these conditions, we could readily demonstrate the presence of Sgt1-13myc in 3HA-Cyr1p immunoprecipitations (Fig. 3A) and the presence of 3HA-Cyr1p in Sgt1-13myc immunoprecipitations (Fig. 3B). Approximately 1% of Sgt1p in the extracts was coprecipitated with overexpressed 3HA-Cyr1p. This relatively low value may reflect the presence of Sgt1p in several different protein complexes (38) or a transient or labile interaction.
The coimmunoprecipitation of Sgt1-13myc and 3HA-Cyr1p indicates that they can form a complex and that they may interact directly. In contrast, we did not observe the coimmunoprecipitation of Skp1p with 3HA-Cyr1p under conditions in which we readily observed the coimmunoprecipitation of Sgt1-13myc (Fig. 3C). This result suggests that Sgt1p forms exclusive complexes with Cyr1p and Skp1p.
We also visualized the intracellular localization of Sgt1-13myc by indirect immunofluorescence with anti-myc antibodies. A stack of optical sections was acquired through fixed yeast cells expressing Sgt1-13myc and stained with DAPI to visualize nuclear DNA. The stack of images was then treated with a deconvolution program to remove out-of-plane fluorescence from each section. Figure 4 shows three consecutive images from a z-section stack in which each image is separated by 150 nm. Sgt1-13myc was found throughout the cytosol as filiform or punctate aggregates. Sgt1-13myc was also observed in the nucleus as filiform or punctate aggregates, although its concentration there might have been lower than that in the cytosol, and it was largely excluded from the vacuole. Similar images were also seen for the immunofluorescent localization of 6HA-Sgt1p (data not shown). No immunofluorescence signal was detected under the same conditions from a congenic strain that did not express epitope-tagged Sgt1p. The observed nuclear and cytosolic localizations of Sgt1p are consistent with its proposed nuclear and cytosolic functions.
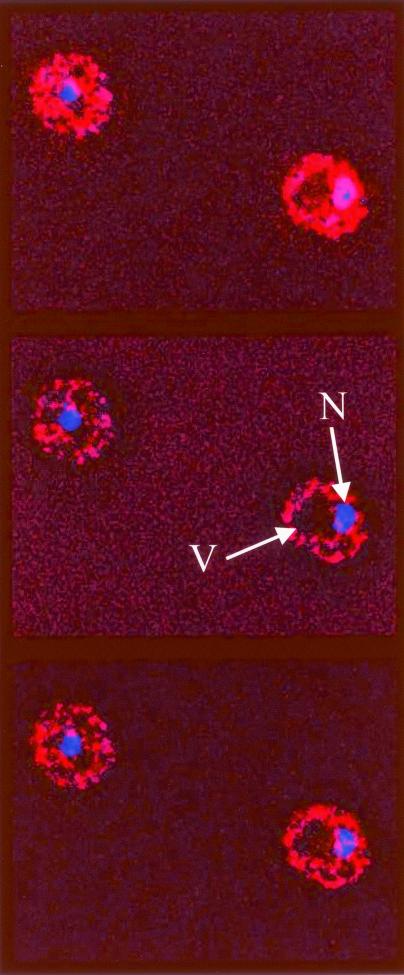
FIG. 4.
Indirect immunofluorescence analysis of strain CDY34 done to visualize the intracellular localization of Sgt1-13myc with purified mouse anti-myc monoclonal antibody 9E10. DNA was visualized with DAPI staining. Three consecutive optical sections separated by 150 nm are shown from a z-section stack after deconvolution of the stack in order to remove out-of-plane fluorescence from each section. Sgt1-13myc fluorescence is shown in red, DNA is shown in blue, andcolocalization is shown in magenta. Under identical conditions, no immunofluorescence signal was observed for a congenic strain that did not express a myc-tagged protein. N, nucleus; V, vacuole. The yeast cells shown are about 5 μm in diameter.
Interaction of Sgt1p with the LRR domain.
The localization of the cdc35-1 mutation to the LRR domain of Cyr1p suggested that Sgt1p may directly or indirectly bind this domain. We compared the ability of wild-type Sgt1p and Sgt1p-S371N to bind the Cdc35-1p mutant. 3HA-Cdc35-1p was overexpressed from the GAL promoter at 24°C in cells expressing only Sgt1p-S371N or in cells expressing Sgt1p-S371N and wild-type Sgt1p. 3HA-Cdc35-1p was then immunoprecipitated from cell extracts with anti-HA antibodies, and coprecipitation of Sgt1p was tested by immunoblotting with anti-Sgt1p rabbit polyclonal antibodies. Sgt1p coprecipitated with Cdc35-1p (Fig. 3D), as it did with wild-type Cdc35p/Cyr1p (Fig. 3A), but Sgt1p-S371N did not coprecipitate with Cdc35-1p (Fig. 3D). The defect in the physical interaction between Cdc35-1p and Sgt1p-S371N correlates with the thermosensitivity of strains containing both of the mutations and supports the idea that the C terminus of Sgt1p may interact with the LRR domain of Cdc35p.
In order to further define its binding specificity, we used Sgt1p as bait in a two-hybrid screen with a library of coding fragments from the yeast genome as interacting prey. Three classes of interacting protein sequences were identified (Table 3). The first class was represented by protein fragments containing the F-box motif (52). Since Skp1p binds the F-box motif (60) and Sgt1p binds Skp1p (38), this class of proteins may indirectly interact with Sgt1p through Skp1p. The second protein class contained well-defined (Sds22p and Ccr4p) or potential (Ydr306c) LRR domains. Sds22p is mainly composed of LRR sequences, and the Sgt1p-interacting region is almost entirely derived from the LRR domain. Thus, Sgt1p may generally bind LRR domains. Another intriguing observation is that Ydr306c is an F-box protein (52), but the interacting region identified in our screen contains the putative LRR sequence and not the F-box motif. Thus, Sgt1p may interact both directly and indirectly with some F-box proteins. The third class of interacting proteins had no shared sequence motifs and is of uncertain functional significance.
TABLE 3.
Sgt1p-interacting protein fragments coded within the yeast genome and identified by a two-hybrid screena
The schematic diagrams show motifs identified by the ProfileScan server (http://hits.isb-sib.ch/cgi-bin/PFSCAN) for each gene product and the position of the two-hybrid-interacting region. Ydr306c was predicted with a low level of confidence to contain an LRR motif (question mark). Symbols represent the interacting region (gray bar), F-box motif (black square), LRR (white bar), WD40 repeat (gray circle), Zn2Cys6 cluster domain (white hexagon), and paired amphipathic helix repeat (black circle within white circle).
Increased intracellular levels of cAMP modify the sgt1-5 phenotype.
If SGT1 is required for adenylyl cyclase function, some sgt1 mutants should show phenotypes similar to those of cdc35/cyr1 mutants. Because SGT1 is essential for S. cerevisiae viability, we studied conditional mutants. The sgt1-5 thermosensitive mutant is affected in its SCF function and is unable to enter S phase at the restrictive temperature like other SCF mutants (38); however, rather than showing a multibud phenotype like other SCF mutants, it instead shows arrested growth with unbudded cells similar to the arrest seen for cdc35/cyr1 mutants (45, 54). Moreover, iodine staining showed that the sgt1-5 strain accumulates glycogen at the restrictive temperature (Fig. 5A), as do cAMP pathway mutants, such as cdc35-1 (22). These phenotypes suggested that the cAMP pathway may be deficient in the sgt1-5 mutant at the restrictive temperature.
We tested a possible contribution of deficient adenylyl cyclase function to the sgt1-5 phenotype by deleting the PDE2 gene, which encodes a high-affinity cAMP phosphodiesterase (57). pde2 mutants have increased intracellular cAMP levels and are phenotypically more sensitive to the addition of cAMP to the growth medium (47, 74). Deletion of PDE2 suppressed the glycogen accumulation of both the sgt1-5 and the cdc35-1 mutants at 37°C (Fig. 5A). Furthermore, when an sgt1-5 pde2 double mutant was shifted to 37°C, it showed arrested growth after two or three cell divisions, whereas the parental sgt1-5 single mutant showed arrested growth under the same conditions after one cell doubling (Fig. 5B). Deletion of PDE2 also partially suppressed the unbudded G1-phase arrest of the sgt1-5 mutant (Fig. 5C to E). Approximately half the cells of the sgt1-5 pde2 double mutant showed arrested division at 37°C, with large buds and a 2C DNA content. About 75% of large-bud cells contained an undivided nucleus, and the remainder were arrested in late nuclear division. These results suggested that inadequate cAMP synthesis partially accounts for the unbudded G1-phase arrest, but they also suggested that the sgt1-5 mutant is deficient in at least one other pathway required for growth and budding, as well as pathways required for nuclear division and cytokinesis. The defect in nuclear division is probably due to a kinetochore assembly problem (38).
The cAMP pathway is affected in a conditional null mutant for SGT1.
The currently available thermosensitive alleles of SGT1 may be deficient only in a subset of its functions (38). To further test the possibility that Sgt1p is an activator of adenylyl cyclase, we constructed a conditional null mutant for SGT1 by using N-degron-Sgt1p expressed from a repressible promoter (Fig. 6A) and the system developed by Moqtaderi et al. (49). In this system, copper induces the expression of Ubr1p and Rox1p. Ubr1p is a ubiquitin ligase that binds proteins containing an N-degron and facilitates their ubiquitination and subsequent destruction by the 26S proteasome. Rox1p represses the transcription of the ANB1 promoter controlling the expression of N-degron-Sgt1p. Thus, the addition of copper both blocks the expression of N-degron-Sgt1p and triggers its proteolysis. As expected, the constructed strain was unable to grow on medium containing 500 μM CuSO4 (Fig. 6B). The levels of N-degron-Sgt1p decreased rapidly after CuSO4 addition and were markedly lower after 4 h of incubation (Fig. 6C). We used a stress response element (STRE)-LacZ reporter system to evaluate the activity of the cAMP pathway (44). STRE-mediated expression is controlled by the Msn2p and Msn4p transcription factors, whose activity is inversely proportional to the activity of the cAMP pathway (7, 24, 27, 65). Induced proteolysis of Sgt1p led to increased β-galactosidase expression from STRE-LacZ (Fig. 6D) and to the accumulation of glycogen, as indicated by iodine staining of cells (Fig. 6E). Combined with our other data, these results suggested that depleting Sgt1p results in reduced cAMP levels, which activate Msn2p and Msn4p, and triggers glycogen accumulation.
Sequence analysis of the CS domain of Sgt1p suggests features of a cochaperone.
An amino-terminal region of Sgt1p was suggested to adopt a TPR fold similar to that of Sti1/Hop (38), a cochaperone for Hsp70 and Hsp90 (11, 58). We obtained similar results by using the 3D-PSSM and FUGUE sequence threading methods (36, 61) to model this region (see Materials and Methods). The central region of Sgt1p contains a cysteine- and histidine-rich domain (CHORD) motif (CS domain) that is shared with the SIP family (46) and some other CHORD-containing proteins (63). The 3D-PSSM and FUGUE methods fitted with high levels of confidence (over 95 and 99%, respectively) the CS motif of S. cerevisiae Sgt1p to the fold of the Hsp90 cochaperone p23 (sequence identity, 18%; PDB no., 1EJF). Similar results were obtained for the CS motifs of H. sapiens Sgt1p and SIP, an Sgt1-related protein that also binds Skp1p (46).
Figure 7A shows a sequence alignment of the CS motifs of Sgt1p, SIP, and p23. The positions of the hydrophobic residues buried in the structure of p23 (73) are highlighted in gray (solvent-accessible surface, below 20%). Figure 7B shows a ribbon representation of the structural model of the CS motif derived from the fold recognition methods. Mutation N213I in the sgt1-3 mutant appears to disrupt the binding of Sgt1p to Skp1p (the other mutations in sgt1-3, L31P and F99L, are located in the TPR domain, which does not appear to be required for Skp1p binding) (38, 46). From the structural model, we found close to conserved N213 several exposed residues that are conserved in S. cerevisiae Sgt1p, H. sapiens Sgt1p, and SIP. Together with N213, they may take part in the binding of Sgt1p or SIP to the Skp1p N-terminal domain (60). Figure 7C shows the accessible surface of this set of residues colored with respect to sequence diversity. Their corresponding positions in the sequence alignment are highlighted. Figure 7D shows the accessible surface of the set of residues close to N213 with a color code related to the amino acid type. This view suggests that the putative binding region may involve both hydrophobic and polar interactions. The remarkable structural similarity of the Sgt1p TPR and CS domains to Hsp90 cochaperones suggests that Sgt1p may have a similar function, although we have not succeeded in immunoprecipitating Hsp90 with Sgt1-13myc (unpublished data).
FIG. 7.
Sequence threading suggesting that the C5 domain of Sgt1p adopts a fold similar to that of the Hsp90 cochaperone p23. (A) Sequences of the CS domains of S. cerevisiae Sgt1 (Sgt1_sc), H. sapiens Sgt1 (Sgt1_hs), and SIP that align with that of the Hsp90 cochaperone p23 (PDB no. 1EJF), validated by using fold recognition methods. The positions of the hydrophobic residues buried in the structure of p23 (73) are highlighted in gray (solvent-accessible surface, below 20%). The arrows indicate the positions of the β strands in the structure of p23, and their colors correspond to those of the β strands in panel B. The red asterisk indicates conserved N213. (B) The structural model derived from the fold recognition methods is shown as a ribbon representation. The colors of the strands indicate the order of the secondary structure elements from the N terminus (blue) to the C terminus (red). (C) The accessible surface of a set of residues close to N213 (implicated in binding Skp1p) is shown and colored with respect to the sequence diversity (blue for conserved, green for similar, and yellow for nonconserved positions). Their corresponding positions in the sequence alignment are indicated. (D) The accessible surface of the set of residues close to N213 is shown with a color code related to the amino acid type (gray for hydrophobic, yellow for polar, and blue for positively charged amino acids).
DISCUSSION
Sgt1p was initially described as an Skp1p-interacting protein involved in kinetochore assembly and SCF activity (38). We demonstrate here that Sgt1p also contributes to the activity of the cAMP pathway and physically interacts with yeast adenylyl cyclase. An initial genetic interaction was based on the presence of a defective allele of SGT1 in the A364a strain background. In an otherwise wild-type background, the A364a sgt1 allele had no evident phenotypic effect. However, when combined with a cdc35-1 mutation, it led to thermosensitive growth due to the inactivation of adenylyl cyclase. The cdc35-1 mutation itself had no evident phenotypic effect in an SGT1+ background. This interaction was allele specific because SGT1+ did not suppress two other thermosensitive adenylyl cyclase mutations, cyr1-2 and cdc35-10. The cdc35-1 mutation is an L901H substitution in the LRR region of adenylyl cyclase, and the defective sgt1 allele contains an S371N mutation affecting a highly conserved residue in the C-terminal region of the protein. We further showed that Sgt1p and Cyr1p/Cdc35p, but not Skp1p and Cyr1p/Cdc35p, could be coimmunoprecipitated from yeast extracts. Furthermore, Sgt1p, but not the Sgt1p-S371N mutant, could be coimmunoprecipitated with the Cdc35-1p mutant. These results suggest that the C-terminal conserved domain of Sgt1 may interact with the LRR domain of adenylyl cyclase and that this interaction may exclude an interaction between Sgt1p and Skp1p. Supporting a possible interaction site on Sgt1p for LRR domains is our observation of two-hybrid interactions between Sgt1p and fragments of Sds22p and Ccr4p that contain LRR motifs.
How might Sgt1p contribute to adenylyl cyclase activity in yeast cells? Biochemical analyses of yeast and fungal adenylyl cyclases are not as advanced as the analysis of mammalian adenylyl cyclases. Like mammalian and fission yeast adenylyl cyclases, budding yeast Cyr1p can be activated by a Gα family protein called Gpa2p (71). However, Cyr1p activity also requires the small G protein Ras, and this property is not conserved in either fission yeast or mammals (4, 23). Immunoaffinity purification of overexpressed, epitope-tagged Cyr1p demonstrated that it was approximately stoichiometrically associated with a 70-kDa protein called cyclase-associated protein (CAP)/Srv2p (18, 19, 26). Other Cyr1p-interacting proteins, including Sgt1p, Ras2p, and Gpa2p, were not identified. This result may mean that these regulators were present in substoichiometric quantities relative to the overexpressed Cyr1p or that their affinity for Cyr1p was lower than that of CAP. The N-terminal domain of CAP was shown to form a coiled-coil interaction with the C terminus of Cyr1p and to be required along with the Cyr1p LRR domain for Ras2p-mediated activation of adenylyl cyclase (20, 51, 67). Nevertheless, the precise mechanistic basis for the activation of Cyr1p by Ras2p and CAP or by Gpa2p is unknown. Adenylyl and guanylyl cyclases generally function as homo- or heterodimers (31). Studies of Gα activation of mammalian adenylyl cyclase suggest that it can both facilitate dimerization and induce a conformational change in the dimerized subunits that increases catalytic activity (70). The immunoaffinity-purified Cyr1p-CAP complex sediments as a dimer on glycerol gradients (19), a finding which would suggest that Sgt1p, Ras2p, and Gpa2p are not required for dimerization. It remains possible that these proteins facilitate the dimerization of the complex in situ within cells or that they act as allosteric activators of the dimerized complex.
Sgt1p and, notably, the C-terminal region that we have implicated in a Cyr1p interaction, are highly conserved in eucaryotes. Adenylyl cyclase catalytic domains are also conserved, but sequences outside of the catalytic domains are not. Remarkably, the C-terminal region of Git7p, the Sgt1p homolog in fission yeast, has also been shown to be required for adenylyl cyclase activity in this yeast (57a). Our results suggest that Sgt1p interacts with the LRR domain of Cyr1p, a domain that is also found in fission yeast and other fungal adenylyl cyclases (42, 75, 76). It is thus likely that Sgt1p is generally required for adenylyl cyclase activity in yeasts and fungi. However, the absence of an LRR domain in metazoan adenylyl cyclases may indicate that Sgt1p is not required for adenylyl cyclase activity in multicellular organisms, although it may well interact with other LRR-containing proteins in metazoans.
Given the role of Sgt1p in several different types of multimeric protein complexes, it is possible that it acts as some sort of protein chaperone, assembly factor, or allosteric activator. This hypothesis is consistent with structure predictions suggesting that the Sgt1p N-terminal region is similar to the TPR regions of the Sti1/Hop cochaperone and that the central CS domain adopts a fold similar to that of the p23 cochaperone. Hop facilitates protein substrate transfer from Hsp70 to Hsp90, whereas p23 stimulates the ATPase activity of Hsp90 and the release of bound substrate (77). Several experimental observations are also consistent with a role for Sgt1p as a cochaperone or assembly factor. First, Sgt1p is required to functionally activate the Skp1p and Ctf13p subunits of the CBF3 kinetochore complex but is not itself a subunit of this complex (38). Second, Sgt1p may be present in substoichiometric quantities in SCF (38) and Cyr1p (this work) complexes. Further work should elucidate the role of Sgt1p in the function of the multiple distinct complexes that are critically involved in the control of cell growth and cell division.
Acknowledgments
We thank Lee Hartwell for supporting the initial stages of this work and Emmanuelle Boy-Marcotte, Michel Jacquet, Georges Renault, Wade Harper, Amit Banerjee, and Jeffrey Field for plasmids, antibodies, and advice. We are grateful to Anne Chevalier for help with the two-hybrid experiments and Charlie Hoffman for the stimulating electronic exchange of information.
C.D. was supported by Allocation Couplée de l'Ecole Normale Supérieure de Paris.
REFERENCES
- 1.Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper, and S. J. Elledge. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86:263-274. [DOI] [PubMed] [Google Scholar]
- 2.Baroni, M. D., P. Monti, and L. Alberghina. 1994. Repression of growth-regulated G1 cyclin expression by cyclic AMP in budding yeast. Nature 371:339-342. [DOI] [PubMed] [Google Scholar]
- 3.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. Sonnhammer. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckner, S. K., S. Hattori, and T. Y. Shih. 1985. The ras oncogene product p21 is not a regulatory component of adenylate cyclase. Nature 317:71-72. [DOI] [PubMed] [Google Scholar]
- 5.Beebe, S. J. 1994. The cAMP-dependent protein kinases and cAMP signal transduction. Semin. Cancer Biol. 5:285-294. [PubMed] [Google Scholar]
- 6.Boutelet, F., A. Petitjean, and F. Hilger. 1985. Yeast cdc35 mutants are defective in adenylate cyclase and are allelic with cyr1 mutants while CAS1, a new gene, is involved in the regulation of adenylate cyclase. EMBO J. 4:2635-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boy-Marcotte, E., G. Lagniel, M. Perrot, F. Bussereau, A. Boudsocq, M. Jacquet, and J. Labarre. 1999. The heat shock response in yeast: differential regulations and contributions of the Msn2p/Msn4p and Hsf1p regulons. Mol. Microbiol. 33:274-283. [DOI] [PubMed] [Google Scholar]
- 8.Boy-Marcotte, E., M. Perrot, F. Bussereau, H. Boucherie, and M. Jacquet. 1998. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J. Bacteriol. 180:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cannon, J. F., and K. Tatchell. 1987. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol. Cell. Biol. 7:2653-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casperson, G. F., N. Walker, and H. R. Bourne. 1985. Isolation of the gene encoding adenylate cyclase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 82:5060-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, H. C., D. F. Nathan, and S. Lindquist. 1997. In vivo analysis of the Hsp90 cochaperone Sti1 (p60). Mol. Cell. Biol. 17:318-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chester, V. E. 1968. Heritable glycogen-storage deficiency in yeast and its induction by ultra-violet light. J. Gen. Microbiol. 51:49-56. [DOI] [PubMed] [Google Scholar]
- 13.Ciechanover, A., A. Orian, and A. L. Schwartz. 2000. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays 22:442-451. [DOI] [PubMed] [Google Scholar]
- 14.Connelly, C., and P. Hieter. 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86:275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deshaies, R. J. 1999. SCF and Cullin/ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 16.Douguet, D., and G. Labesse. 2001. Easier threading through web-based comparisons and cross-validations. Bioinformatics 17:752-753. [DOI] [PubMed]
- 17.D'Souza, C. A., and J. Heitman. 2001. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25:349-364. [DOI] [PubMed]
- 18.Fedor-Chaiken, M., R. J. Deschenes, and J. R. Broach. 1990. SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell 61:329-340. [DOI] [PubMed] [Google Scholar]
- 19.Field, J., J. Nikawa, D. Broek, B. MacDonald, L. Rodgers, I. A. Wilson, R. A. Lerner, and M. Wigler. 1988. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol. 8:2159-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Field, J., H. P. Xu, T. Michaeli, R. Ballester, P. Sass, M. Wigler, and J. Colicelli. 1990. Mutations of the adenylyl cyclase gene that block RAS function in Saccharomyces cerevisiae. Science 247:464-467. [DOI] [PubMed] [Google Scholar]
- 21.Flores, A., J. F. Briand, O. Gadal, J. C. Andrau, L. Rubbi, V. Van Mullem, C. Boschiero, M. Goussot, C. Marck, C. Carles, P. Thuriaux, A. Sentenac, and M. Werner. 1999. A protein-protein interaction map of yeast RNA polymerase III. Proc. Natl. Acad. Sci. USA 96:7815-7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Francois, J., and J. L. Parrou. 2001. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:125-145. [DOI] [PubMed] [Google Scholar]
- 23.Fukui, Y., T. Kozasa, Y. Kaziro, T. Takeda, and M. Yamamoto. 1986. Role of a ras homolog in the life cycle of Schizosaccharomyces pombe. Cell 44:329-336. [DOI] [PubMed] [Google Scholar]
- 24.Garreau, H., R. N. Hasan, G. Renault, F. Estruch, E. Boy-Marcotte, and M. Jacquet. 2000. Hyperphosphorylation of Msn2p and Msn4p in response to heat shock and the diauxic shift is inhibited by cAMP in Saccharomyces cerevisiae. Microbiology 146:2113-2120. [DOI] [PubMed] [Google Scholar]
- 25.Gerisch, G. 1987. Cyclic AMP and other signals controlling cell development and differentiation in Dictyostelium. Annu. Rev. Biochem. 56:853-879. [DOI] [PubMed] [Google Scholar]
- 26.Gerst, J. E., K. Ferguson, A. Vojtek, M. Wigler, and J. Field. 1991. CAP is a bifunctional component of the Saccharomyces cerevisiae adenylyl cyclase complex. Mol. Cell. Biol. 11:1248-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorner, W., E. Durchschlag, M. T. Martinez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schuller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guarente, L., and M. Ptashne. 1981. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 78:2199-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall, D. D., D. D. Markwardt, F. Parviz, and W. Heideman. 1998. Regulation of the Cln3-Cdc28 kinase by cAMP in Saccharomyces cerevisiae. EMBO J. 17:4370-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanoune, J., and N. Defer. 2001. Regulation and role of adenylyl cyclase isoforms. Annu. Rev. Pharmacol. Toxicol. 41:145-174. [DOI] [PubMed] [Google Scholar]
- 31.Hurley, J. H. 1998. The adenylyl and guanylyl cyclase superfamily. Curr. Opin. Struct. Biol. 8:770-777. [DOI] [PubMed] [Google Scholar]
- 32.Irniger, S., M. Baumer, and G. H. Braus. 2000. Glucose and ras activity influence the ubiquitin ligases APC/C and SCF in Saccharomyces cerevisiae. Genetics 154:1509-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 34.Jones, D. T. 2000. A practical guide to protein structure prediction. Methods Mol. Biol. 143:131-154. [DOI] [PubMed] [Google Scholar]
- 35.Kataoka, T., D. Broek, and M. Wigler. 1985. DNA sequence and characterization of the S. cerevisiae gene encoding adenylate cyclase. Cell 43:493-505. [DOI] [PubMed] [Google Scholar]
- 36.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 37.King, R. W., R. J. Deshaies, J. M. Peters, and M. W. Kirschner. 1996. How proteolysis drives the cell cycle. Science 274:1652-1659. [DOI] [PubMed] [Google Scholar]
- 38.Kitagawa, K., D. Skowyra, S. J. Elledge, J. W. Harper, and P. Hieter. 1999. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 4:21-33. [DOI] [PubMed] [Google Scholar]
- 39.Labesse, G., and J. Mornon. 1998. Incremental threading optimization (TITO) to help alignment and modelling of remote homologues. Bioinformatics 14:206-211. [DOI] [PubMed] [Google Scholar]
- 40.Lockwood, A. H., S. K. Murphy, S. Borislow, A. Lazarus, and M. Pendergast. 1987. Cellular signal transduction and the reversal of malignancy. J. Cell. Biochem. 33:237-255. [DOI] [PubMed] [Google Scholar]
- 41.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 42.Mallet, L., G. Renault, and M. Jacquet. 2000. Functional cloning of the adenylate cyclase gene of Candida albicans in Saccharomyces cerevisiae within a genomic fragment containing five other genes, including homologues of CHS6 and SAP185. Yeast 16:959-966. [DOI] [PubMed] [Google Scholar]
- 43.Mann, C., J. Y. Micouin, N. Chiannilkulchai, I. Treich, J. M. Buhler, and A. Sentenac. 1992. RPC53 encodes a subunit of Saccharomyces cerevisiae RNA polymerase C (III) whose inactivation leads to a predominantly G1 arrest. Mol. Cell. Biol. 12:4314-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez-Pastor, M. T., G. Marchler, C. Schuller, A. Marchler-Bauer, H. Ruis, and F. Estruch. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15:2227-2235. [PMC free article] [PubMed] [Google Scholar]
- 45.Matsumoto, K., I. Uno, and T. Ishikawa. 1983. Control of cell division in Saccharomyces cerevisiae mutants defective in adenylate cyclase and cAMP-dependent protein kinase. Exp. Cell Res. 146:151-161. [DOI] [PubMed] [Google Scholar]
- 46.Matsuzawa, S. I., and J. C. Reed. 2001. Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses Mol. Cell 7:915-926. [DOI] [PubMed] [Google Scholar]
- 47.Mitsuzawa, H. 1993. Responsiveness to exogenous cAMP of a Saccharomyces cerevisiae strain conferred by naturally occurring alleles of PDE1 and PDE2. Genetics 135:321-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montminy, M. 1997. Transcriptional regulation by cyclic AMP. Annu. Rev. Biochem. 66:807-822. [DOI] [PubMed] [Google Scholar]
- 49.Moqtaderi, Z., Y. Bai, D. Poon, P. A. Weil, and K. Struhl. 1996. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 383:188-191. [DOI] [PubMed] [Google Scholar]
- 50.Morishita, T., A. Matsuura, and I. Uno. 1993. Characterization of the cyr1-2 UGA mutation in Saccharomyces cerevisiae. Mol. Gen. Genet. 237:463-466. [DOI] [PubMed] [Google Scholar]
- 51.Nishida, Y., F. Shima, H. Sen, Y. Tanaka, C. Yanagihara, Y. Yamawaki-Kataoka, K. Kariya, and T. Kataoka. 1998. Coiled-coil interaction of N-terminal 36 residues of cyclase-associated protein with adenylyl cyclase is sufficient for its function in Saccharomyces cerevisiae ras pathway. J. Biol. Chem. 273:28019-28024. [DOI] [PubMed] [Google Scholar]
- 52.Patton, E. E., A. R. Willems, and M. Tyers. 1998. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 14:236-243. [DOI] [PubMed] [Google Scholar]
- 53.Pringle, J. R., A. E. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565-602. [DOI] [PubMed] [Google Scholar]
- 54.Pringle, J. R., and L. H. Hartwell. 1981. The Saccharomyces cerevisiae cell cycle. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 55.Rose, M. D., P. Novick, J. H. Thomas, D. Botstein, and G. R. Fink. 1987. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60:237-243. [DOI] [PubMed] [Google Scholar]
- 56.Russell, L. C., S. R. Whitt, M. S. Chen, and M. Chinkers. 1999. Identification of conserved residues required for the binding of a tetratricopeptide repeat domain to heat shock protein 90. J. Biol. Chem. 274:20060-20063. [DOI] [PubMed] [Google Scholar]
- 57.Sass, P., J. Field, J. Nikawa, T. Toda, and M. Wigler. 1986. Cloning and characterization of the high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 83:9303-9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57a.Schadick, K., H. M. Fourcade, P. Boumenot, J. J. Seitz, J. L. Morrell, L. Chang, K. L. Gould, J. F. Partridge, R. C. Allshire, K. Kitagawa, P. Hieter, and C. S. Hofman. 2002. Schizosaccharomyces pombe Git7p, a member of the Saccharomyces cerevisiae Sgt1p family, is required for glucose and cyclic AMP signaling, cell wall integrity, and septation. Eukaryot. Cell 1:558-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheufler, C., A. Brinker, G. Bourenkov, S. Pegoraro, L. Moroder, H. Bartunik, F. U. Hartl, and I. Moarefi. 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101:199-210. [DOI] [PubMed] [Google Scholar]
- 59.Schmitt, A. P., and K. McEntee. 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5777-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schulman, B. A., A. C. Carrano, P. D. Jeffrey, Z. Bowen, E. R. Kinnucan, M. S. Finnin, S. J. Elledge, J. W. Harper, M. Pagano, and N. P. Pavletich. 2000. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408:381-386. [DOI] [PubMed] [Google Scholar]
- 61.Shi, J., T. L. Blundell, and K. Mizuguchi. 2001. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310:243-257. [DOI] [PubMed] [Google Scholar]
- 62.Shima, F., T. Okada, M. Kido, H. Sen, Y. Tanaka, M. Tamada, C. D. Hu, Y. Yamawaki-Kataoka, K. Kariya, and T. Kataoka. 2000. Association of yeast adenylyl cyclase with cyclase-associated protein CAP forms a second Ras-binding site which mediates its Ras-dependent activation. Mol. Cell. Biol. 20:26-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shirasu, K., T. Lahaye, M. W. Tan, F. Zhou, C. Azevedo, and P. Schulze-Lefert. 1999. A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99:355-366. [DOI] [PubMed] [Google Scholar]
- 64.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith, A., M. P. Ward, and S. Garrett. 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17:3556-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stemmann, O., and J. Lechner. 1996. The Saccharomyces cerevisiae kinetochore contains a cyclin-CDK complexing homologue, as identified by in vitro reconstitution. EMBO J. 15:3611-3620. [PMC free article] [PubMed] [Google Scholar]
- 67.Suzuki, N., H. R. Choe, Y. Nishida, Y. Yamawaki-Kataoka, S. Ohnishi, T. Tamaoki, and T. Kataoka. 1990. Leucine-rich repeats and carboxyl terminus are required for interaction of yeast adenylate cyclase with RAS proteins. Proc. Natl. Acad. Sci. USA 87:8711-8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sy, J., and Y. Tamai. 1986. An altered adenylate cyclase in cdc35-1 cell division cycle mutant of yeast. Biochem. Biophys. Res. Commun. 140:723-727. [DOI] [PubMed] [Google Scholar]
- 69.Tasken, K., B. S. Skalhegg, K. A. Tasken, R. Solberg, H. K. Knutsen, F. O. Levy, M. Sandberg, S. Orstavik, T. Larsen, A. K. Johansen, T. Vang, H. P. Schrader, N. T. Reinton, K. M. Torgersen, V. Hansson, and T. Jahnsen. 1997. Structure, function, and regulation of human cAMP-dependent protein kinases. Adv. Second Messenger Phosphoprot. Res. 31:191-204. [DOI] [PubMed] [Google Scholar]
- 70.Tesmer, J. J., and S. R. Sprang. 1998. The structure, catalytic mechanism and regulation of adenylyl cyclase. Curr. Opin. Struct. Biol. 8:713-719. [DOI] [PubMed] [Google Scholar]
- 71.Thevelein, J. M., and J. H. de Winde. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33:904-918. [DOI] [PubMed] [Google Scholar]
- 72.Tokiwa, G., M. Tyers, T. Volpe, and B. Futcher. 1994. Inhibition of G1 cyclin activity by the Ras/cAMP pathway in yeast. Nature 371:342-345. [DOI] [PubMed] [Google Scholar]
- 73.Weaver, A. J., W. P. Sullivan, S. J. Felts, B. A. Owen, and D. O. Toft. 2000. Crystal structure and activity of human p23, a heat shock protein 90 co-chaperone. J. Biol. Chem. 275:23045-23052. [DOI] [PubMed] [Google Scholar]
- 74.Wilson, R. B., G. Renault, M. Jacquet, and K. Tatchell. 1993. The pde2 gene of Saccharomyces cerevisiae is allelic to rca1 and encodes a phosphodiesterase which protects the cell from extracellular cAMP. FEBS Lett. 325:191-195. [DOI] [PubMed] [Google Scholar]
- 75.Yamawaki-Kataoka, Y., T. Tamaoki, H. R. Choe, H. Tanaka, and T. Kataoka. 1989. Adenylate cyclases in yeast: a comparison of the genes from Schizosaccharomyces pombe and Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 86:5693-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Young, D., M. Riggs, J. Field, A. Vojtek, D. Broek, and M. Wigler. 1989. The adenylyl cyclase gene from Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 86:7989-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Young, J. C., I. Moarefi, and F. U. Hartl. 2001. Hsp90: a specialized but essential protein-folding tool. J. Cell Biol. 154:267-273. [DOI] [PMC free article] [PubMed] [Google Scholar]