Abstract
The genome of the filamentous fungus Neurospora crassa contains a single gene encoding a heterotrimeric G-protein β subunit, gnb-1. The predicted GNB-1 protein sequence is most identical to Gβ proteins from the filamentous fungi Cryphonectria parasitica and Aspergillus nidulans. N. crassa GNB-1 is also 65% identical to the human GNB-1 protein but only 38 and 45% identical to Gβ proteins from budding and fission yeasts. Previous studies in animal and fungal systems have elucidated phenotypes of Gβ null mutants, but little is known about the effects of Gβ loss on Gα levels. In this study, we analyzed a gnb-1 deletion mutant for cellular phenotypes and levels of the three Gα proteins. Δgnb-1 strains are female-sterile, with production of aberrant fertilized reproductive structures. Δgnb-1 strains conidiate more profusely and have altered mass on solid medium. Loss of gnb-1 leads to inappropriate conidiation and expression of a conidiation-specific gene during growth in submerged culture. Intracellular cyclic AMP levels are reduced by 60% in vegetative plate cultures of Δgnb-1 mutants. Loss of gnb-1 leads to lower levels of the three Gα proteins under a variety of conditions. Analysis of transcript levels for the gna-1 and gna-2 Gα genes in submerged cultures indicates that regulation of Gα protein levels by gnb-1 is posttranscriptional. The results suggest that GNB-1 directly regulates apical extension rate and mass accumulation. In contrast, many other Δgnb-1 phenotypes, including female sterility and defective conidiation, can be explained by altered levels of the three N. crassa Gα proteins.
Heterotrimeric G proteins (Gαβγ) transmit external signals sensed by seven-helix transmembrane receptors, leading to a variety of physiological responses (reviewed in references 12, 17, and 38). In the inactive state, Gα, Gβ, and Gγ subunits are in association, with GDP bound to Gα. Ligand-induced conformational changes in its coupled receptor cause the G protein to dissociate into a GTP-bound Gα and the Gβγ heterodimer. Both of these complexes can activate or inhibit downstream effectors, thus triggering an array of cellular responses (reviewed in reference 17). Characterized Gβγ effectors include adenylyl cyclases, phospholipase A2, phospholipase Cβ, Na+, Ca2+, and K+ channels, and tyrosine and serine/threonine protein kinases (reviewed in references 8 and 17). Hydrolysis of GTP by the Gα subunit leads to reformation of the inactive heterotrimeric form.
Gβ proteins are important for environmental and cell-type signaling in yeasts and filamentous fungi. In the budding yeast Saccharomyces cerevisiae, the Gβ subunit Ste4p functions as a positive regulator of the pheromone response in haploid cells by recruiting the Ste20p mitogen-activated protein kinase kinase kinase kinase (MAPKKKK) to the Ste11p MAPKKK on the Ste5p scaffold (reviewed in reference 14). The Git5 Gβ protein from Schizosaccharomyces pombe was originally thought to participate in the mating pathway through its association with the Gα Gpa1 (25). However, accumulating evidence now suggests that Git5 is coupled to the Gpa2 Gα subunit and is required for the increased cyclic AMP (cAMP) levels observed during transfer from glucose-starved to adequate glucose conditions in S. pombe; basal steady-state cAMP levels in adequate glucose medium are normal in git5 mutants (28). In the filamentous fungus Aspergillus nidulans, mutation of the Gβ-encoding gene sfaD results in hyperactive asexual sporulation (conidiation) and slowed vegetative growth; genetic evidence suggests that SfaD may be coupled to the FadA Gα protein (48). Disruption of the cpgb-1 Gβ subunit from the filamentous fungus Cryphonectria parasitica leads to reduced pigmentation, conidiation, hyphal tip branching, and virulence while causing increased growth on vegetative solid medium (22). In the basidiomycete Cryptococcus neoformans, deletion of the Gβ gene GPB-1 leads to sterility and defective monokaryotic fruiting (57).
Mutational inactivation of G-protein subunit genes has been demonstrated to affect expression of other associated subunits and regulatory proteins in both fungal and animal systems. For example, C. parasitica strains that lack the cpgb-1 Gβ gene have greatly reduced levels of the CPG-1 Gα protein (23). The levels of Gβ proteins are reduced 68% in Gαo null mutant mice (34). In the nematode Caenorhabditis elegans, loss of the GPB-2 Gβ gene leads to reduced protein levels for the EGL-10 regulator of G protein signaling (6). In contrast, Gαo protein levels are normal in gpb-2 mutants (6). Thus, only in the case of C. parasitica has it been reported that loss of a Gβ gene influences the level of a Gα protein.
It was previously demonstrated that levels of a Gβ protein (GNB-1) are not affected by deletion of any one of the three Gα genes (gna-1, gna-2, and gna-3) in the filamentous fungus Neurospora crassa (21, 24). However, GNB-1 amount is reduced by approximately 50% in Δgna-1 Δgna-2 double mutants (21). Levels of the two remaining Gα proteins are unaffected in N. crassa strains lacking a single Gα subunit gene (2, 24; A. M. Kays and K. A. Borkovich, unpublished data).
N. crassa strains containing null or constitutively activated gna-1 alleles exhibit different phenotypes for several cellular functions. The results demonstrated that gna-1 positively regulates apical extension rates on normal and hyperosmotic medium, aerial hypha height, and female fertility but is a negative regulator of conidium production, thermotolerance, and resistance to oxidative stress (20, 63). Since Gβγ is predicted to be free to signal in strains with null or activated gna-1, the observation of different phenotypes in strains with these two alleles supports an active role in signaling for GNA-1, independent of Gβγ. This result is also consistent with Gβγ serving as a negative regulator of GNA-1, through formation of the inactive heterotrimeric complex.
Although Δgna-2 strains do not have detectable defects, Δgna-1 Δgna-2 mutants are more affected in Δgna-1 phenotypes, suggesting that gna-1 and gna-2 possess overlapping functions (2). Δgna-3 strains share several phenotypes with the cr-1 adenylyl cyclase mutant (52), including premature conidiation, short aerial hyphae, and inappropriate conidiation in submerged culture (24). Δgna-3 mutants also have reduced ascospore viability during homozygous crosses (24).
Here we report the cloning and characterization of the only predicted heterotrimeric Gβ gene, gnb-1, from N. crassa. gnb-1 transcripts were identified, and GNB-1 protein levels were measured in the wild type throughout the life cycle. A gnb-1 deletion mutant strain was constructed and analyzed for morphological phenotypes and levels of cAMP and adenylyl cyclase protein. Transcript and/or protein levels for the Gα genes were also determined. The three Gα genes previously cloned and mutated in our laboratory are the only Gα genes in N. crassa (http://www-genome.wi.mit.edu/annotation/fungi/neurospora/). Thus, with this study, we have now compared the characteristics of single mutations in all Gα and Gβ genes in a given organism. Our findings suggest that GNB-1 is an active and direct regulator of at least two processes in N. crassa. However, many cellular and developmental phenotypes of Δgnb-1 mutants can be accounted for by reduced levels of Gα proteins.
MATERIALS AND METHODS
Strains and growth conditions.
N. crassa strains used in this study are listed in Table 1. Media supporting vegetative growth are Vogel's minimal medium (VM) (56) and sorbose-containing medium (to facilitate formation of colonies on plates) (10). Synthetic crossing medium (SCM) (59), containing low levels of nitrogen sources, was used to induce development of female reproductive structures. Hygromycin B was used at 200 μg/ml in media as indicated. Where indicated, cultures were supplemented with peptone (2%, wt/vol) or cAMP (1 mM). Five- to seven-day-old conidia were used to inoculate all cultures. Plasmids were maintained in Escherichia coli strain DH5α (18).
TABLE 1.
N. crassa strains
| Strain | Relevant genotype | Comments | Source or reference |
|---|---|---|---|
| 74-OR23-1A | Wild type, mat A | 74A (wild type) | R. L. Weiss, UCLA |
| OR8-1a | Wild type, mat a | 74a (wild type) | FGSCa |
| 3B10 | Δgna-1::hph mat a | Δgna-1 gene replacement | 21 |
| 21c | Δgna-2::pyrG+mat a | Δgna-2 gene replacement | 2 |
| Sta(73a) | Wild type, mat a | 73a (wild type) | R. L. Weiss |
| 42-5-1 | Δgnb-1::hph mat a | Δgnb-1 homokaryon | This study |
| 42-5-18 | Δgnb-1::hph mat A | Δgnb-1 homokaryon | This study |
| 42-8-3 | Δgnb-1::hph mat A | Δgnb-1 homokaryon | This study |
| 42-8-5 | Δgnb-1::hph mat a | Δgnb-1 homokaryon | This study |
| 42-31-2 | Δgnb-1::hph mat a | Δgnb-1 homokaryon | This study |
| A16 | Δgnb-1::hph gnb-1+::benR mat A | Complemented mutant | This study |
| G10 | Δgnb-1::hph gnb-1+::benR mat A | Complemented mutant | This study |
FGSC, Fungal Genetics Stock Center, Kansas City, Mo.
Cloning, sequencing, and restriction fragment length polymorphism (RFLP) mapping of the gnb-1 gene.
A G-protein β gene was isolated during low-stringency hybridization screening of an N. crassa mycelial cDNA library (41) using a degenerate oligonucleotide corresponding to a conserved region of mammalian Gβ subunits (primer BETA-03) (31). Sequence analysis of the 1.7-kb insert in hybridizing clone M1-1 showed high identity to Gβ proteins from other organisms (data not shown). The M1-1 cDNA clone was then used to screen an N. crassa BARGEM7 λ genomic library (42). Four hybridizing clones were purified; one of these (BG3) contained the gnb-1 coding sequence with significant 5′ and 3′ flanking regions. The entire 7.65-kb genomic fragment from BG3 was excised and inserted into pGEM7Zf to yield plasmid pKB60 (Fig. 1A). Sequencing of the gnb-1 cDNA and a region corresponding to 3.44 kb of the gnb-1 genomic fragment in pKB60 (Fig. 1A) was performed as described elsewhere (54).
FIG. 1.
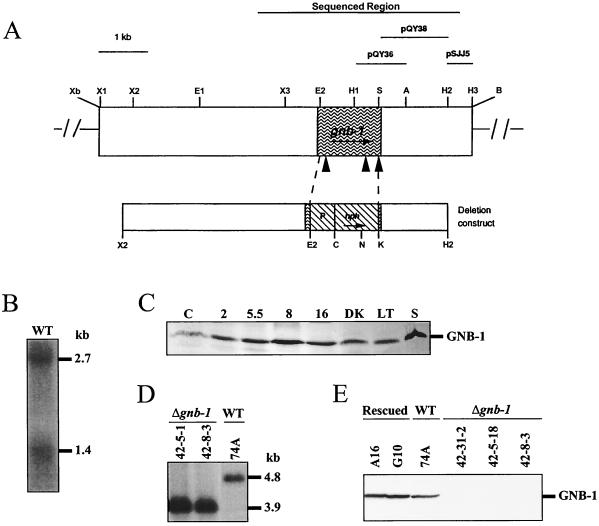
Structure of the N. crassa gnb-1 genomic region and construction of Δgnb-1 and rescued strains. (A) gnb-1 genomic clone. The unique XbaI and BamHI sites are artifacts of cloning. The region that was sequenced, as well as the fragments used as probes for Southern and Northern analysis, is shown at the top of the figure. The shaded area depicts the ORF, and the closed triangles correspond to introns which are (left to right) 106, 80, and 67 bp in length. The gene replacement construct using the hph marker is shown below the genomic clone. P, A. nidulans trpC promoter. Enzyme abbreviations: A, AatII; B, BamHI, C, ClaI; E, EcoRI; H, HindIII; K, KpnI; N, NcoI; S, StyI; X, XhoI; Xb, XbaI. In the case of multiple sites for a given enzyme, the number following the abbreviation indicates order (from left to right) in the figure. (B) gnb-1 mRNA species. Samples containing 20 μg of total RNA isolated from 5.5-h-submerged cultures inoculated at 107 conidia of wild type (WT) strain 74A per ml were subjected to Northern analysis using a 1.0-kb HindIII-AatII fragment from pQY36 as a probe (see panel A). (C) Expression of GNB-1 during the N. crassa life cycle. Samples from wild-type strain 74A (30 μg of the plasma membrane protein fraction) were conidia (C); 2-, 5.5-, 8-, and 16-h-submerged cultures (inoculated at 107 conidia/ml); cultures grown at 30°C on solid VM in the dark (DK); cultures grown at room temperature on solid VM under light (LT); and cultures grown at room temperature on SCM under light (S). (D) Southern analysis. Genomic DNA was digested with NcoI and HindIII. The 600-bp HindIII fragment from plasmid pSJJ5 was used as a probe (see panel A). 42-5-1 and 42-8-3 are purified homokaryotic Δgnb-1 mutants, while 74A is the wild-type (WT) strain. (E) GNB-1 protein levels. Samples containing 30 μg of protein from plasma membrane fractions of 16-h-submerged cultures inoculated at 3 × 106 conidia/ml were subjected to Western analysis using the GNB-1 antibody. Rescued, Δgnb-1, gnb-1+ complemented strains; WT, wild type.
RFLP mapping was performed as described previously (36). Digestion with either EcoRV or XhoI revealed RFLP patterns when the M1-1 cDNA insert was used as a probe. The RFLP pattern observed in the 18 progeny was compared to that of other mapped genes, allowing assignment of the gnb-1 gene to an N. crassa linkage group (35).
Construction of Δgnb-1 and complemented Δgnb-1, gnb-1+ strains and Southern analysis.
Plasmid pCSN44 contains the dominant drug resistance marker, E. coli hygromycin B phosphotransferase (hph), under the control of the A. nidulans trpC promoter, which is expressed in N. crassa (50). The gnb-1 gene replacement construct, pQY42, was made by replacing the EcoRI-StyI fragment from the gnb-1 open reading frame (ORF) with the HindIII-BamHI fragment from pCSN44 (containing the hph gene and A. nidulans promoter). pQY42 contains 3.8 kb of 5′ and 1.4 kb of 3′ flanking DNA, extending from the second XhoI site to the second HindIII site in the gnb-1 genomic clone (Fig. 1A). To obtain the gnb-1 deletion strain, 2 μg of plasmid pQY42 was electroporated into wild-type strain 74A (20, 55), with selection on sorbose medium (5) containing hygromycin B. Genomic DNA was isolated from the hygromycin B-resistant transformants, digested with NcoI and XhoI, and subjected to Southern analysis as described elsewhere (20), using the 0.6-kb HindIII fragment from pSJJ5 as a probe (Fig. 1A). Ectopic integration events of the deletion construct were detected by Southern analysis using both the 1.4-kb XbaI-BamHI insert from pQY38 (Fig. 1A) and the 1.4-kb SalI-BamHI insert from pCSN44 (containing the hph gene) as probes. Heterokaryotic Δgnb-1 strains (with no ectopic integration events) were subsequently crossed to wild-type strain 74a. Hygromycin-resistant progeny were selected and tested for homokaryon status by Southern analysis using the pSJJ5 and pCSN44 probes described above.
The Δgnb-1 mutation was complemented in trans by using the 7.65-kb gnb-1 genomic clone in a vector containing the benomyl resistance gene from N. crassa (plasmid pBT6) (40). Briefly, plasmid pKB60 was digested with BamHI and the ends blunted with Klenow fragment, followed by digestion with XbaI. The 7.65-kb gnb-1 fragment was isolated and ligated with plasmid pBT3 digested with XbaI and SmaI to yield pQY52-4, the complementation construct. Δgnb-1 strain 42-8-3 was transformed by electroporation using pQY52-4, with selection on sorbose plates containing 500 ng of benomyl (Dupont, Wilmington, Del.) per ml. Genomic DNA was isolated from benomyl-resistant transformants, digested with XhoI, and subjected to Southern analysis using the insert from plasmid pBT6 as a probe. Heterokaryons containing a single copy of the pQY52-4 DNA were identified, and homokaryons were obtained from these strains by repeated plating on sorbose plates containing 500 ng of benomyl per ml, followed by isolation of microconidia (13) and plating on benomyl-containing plates (data not shown).
Western and Northern analysis.
Samples for Western and Northern analysis were obtained from the following tissues. For submerged cultures, conidia were inoculated into liquid VM (with or without 2% [wt/vol] peptone) at a final concentration of 3 × 106 or 1 × 107 cells/ml, as indicated in the figure legends. Cultures were incubated in the dark at 30°C with shaking for 2, 5.5, 8, or 16 h. For cultures growing on solid medium, 1 μl of a conidial suspension was inoculated onto the center of VM or SCM plates overlaid with cellophane (Bio-Rad Laboratories, Hercules, Calif.). VM plates were grown either in the dark at 30°C or under light at room temperature for 3 days, while SCM plates were grown for 6 days at room temperature under constant light.
The M1-1 cDNA clone is truncated at the 5′ end, missing the first 46 bp of the amino acid coding region. In order to construct a vector for overexpression of GNB-1 in E. coli, the first exon of the gnb-1 coding region (127 bp) was amplified from a gnb-1 genomic subclone plasmid using the PCR. The primers were designed to generate a SpeI site at the 3′ end of the first exon (creating a silent mutation) and a NdeI site at the translational initiation methionine. This step facilitated cloning of the amino-terminal fragment and the remainder of the ORF into the E. coli overexpression vector pET11a (Novagen, Madison, Wis.). The resultant overexpression construct (pQY49) was transformed into E. coli strain HMS 174 plysS (51). Growth of E. coli cells, purification of the expressed 38-kDa GNB-1 protein, and antiserum generation were as previously described (2, 20).
For Western analysis, extraction of the plasma membrane fraction, determination of protein concentration, gel electrophoresis, and blotting were essentially as described previously (20, 54). GNB-1, GNA-1, GNA-2, GNA-3, and CR-1 antisera were used at dilutions of 1:5,000, 1:1,000 or 1:5,000, 1:3,000, 1:1,000 and 1:10,000, respectively. The two alternatives for secondary antibody and signal detection have been previously described (20, 24).
Isolation of total RNA and Northern analysis of samples containing 20 μg of total RNA were performed as previously described (62). Δgna-1 and Δgna-2 gene replacement mutants were used as negative controls for the presence of the gna-1 and gna-2 transcripts. A 1.0-kb AatII-XbaI fragment from pQY36 (Fig. 1A), a 5.6-kb EcoRI-ClaI gna-1 genomic fragment from pPNO5 (20), the 2.0-kb BamHI gna-2 cDNA insert from plasmid 13M2A5-2 (54), and the 200-bp BamHI-EcoRI insert from pBW100 (47) were used as probes to detect expression of gnb-1, gna-1, gna-2, and con-10 mRNA species, respectively. A cosmid containing the rRNA gene (obtained from D. E. Ebbole, Texas A&M University) was labeled and used as a probe to control for mRNA transfer.
Phenotypic analysis and determination of intracellular cAMP levels.
Phenotypic studies, including determination of apical extension rate on normal and hyperosmotic medium, measurements of dry weight accumulation on cellophane-overlaid plates, and assessment of female fertility, were performed as described elsewhere (63), with details given in the figure legends. Microscopic observation of submerged cultures was performed using an Olympus BH-2 microscope outfitted with an OM-2 camera (Olympus America, Lake Success, N.Y.). Intracellular cAMP was extracted (21) and assayed by a binding protein method according to the manufacturer's recommendations (kit TRK432; Amersham Pharmacia Biotech, Piscataway, N.J.). For these determinations, protein was quantitated with the BCA protein assay (Pierce, Rockford, Ill.), as previously described (21).
Nucleotide sequence accession number.
The GenBank accession number for the gnb-1 sequence is AF491286.
RESULTS
gnb-1 gene structure and expression of mRNA and protein.
An N. crassa mycelial cDNA library was screened by using a degenerate oligonucleotide primer corresponding to a conserved region of Gβ genes (BETA-03) (31) under low-stringency hybridization conditions. The sequence of the insert in the isolated cDNA clone, M1-1, showed high identity to reported Gβ subunits (data not shown), and the gene was designated gnb-1. RFLP mapping using the M1-1 cDNA as a probe demonstrated that gnb-1 gene resides on the right arm of chromosome III, linked to the con-7 and trp-1 genes (data not shown).
In order to isolate a gnb-1 genomic clone, the M1-1 cDNA was used as a probe to screen the N. crassa BARGEM-7 λ genomic DNA library (42). A clone containing a 7.65-kb insert was isolated, and 3.44 kb of sequence encompassing the ORF and 5′ and 3′ flanking regions was determined (Fig. 1A). The gnb-1 ORF is 1.33 kb in length and contains three introns (Fig. 1A). Subsequent to this work, the entire genomic sequence of N. crassa was determined (http://www-genome.wi.mit.edu/annotation/fungi/neurospora/). BLAST (1) searches revealed that GNB-1 is the only Gβ protein in the genome.
Analysis of the sequence upstream of the gnb-1 coding region reveals several putative transcriptional regulatory motifs, including two pyrimidine-rich segments at −686 to −664 and −405 to −394, two CTTTG motifs at −224 and −139 and two CCAAT boxes located at −395 and −57. The pyrimidine-rich segment is important for efficient transcription in S. cerevisiae (16). SRY-family high-mobility-group (HMG)-box proteins recognize CTTTG or CAAAG motifs in a sequence-specific but not conformation-specific pattern (3). An example of an N. crassa SRY-HMG box protein is the MATa-1 mating type protein (44). The CCAAT motif is a transcriptional regulatory sequence found in eukaryotes (16).
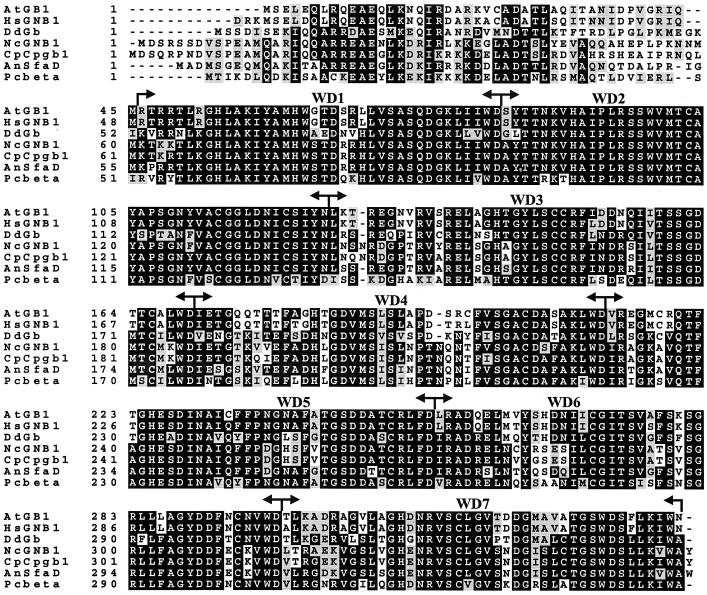
The predicted GNB-1 protein contains 358 amino acid residues and has a molecular mass of 39.7 kDa (Fig. 2). GNB-1 is 91% identical to the C. parasitica Gβ subunit CPGB-1 (22) and 82% identical to A. nidulans SfaD (48). GNB-1 is almost equally related (65 to 68% identical) to Gβ proteins from the fungus Pneumocystis carinii, the slime mold Dictyostelium discoideum, the salamander Ambystoma tigrinum and to a human Gβ subunit. Interestingly, N. crassa GNB-1 shows only 38 and 45% identity to Gβ proteins from S. cerevisiae and S. pombe, respectively (25, 60), suggesting evolutionary divergence between Gβ genes from filamentous fungi and yeasts.
FIG. 2.
Alignment of GNB-1 with its five closest Gβ relatives. The amino acid sequence of GNB-1 (NcGNB1) was aligned with Gβ proteins from A. tigrinum (AtGB1; accession no. AF277161), Homo sapiens (HsGNB1; XM 010575.1), D. discoideum (DdGb; DDGPBS), C. parasitica (CpCpgb1; CPU95139), A. nidulans (AnSfaD; AF056182), and P. carinii (Pcbeta; AF306565) using the Genetics Computer Group PILEUP program, followed by shading using Boxshade. Black shading indicates identical sequences. The positions of the seven WD repeats are labeled and indicated by arrows.
Using Northern analysis, two gnb-1 specific transcripts of 2.7 and 1.4 kb were detected in wild-type N. crassa (Fig. 1B). Two different-sized transcripts have also been reported for the C. parasitica cpgb-1 gene (22). In addition, expression of different-sized mRNA species seems to be a common theme for mammalian Gβ genes (30, 58). In contrast, only one Gβ-specific mRNA species has been detected in S. cerevisiae, S. pombe, and D. discoideum (25, 31, 60).
To determine the expression pattern of the GNB-1 protein during the N. crassa life cycle, plasma membranes were isolated from a variety of culture conditions and subjected to Western blot analysis using antiserum raised against GNB-1 protein heterologously expressed and purified from E. coli (Fig. 1C). The antiserum recognized a single protein species in all samples analyzed. GNB-1 levels are relatively low in conidia isolated from solid VM. The amount of GNB-1 increases within 2 h after conidia are transferred to liquid VM, peaking at 8 h of germination and then decreasing at 16 h. Cells grown on solid VM (under light or dark conditions) display the same relative amount of GNB-1 protein as 2-h germlings, while SCM cultures have the same high level of GNB-1 as 8-h germlings. These results demonstrate that a single protein species is recognized by antiserum directed against the gnb-1 gene product and that this protein is expressed throughout the life cycle. A similar observation has been reported for the single Gβ protein in D. discoideum (31). In contrast, the Ste4p Gβ protein is expressed in haploid but not diploid cells of S. cerevisiae (60).
Targeted deletion of gnb-1 and construction of Δgnb-1, gnb-1+ complemented strains.
A Δgnb-1 mutant was created by electroporation of the wild type with a construct in which most of the gnb-1 coding region is replaced by the hygromycin B resistance marker, hph (50) (Fig. 1A). Using the 600-bp insert from pSJJ5 as the probe (Fig. 1A), wild-type genomic DNA digested with XhoI and NcoI yields a 4.8-kb hybridizing species during Southern analysis (Fig. 1D). Because the hph gene contains a unique NcoI site, digestion of DNA from Δgnb-1 strains with XhoI and NcoI yields a 3.9-kb hybridizing fragment (Fig. 1D). Heterokaryotic primary transformants that contained both hybridizing fragments were identified (data not shown). Homokaryotic Δgnb-1 mutants were obtained by crossing the heterokaryons to a wild-type strain, with selection of progeny on hygromycin-containing medium (data not shown). Southern analysis using the same probe and restriction enzyme combination demonstrated that the hygromycin-resistant progeny strains contain only the 3.9-kb hybridizing fragment and no extra ectopic copies of the gene replacement construct (Fig. 1D; Table 1; data not shown). Northern and Western analysis showed that Δgnb-1 strains lack the two gnb-1 mRNA species (data not shown) and the GNB-1 protein (Fig. 1E). The Δgnb-1 mutation was complemented in trans with the original 7.65-kb gnb-1 genomic fragment subcloned into a plasmid containing the benomyl resistance selectable marker. Such Δgnb-1, gnb-1+ rescued strains have essentially normal levels of GNB-1 protein (Fig. 1E).
Δgnb-1 strains are male-fertile but female-sterile.
When cultured under nitrogen-starved conditions, N. crassa initiates the sexual cycle with production of female reproductive structures, termed protoperithecia (reviewed in reference 46). Fertilization is accomplished when a male gamete (usually a conidium or other vegetative cell) of opposite mating type comes in contact with the trichogyne tube emanating from the protoperithecium. The protoperithecium then enlarges to become a perithecium, the structure in which both meiosis and sexual spore (ascospore) formation take place.
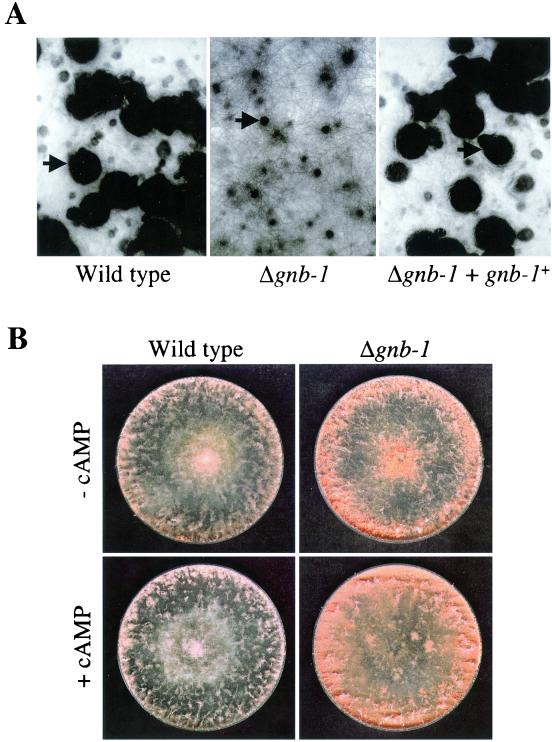
Previous work from our laboratory has uncovered roles for all three Gα genes in sexual fertility. Therefore, Δgnb-1 strains were analyzed for possible defects during the sexual cycle. Δgnb-1 strains (mating type mat a or mat A) are able to function as males during crosses with wild-type strains (74A or 74a; data not shown). However, when used as female parents, Δgnb-1 strains are sterile (Fig. 3A). Δgnb-1 strain protoperithecia do not develop normally after fertilization using wild-type conidia as males, with production of small perithecia and no ascospore ejection; this phenotype is identical to that observed for Δgna-1 strains (2, 20). The Δgnb-1 defects are common to both mating types. Homozygous sexual crosses between Δgnb-1 strains were not successful using either mat A or mat a mating types as female parents. cAMP supplementation did not restore female fertility to Δgnb-1 mutants, consistent with previous results indicating that mating is largely cAMP-independent in N. crassa (data not shown) (20, 24, 43).
FIG. 3.
Fertility and conidiation on solid medium. (A) Strains 42-8-3 (Δgnb-1), 74A (wild type), and A16 (Δgnb-1, gnb-1+) were cultured on SCM plates for 6 days in light prior to fertilization with wild-type strain 73a conidia. Plates were photographed 6 days after fertilization. Arrows indicate normal perithecia in wild-type and Δgnb-1, gnb-1+ strains and an aberrant perithecium in the Δgnb-1 strain. (B) Strains 42-8-3 and 74A were cultured on solid VM plates in the absence or presence of 1 mM cAMP for 3 days at 30°C in the dark. The more abundant conidium production in the Δgnb-1 strain is visible as the darker-orange, dense area at the edge of the plate.
Deletion of gnb-1 does not greatly impact apical extension rate but influences mass accumulation and conidiation on solid medium.
N. crassa grows vegetatively by apical extension and fusion of septated hyphae to form the basic body structure of the organism, the mycelium (reviewed in reference 49). We have previously shown that deletion of gna-1 leads to a lower apical extension rate on nitrogen-sufficient VM, with a more pronounced reduction on hyperosmotic media at 30°C in the dark (2, 20). Δgna-2 strains have essentially wild-type apical extension rates (2). However, deletion of gna-2 in the Δgna-1 background enhances the growth rate phenotypes, with levels at ∼15 to 20% of the wild-type level under hyperosmotic conditions (2). In contrast, loss of gna-3 does not greatly impact apical extension rates on normal and hyperosmotic solid media (24). Analysis of apical extension rates for the Δgnb-1 mutant demonstrates similarities to Δgna-3 strains, with nearly wild-type growth rates on normal and hyperosmotic medium (0.75 M NaCl, 0.75 M KCl, or 1.5 M sorbitol) (Table 2).
TABLE 2.
Apical extension rates on solid mediuma
| Strain | Apical extension rate (mm/h)b
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Light, RTc | % of WTd | Dark, RTc | % of WT | Dark, 30°Cc | % of WT | 0.75 M NaCle | % of WT | 0.75 M KCle | % of WT | 1.5 M sorbitole | % of WT | |
| 74A | 4.32 ± 0.07 | 100 | 4.02 ± 0.06 | 100 | 4.57 ± 0.21 | 100 | 2.63 ± 0.10 | 100 | 3.06 ± 0.14 | 100 | 2.52 ± 0.12 | 100 |
| 42-8-3 | 3.60 ± 0.11 | 83 | 3.18 ± 0.07 | 79 | 4.53 ± 0.07 | 99 | 2.72 ± 0.05 | 103 | 2.84 ± 0.12 | 93 | 2.52 ± 0.10 | 100 |
| A16 | 4.11 ± 0.26 | 95 | 4.31 ± 0.02 | 107 | 5.05 ± 0.03 | 111 | 2.55 ± 0.17 | 97 | 3.06 ± 0.17 | 100 | 2.06 ± 0.07 | 82 |
| G10 | 4.11 ± 0.21 | 95 | 4.34 ± 0.04 | 108 | 4.72 ± 0.13 | 103 | 2.29 ± 0.01 | 87 | 2.89 ± 0.05 | 94 | 2.07 ± 0.04 | 82 |
Plates were inoculated in the center with 1 μl of a conidial suspension. Growth rates were determined from measurements of colony diameters at various times.
Values are means ± standard errors of the means.
Cells were inoculated on VM plates with no additions and incubated in the indicated conditions of light and temperature. RT, room temperature.
WT, wild type.
Cells were cultured on VM plates with the indicated addition at 30°C in darkness.
Previous studies from our laboratory indicated that GNA-1 plays a positive role in regulation of mass accumulation under both light and dark conditions (20). The mass accumulation of Δgnb-1 strains was explored using cultures grown under two different conditions (Table 3). Δgnb-1 strains have significantly greater mass than the wild type (122 to 129%) at 30°C in the dark. However, the mass of Δgnb-1 mutants is only 54 to 57% that of the wild type at room temperature in light. The difference in mass accumulation is most influenced by the two growth temperatures (data not shown).
TABLE 3.
Mass accumulation on solid medium
| Strain | Dry wt (mg)a
|
|||
|---|---|---|---|---|
| Light, RT | % of WT | Dark, 30°C | % of WT | |
| 74A | 327 ± 8.1 | 100 | 300 ± 1.9 | 100 |
| 42-8-5 | 175 ± 18 | 54 | 366 ± 14 | 122 |
| 42-31-2 | 188 ± 12 | 57 | 387 ± 34 | 129 |
Cells were grown on VM plates with a cellophane overlay under the indicated conditions. Values are means ± standard errors of the means. RT, room temperature; WT, wild type.
We and others have shown that cr-1 and Δgna-3 N. crassa strains produce short aerial hyphae and conidiate prematurely on solid medium (24, 52). Addition of cAMP rescues the premature-conidiation but not the short-aerial-hypha defect (24). Δgna-1 strains produce short aerial hyphae and exhibit delayed but abundant conidiation; these defects are not reversed by cAMP supplementation (20). Based upon these previous observations, Δgnb-1 strains were analyzed during growth on solid medium in the absence and presence of cAMP (Fig. 3B). In the absence of cAMP, Δgnb-1 strains have normal-height aerial hyphae but produce more conidia than the wild type, particularly around the edge of the plate (Fig. 3B; data not shown). The abundant conidiation phenotype is a less severe version of that noted previously for Δgna-3 and cr-1 mutants. The observed abundant conidiation, coupled with the normal apical extension rate, may explain the higher mass accumulation of Δgnb-1 strains under these conditions. Addition of cAMP does not revert the appearance of Δgnb-1 strains to that of wild-type, and instead, Δgnb-1 strains exhibit mat-like growth, with more abundant aerial hyphae around the edges of the plate.
Δgnb-1 strains conidiate inappropriately in submerged culture.
At normal growth temperatures, wild-type N. crassa produces asexual spores (conidia) only on solid medium. However, N. crassa can be induced to conidiate in submerged culture by starvation for carbon or nitrogen or by exposure to high temperatures or cellular stress (9, 15, 45, 53). It was previously shown that loss of gna-3 leads to inappropriate conidiation in submerged culture (24). Recent results from our laboratory demonstrate that Δgna-1 strains exhibit cell density-dependent conidiation in submerged culture, with formation of conidiophores in cultures inoculated at 3 × 106 but not 1 × 106 cells/ml (F. D. Ivey, A. M. Kays, and K. A. Borkovich, unpublished observations). Deletion of gna-2 in a wild-type or Δgna-1 background does not affect submerged conidiation (data not shown). Addition of 2% peptone, but not 1 mM cAMP, to cultures of Δgna-3 and Δgna-1 strains suppresses the submerged conidiation phenotype (24; F. D. Ivey and K. A. Borkovich, unpublished observations).
Based on the connection between Gα genes and inappropriate conidiation, we analyzed submerged cultures of Δgnb-1 mutants and controls for the presence of conidiophores and conidia. Similar to previous results, wild-type strains did not produce conidiophores in 16-h-submerged cultures (Fig. 4A). Rescued Δgnb-1, gnb-1+ strains were phenotypically wild type. In contrast, loss of gnb-1 led to production of conidiophores and free conidia in submerged cultures at all cell densities tested (Fig. 4A; data not shown). Addition of 2% peptone to the medium caused all strains to produce swollen hyphae and suppressed the conidiation phenotype of Δgnb-1 strains (Fig. 4A).
FIG. 4.
Growth in submerged culture. (A) Microscopic observation. Wild-type (74A), Δgnb-1 (42-8-3 and 42-5-18), and Δgnb-1, gnb-1+ (A16) strains were cultured for 16 h in liquid VM in the absence or presence of 2% peptone. Representative conidiophores in the Δgnb-1 (no peptone) culture are indicated by arrows. (B) Levels of con-10 message. Samples containing 20 μg of total RNA obtained from the cultures in panel A were subjected to Northern analysis using the 250-bp BamHI-EcoRI insert from pW100 (con-10 gene) as a probe. Blots were reprobed with an rRNA gene to check for equal loading and transfer of RNA.
The submerged-conidiation phenotype of Δgnb-1 mutants was further examined by analysis of message levels for a conidiation-specific gene, con-10 (47). The results show that submerged conidiation of Δgnb-1 strains is correlated with expression of con-10 (Fig. 4B). Supplementation with 2% peptone eliminated most, if not all, of the con-10 expression in submerged cultures of Δgnb-1 mutants (Fig. 4B). Peptone also suppresses the submerged conidiation phenotype of other N. crassa mutants, including cr-1 and rco-3, the latter encoding a putative glucose sensor (33).
Δgnb-1 mutants have lower levels of intracellular cAMP in VM plate cultures.
We have previously demonstrated roles for all three Gα proteins in cAMP metabolism in N. crassa (21, 24, 63). Submerged cultures of Δgna-1 mutants lack GTP-stimulated adenylyl cyclase activity but have near-normal levels of adenylyl cyclase protein and steady-state cAMP (21; Ivey et al., unpublished). Observation of normal cAMP amounts presumably results from a compensatory mechanism involving reduced cAMP-phosphodiesterase activity in submerged cultures of Δgna-1 (and Δgna-2) mutants. GNA-1 antibody can inhibit adenylyl cyclase activity in wild-type extracts, while a GNA-2 antibody has no effect (21). Δgna-1 strains have reduced cAMP amounts in VM and SCM plate cultures (67 and 53% of wild type, respectively) (21). Δgna-2 mutants have normal levels of adenylyl cyclase activity and protein and intracellular cAMP during submerged growth, wild-type cAMP amounts in VM plate cultures, and levels that are 61% of wild-type levels on SCM (21). Δgna-1 Δgna-2 mutants have normal cAMP during submerged growth, but levels on VM and SCM plates are only 40 and 33% of wild-type levels, respectively (21). Submerged cultures of Δgna-3 mutants have lower levels of intracellular cAMP (∼32% of wild type) and adenylyl cyclase protein but normal GTP stimulation of remaining adenylyl cyclase activity (24). Δgna-3 strains have greatly reduced cAMP amounts when cultured on VM and SCM plate cultures (∼9 and 11% of wild type, respectively) (24). Results from analysis of adenylyl cyclase in all mutants support the hypothesis that GNA-1 functions as a GTP-dependent, stimulatory Gα, while GNA-3 regulates the levels of adenylyl cyclase protein.
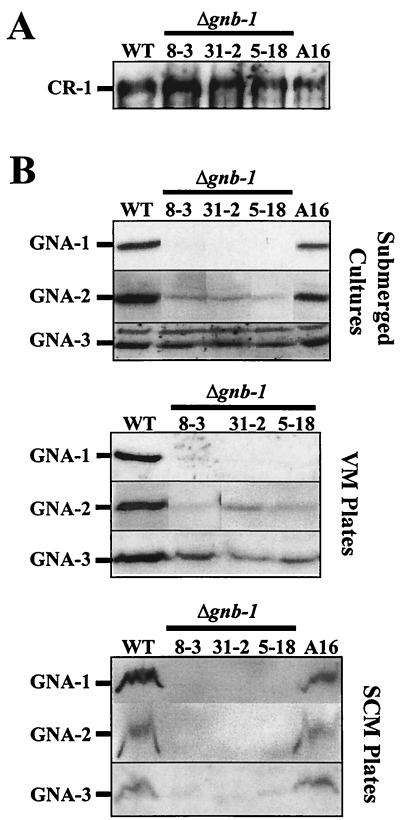
Work from our laboratory and others has demonstrated that sexual fertility is largely cAMP independent (20, 24). This observation, coupled with the known connection between Gα proteins and cAMP metabolism in N. crassa, prompted measurement of intracellular cAMP and/or adenylyl cyclase protein in Δgnb-1 strains cultured in submerged liquid and solid VM cultures. The results show that Δgnb-1 mutants have normal or slightly elevated cAMP levels in submerged culture (Table 4), similar to Δgna-1 and Δgna-2 strains. Furthermore, like Δgna-1 and Δgna-2 strains, Δgnb-1 mutants have essentially normal levels of the CR-1 protein in submerged cultures (Fig. 5A). Finally, similar to Δgna-1 mutants, Δgnb-1 strains have normal Mn2+ ATP adenylyl cyclase activity but drastically reduced Mg2+ ATP-dependent activity that is refractory to stimulation by GTP (data not shown).
TABLE 4.
Steady-state intracellular cAMP levels
| Genotype (strain) | Culture conditions | Intracellular cAMP levela
|
|
|---|---|---|---|
| pmol of cAMP/mg of protein | % of wild type | ||
| Wild type (74A) | Submerged culture | 4.50 ± 0.06 | 100 |
| Δgnb-1 (42-8-3) | Submerged culture | 5.35 ± 0.11 | 119 |
| Δgnb-1 (42-5-18) | Submerged culture | 5.54 ± 0.50 | 123 |
| Δgnb-1 (42-31-2) | Submerged culture | 4.40 ± 1.16 | 98 |
| Wild type (74A) | VM plates | 2.68 ± 0.36 | 100 |
| Δgnb-1 (42-8-3) | VM plates | 1.08 ± 0.02 | 40 |
| Δgnb-1 (42-5-18) | VM plates | 1.10 ± 0.34 | 41 |
Values are means ± standard errors of the means.
FIG. 5.
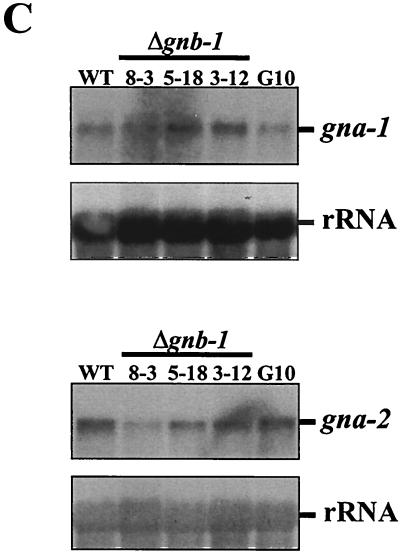
Analysis of Gα and adenylyl cyclase levels. (A) CR-1 protein levels in submerged cultures. Whole-cell extracts were prepared from 16-h-submerged cultures of the indicated strains, and 30 μg of protein was examined by Western analysis with CR-1 antiserum. (B) GNA-1, GNA-2 and GNA-3 protein levels under various growth conditions. Plasma membrane fractions were obtained from 16-h-submerged (inoculated at 3 × 106 conidia/ml), VM plate, or SCM plate cultures. Samples containing 30 μg (submerged or VM plate cultures) or 15 μg (SCM plate cultures) of protein were subjected to Western analysis with specific antisera. The curved nature of the cross-reacting protein bands in the SCM preparations presumably results from residual cell wall components that are not removed during the plasma membrane isolation procedure. (C) Levels of the gna-1 and gna-2 messages. Total RNA was extracted from 16-h-submerged cultures, and 20 μg was subjected to Northern analysis using the EcoRI-ClaI fragment of pPNO5 or the BamHI insert of p13 M2A5-2 as a probe to detect gna-1 and gna-2 expression, respectively. The N. crassa rRNA gene probe was used as an internal standard to check for relative amounts of RNA blotted onto the membrane. WT, wild-type strain 74A.
In VM plate cultures, strains lacking gnb-1 have significantly reduced levels of cAMP (40 to 41% of wild-type levels) (Table 4). This value is identical to that previously measured in Δgna-1 Δgna-2 double mutants (40% of wild type) and intermediate between those of Δgna-3 and Δgna-1 single mutants (∼11 and 67% of wild-type levels, respectively). Since the CR-1 protein cannot be reliably detected or assayed in extracts from VM plate cultures (data not shown), it is not known whether the reduced cAMP results from loss of stimulation or lowered levels of the enzyme.
Loss of gnb-1 impacts levels of the three N. crassa Gα proteins.
It was previously shown that levels of GNB-1 are normal in N. crassa strains lacking any one of the three Gα genes (21, 24). Similarly, deletion of individual Gα gene(s) does not influence levels of the remaining Gα proteins (2, 24). Therefore, it was of interest to see if loss of gnb-1 affects expression of the three Gα proteins, GNA-1, GNA-2, and GNA-3. Western analysis was conducted with the plasma membrane fraction isolated from 16-h-submerged VM cultures, VM plates, and SCM plates of wild-type and Δgnb-1 strains. The results show that in general, loss of gnb-1 leads to lower levels of Gα proteins (Fig. 5B). Similar results were obtained with whole-cell extracts, suggesting that the lower levels observed in the plasma membrane are not caused by free Gα proteins being released into the soluble fraction (data not shown). GNA-1 is almost undetectable in 16-h-submerged cultures, and levels of GNA-2 are greatly reduced. However, the effect on GNA-3 levels is much more subtle, with perhaps a 50% decrease in Δgnb-1 strains. The expression trend for Gα proteins in VM plate cultures is almost identical to that observed in submerged cultures. In SCM cultures, levels of all three Gα proteins are greatly reduced (Fig. 5B).
Altered expression of Gα proteins in the Δgnb-1 strain background could result from several mechanisms, including regulation of Gα mRNA expression by GNB-1. Because levels of GNA-1 and GNA-2 are most affected by loss of gnb-1, and the mRNA species for gna-1 and gna-2 are easily detected during Northern analysis of submerged culture mRNA, we analyzed the levels of gna-1 and gna-2 transcripts in submerged cultures of wild-type and Δgnb-1 strains. The results demonstrate that expression of the gna-1 and gna-2 mRNA species are normal in the Δgnb-1 mutant, consistent with posttranscriptional regulation of GNA-1 and GNA-2 levels in this tissue (Fig. 5C). mRNA species for gna-1 and gna-2 were absent from Δgna-1 and Δgna-2 mutants, respectively, but were present at normal levels in all other strains (24; data not shown). Since posttranscriptional control of a single Gα protein in the absence of the Gβ subunit has also been reported for C. parasitica (23), our results with multiple Gα proteins in N. crassa may reflect a conserved regulatory mechanism in filamentous fungi.
DISCUSSION
We have analyzed the contribution of the only known Gβ protein gene, gnb-1, to signal transduction during the life cycle of N. crassa. The predicted GNB-1 amino acid sequence shares the greatest homology with corresponding genes from filamentous fungi, followed by protist slime mold, amphibian, and human Gβ subunits. The GNB-1 protein is constitutively expressed throughout the N. crassa life cycle. The most severe phenotypes of Δgnb-1 mutants are manifested during the sexual cycle and in the control of conidiation in solid and liquid medium. A model summarizing our data is shown in Fig. 6.
FIG. 6.
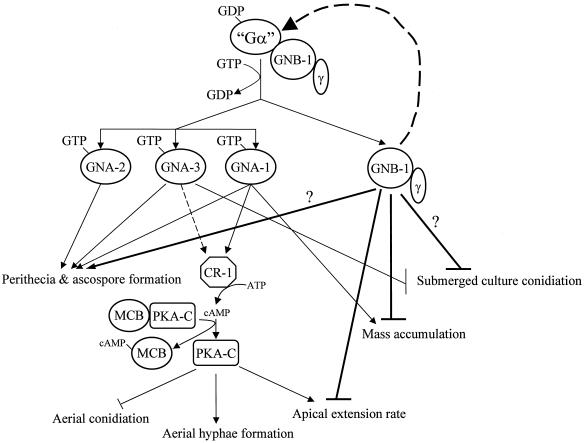
Contributions of GNB-1 and the three Gα subunits to N. crassa growth and development. It is presumed that each of the three GDP-bound Gα subunits (GNA-1, -2, and -3) can form a complex with GNB-1 and a yet-uncharacterized Gγ subunit in N. crassa; however, physical association has not been directly demonstrated. Ligand binding to a receptor(s) triggers GDP/GTP exchange on a Gα protein(s) and dissociation from the GNB-1/Gγ heterodimer. GNB-1 plays a global, positive role in maintaining normal levels of all three Gα proteins. During sexual development, the three Gα proteins (and perhaps GNB-1) regulate perithecial and ascospore formation via a cAMP-independent mechanism. GNA-1 and GNA-3 positively regulate adenylyl cyclase (CR-1) activity and protein levels, respectively. cAMP binds the regulatory subunit of protein kinase A (MCB) (4), leading to dissociation of MCB from the catalytic subunit (PKA-C). PKA-C activity promotes aerial hypha formation and apical extension while negatively regulating aerial conidiation. Apical extension rate and mass accumulation are each negatively regulated by GNB-1. Both GNA-3 and GNB-1 block conidiation in submerged culture. Bold lines indicate pathways regulated by GNB-1. Dashed lines depict regulation of Gα or CR-1 protein levels. “Gα” is GNA-1, -2, and -3. A question mark represents an uncertain contribution.
Analysis of Gα proteins in the Δgnb-1 mutant indicates a profound role for GNB-1 in regulating their expression level. Many of the cellular phenotypes of Δgnb-1 mutants can be explained by altered Gα protein amounts. Decreased Gα levels in the absence of Gβγ may reflect a cellular mechanism for preventing undesired activities of free Gα subunits. Comparison of RNA and protein levels for gna-1 and gna-2 in Δgnb-1 and wild-type hyphal submerged cultures suggests that GNB-1 regulates Gα expression at a posttranscriptional step in this cell type. Possibilities for posttranscriptional regulation include altered translation of Gα mRNA species and/or accelerated degradation of Gα proteins in the absence of the Gβγ tether. In mammals, loss of the Gαo gene leads to lower levels of Gβ proteins, via a posttranscriptional mechanism (34). In contrast, deletion of a single Gα gene or activation of GNA-1 does not influence GNB-1 levels in N. crassa (21).
The female sterility defect of Δgnb-1 strains is identical to that of Δgna-1 mutants (20). It is reasonable to speculate that the sexual defects of Δgnb-1 strains result from the drastically reduced GNA-1 protein levels in SCM cultures. On the other hand, because the GNB-1 protein level is relatively high in protoperithecial SCM cultures, GNB-1 itself could play a role in regulating female fertility synergistically with, or independently of, GNA-1. This hypothesis is supported by the finding that Gβ proteins play a positive role during mating and/or meiosis in all fungal species analyzed, with the exception of the fission yeast S. pombe. A Ste4p-regulated MAPK cascade that modulates the pheromone response has been well-characterized in the budding yeast S. cerevisiae (reviewed in reference 14), but less is known about Gβ signaling during sexual differentiation in filamentous fungi. In both A. nidulans (48) and N. crassa, mutation of the Gβ gene blocks normal fruiting body development (cleistothecia and perithecia, respectively), which normally follows mating in these organisms. The mode of regulation differs between these species, however, in that activating mutations in the Gα most similar to N. crassa gna-1 (fadA) leads to loss of cleistothecium formation in A. nidulans (48) while N. crassa strains containing the corresponding gna-1 alleles produce fertile perithecia (albeit fewer and larger ones) (63). In C. neoformans, the mating defect of Gβ mutants is not corrected by cAMP (57), similar to N. crassa. Instead, evidence has been presented that Gpb-1 regulates a MAPK to control sexual development, analogous to S. cerevisiae. Several MAPKs have now been identified in the N. crassa genome sequence (data not shown), including NRC-1 (26). NRC-1 is most similar to the MAPKKK proteins S. cerevisiae Ste11p, a component of MAPK cascades required for mating, haploid invasive growth, and diploid pseudohyphal growth, and S. pombe Byr2, required for mating (reviewed in reference 9). nrc-1 and gnb-1 mutants share the phenotypes of inappropriate submerged conidiation and female sterility. However, it is not clear that GNB-1 functions upstream of NRC-1, in that nrc-1 mutants do not produce protoperithecia (26), unlike gnb-1 null strains. Alternatively, NRC-1 may be regulated by multiple input pathways, including one containing GNB-1.
The submerged conidiation phenotype of Δgnb-1 strains is similar to that previously observed for Δgna-3 mutants (24). Since submerged cultures of Δgnb-1 strains have lower levels of GNA-3 than wild type, it is possible that this reduction is responsible for the inappropriate conidiation. However, GNB-1 may itself negatively regulate conidiation in submerged culture, independent of GNA-3. The latter hypothesis is supported by the fluctuation in GNB-1 levels observed in wild-type cells during early germination in submerged culture. Inappropriate submerged conidiation has also been reported for A. nidulans sfaD mutants (48). However, an interesting difference between the two species is that rich nutrient sources suppress the submerged conidiation of N. crassa Δgnb-1 mutants but actually accentuate this phenotype in A. nidulans sfaD strains (48). This divergence may reflect a difference in the relative contributions of G proteins to growth in the two species.
The reduced levels of GNA-1 and GNA-2 in Δgnb-1 mutants can account for the observation that the cAMP metabolism profile of Δgnb-1 strains is most similar to that of Δgna-1 or Δgna-1 Δgna-2 strains. Thus, at this time, there is no evidence for direct regulation of adenylyl cyclase activity by GNB-1 in N. crassa. This hypothesis is supported by results from a previous study comparing strains with wild-type, null, or activated gna-1 alleles, in which it was shown that the GTP-bound state of GNA-1 is positively correlated with adenylyl cyclase activity but does not influence levels of GNB-1 (63; F. D. Ivey, Q. Yang, and K. A. Borkovich, unpublished data). This scenario can be contrasted with the situation in D. discoideum, in which Gβ, in concert with other proteins, serves as a positive regulator of adenylyl cyclase activity (7, 19, 61). Finally, S. pombe strains lacking git5 exhibit altered cAMP amounts only transiently during the shift from glucose-starved to glucose-rich conditions in liquid culture (28). This observation is in keeping with the finding that Δgnb-1 mutants have essentially normal steady-state cAMP levels during submerged growth in liquid medium.
The aerial-hypha phenotype of Δgnb-1 mutants on cAMP-containing medium is a less severe version of that observed for strains containing constitutively activated gna-1 alleles (Q204L and R179C) (63) and, to a more modest degree, for corresponding activated gna-2 alleles (2). Strains carrying the gna-1Q204L or gna-1R179C allele have normal amounts of the respective mutant GNA-1 protein and elevated levels of cAMP on VM plates (63). Δgnb-1 strains contain lower levels of all three Gα proteins, with the greatest reduction in GNA-1. A possible explanation for these results is that strains with activated gna-1 alleles have elevated cAMP levels and free GNA-1, both of which are necessary for production of abundant aerial hyphae. The low level of free GNA-1 in Δgnb-1 strains can only increase aerial-hypha amount when cAMP is supplied exogenously. This model is consistent with our previous observation that the short-aerial-hypha phenotype of Δgna-1, cr-1, and Δgna-3 mutants on solid VM cannot be corrected by exogenous cAMP (20, 24), presumably due to the absence of GNA-1 protein or normal tethering of GNA-1 in these strains. The apparent requirement for free GNA-1 in addition to elevated cAMP implies that GNA-1 regulates a cAMP-independent effector pathway responsible for aerial-hypha formation in N. crassa. One possibility is that GNA-1 acts upstream of a MAPK pathway that includes NRC-1 and regulates aerial-hypha production.
There are several Δgnb-1 phenotypes that cannot be accounted for by loss of a Gα subunit(s). The near-wild-type growth rates of Δgnb-1 strains on normal and hyperosmotic medium cannot easily be explained by a mechanism solely involving regulation of apical extension rates by Gα proteins, as VM plate cultures of Δgnb-1 strains have greatly reduced levels of GNA-1 and lower levels of GNA-2 and GNA-3, yet the apical extension rate on this medium is normal. Rather, these observations suggest that GNB-1 may have a negative effect on growth rate, independent of GNA-1 and GNA-2. A similar argument can be made for the increased mass accumulation of Δgnb-1 cultures grown in the dark at 30°C. An alternative explanation is that loss of GNB-1 leads to constitutive activation of residual Gα subunits. There are examples of partnerless Gα proteins in several organisms as well as evidence that they perform cellular functions. For example, Gα, but not Gβγ, proteins can be detected in Golgi fractions in rat exocrine pancreas (11). S. cerevisiae Gpa2p is expressed in diploids where Ste4p is not and is required for proper regulation of pseudohyphal development (27, 32, 60). Similarly, evidence shows that S. pombe Gpa1, a positive regulator of sexual differentiation (39), is not coupled to the only known Gβ protein in this organism (28). A possible mechanism for signaling by Gα in the absence of Gβγ is provided by evidence that Gα proteins can form oligomers (37). Oligomerization of Gα subunits may facilitate the GDP/GTP exchange cycle without the requirement for Gβγ. Additionally, polymerization of Gα allows higher local concentration so that activated receptor can transduce the signal to Gα in a faster kinetic status (37).
Maintenance of appropriate interactions between Gα and Gβγ subunits is critical for normal G protein signaling in eukaryotic organisms. Loss of a subunit can lead to activation or to inhibition and/or destabilization of its corresponding partner. Our work suggests that Gβ proteins are necessary for maintaining normal levels of associated Gα subunits in filamentous fungi. Regardless of the specific mechanism, assignment of functions to Gβ is complicated by the observation that levels of all three Gα proteins are aberrant in a Δgnb-1 strain. However, we have the advantage of having previously analyzed the three Gα subunits and characterized βγ-independent functions for GNA-1 in N. crassa. Future studies will be focused on probing the mechanism of G protein activation and the relative contribution of Gα and Gβγ to signaling, using genetic and biochemical tools.
Acknowledgments
We thank Simon Jakubowski and Wafa Elbjeirami for assistance with library screening and subcloning; Gloria Turner for Northern analysis; Thomas Vida and Dale Hereld for help with microscopy; Dupont Chemical Company for the gift of benomyl, and Jennifer Bieszke, Carmen Dessauer, Douglas Ivey, Ann Kays, Svetlana Krystofova, Gloria Turner, Thomas Wilkie, and Lynn Zechiedrich for comments on the manuscript and/or helpful discussions. We acknowledge the Center for Genome Research at the Whitehead Institute for Biomedical Research (Cambridge, Mass.) for release of the N. crassa genomic sequence (http://www-genome.wi.mit.edu/annotation/fungi/neurospora/).
This work was supported by Public Health Service grant GM-48626 from the National Institutes of Health (to K.A.B.).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baasiri, R. A., X. Lu, P. S. Rowley, G. E. Turner, and K. A. Borkovich. 1997. Overlapping functions for two G protein α subunits in Neurospora crassa. Genetics 147:137-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxevanis, A. D., and D. Landsman. 1995. The HMG-1 box protein family: classification and functional relationships. Nucleic Acids Res. 23:1604-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruno, K. S., R. Aramayo, P. F. Minke, R. L. Metzenberg, and M. Plamann. 1996. Loss of growth polarity and mislocalization of septa in a Neurospora mutant altered in the regulatory subunit of cAMP-dependent protein kinase. EMBO J. 15:5772-5782. [PMC free article] [PubMed] [Google Scholar]
- 5.Case, M. E., M. Schweizer, S. R. Kushner, and N. H. Giles. 1979. Efficient transformation of Neurospora crassa by utilizing hybrid plasmid DNA. Proc. Natl. Acad. Sci. USA 76:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chase, D. L., G. A. Patikoglou, and M. R. Koelle. 2001. Two RGS proteins that inhibit Gαo and Gαq signaling in C. elegans neurons require a Gβ5-like subunit for function. Curr. Biol. 11:222-231. [DOI] [PubMed] [Google Scholar]
- 7.Chen, M. Y., Y. Long, and P. N. Devreotes. 1997. A novel cytosolic regulator, Pianissimo, is required for chemoattractant receptor and G protein-mediated activation of the 12 transmembrane domain adenylyl cyclase in Dictyostelium. Genes Dev. 11:3218-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clapham, D. E., and E. J. Neer. 1997. G protein beta gamma subunits. Annu. Rev. Pharmacol. Toxicol. 37:167-203. [DOI] [PubMed] [Google Scholar]
- 9.Cortat, M., and G. Turian. 1974. Conidiation of Neurospora crassa in submerged culture without mycelial phase. Arch. Mikrobiol. 95:305-309. [DOI] [PubMed] [Google Scholar]
- 10.Davis, R. H., and F. J. deSerres. 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 71A:79-143. [Google Scholar]
- 11.Denker, S. P., J. M. McCaffery, G. E. Palade, P. A. Insel, and M. G. Farquhar. 1996. Differential distribution of alpha subunits and beta gamma subunits of heterotrimeric G proteins on Golgi membranes of the exocrine pancreas. J. Cell Biol. 133:1027-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Downes, G. B., and N. Gautam. 1999. The G protein subunit gene families. Genomics 62:544-552. [DOI] [PubMed] [Google Scholar]
- 13.Ebbole, D., and M. S. Sachs. 1990. A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newsl. 37:17-18. [Google Scholar]
- 14.Elion, E. A. 2000. Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 3:573-581. [DOI] [PubMed] [Google Scholar]
- 15.Guignard, R., F. Grange, and G. Turian. 1974. Microcycle conidiation induced by partial nitrogen deprivation in Neurospora crassa. Can. J. Microbiol. 30:1210-1215. [Google Scholar]
- 16.Gurr, S. J., S. E. Unkles, and J. R. Kinghorn. 1987. The structure and organization of nuclear genes of filamentous fungi, p. 93-139. In J. R. Kinghorn (ed.), Gene structure in eukaryotic microbes, vol. 22. IRL Press Ltd., Oxford, United Kingdom. [Google Scholar]
- 17.Hamm, H. E. 1998. The many faces of G protein signaling. J. Biol. Chem. 273:669-672. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557. [DOI] [PubMed] [Google Scholar]
- 19.Insall, R., A. Kuspa, P. J. Lilly, G. Shaulsky, L. R. Levin, W. F. Loomis, and P. Devreotes. 1994. CRAC, a cytosolic protein containing a pleckstrin homology domain, is required for receptor and G protein-mediated activation of adenylyl cyclase in Dictyostelium. J. Cell Biol. 126:1537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivey, F. D., P. N. Hodge, G. E. Turner, and K. A. Borkovich. 1996. The Gαi homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol. Biol. Cell 7:1283-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivey, F. D., Q. Yang, and K. A. Borkovich. 1999. Positive regulation of adenylyl cyclase activity by a Gαi homologue in Neurospora crassa. Fungal Genet. Biol. 26:48-61. [DOI] [PubMed] [Google Scholar]
- 22.Kasahara, S., and D. L. Nuss. 1997. Targeted disruption of a fungal G-protein β subunit gene results in increased vegetative growth but reduced virulence. Mol. Plant-Microbe Interact. 10:984-993. [DOI] [PubMed] [Google Scholar]
- 23.Kasahara, S., P. Wang, and D. L. Nuss. 2000. Identification of bdm-1, a gene involved in G protein beta-subunit function and alpha-subunit accumulation. Proc. Natl. Acad. Sci. USA 97:412-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kays, A. M., P. S. Rowley, R. A. Baasiri, and K. A. Borkovich. 2000. Regulation of conidiation and adenylyl cyclase levels by the Gα protein GNA-3 in Neurospora crassa. Mol. Cell. Biol. 20:7693-7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, D.-U., S.-K. Park, K.-S. Chung, M.-U. Choi, and H.-S. Yoo. 1996. The G protein β subunit Gpb1 of Schizosaccharomyces pombe is a negative regulator of sexual development. Mol. Gen. Genet. 252:20-32. [DOI] [PubMed] [Google Scholar]
- 26.Kothe, G. O., and S. J. Free. 1998. The isolation and characterization of nrc-1 and nrc-2, two genes encoding protein kinases that control growth and development in Neurospora crassa. Genetics 149:117-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubler, E., H. U. Mosch, S. Rupp, and M. P. Lisanti. 1997. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J. Biol. Chem. 272:20321-20323. [DOI] [PubMed] [Google Scholar]
- 28.Landry, S., M. T. Pettit, E. Apolinario, and C. S. Hoffman. 2000. The fission yeast git5 gene encodes a Gβ subunit required for glucose-triggered adenylate cyclase activation. Genetics 154:1463-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine, M. A., P. M. Smallwood, P. T. Moen, L. J. Helman, and T. G. Ahn. 1990. Molecular cloning of β3 subunit, a third form of the G protein β-subunit polypeptide. Proc. Natl. Acad. Sci. USA 87:2329-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lilly, P., L. Wu, D. L. Welker, and P. N. Devreotes. 1993. A G-protein β-subunit is essential for Dictyostelium development. Genes Dev. 7:986-995. [DOI] [PubMed] [Google Scholar]
- 32.Lorenz, M. C., and J. Heitman. 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J. 16:7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madi, L., S. A. McBride, L. A. Bailey, and D. J. Ebbole. 1997. rco-3, a gene involved in glucose transport and conidiation in Neurospora crassa. Genetics 146:499-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mende, U., B. Zagrovic, A. Cohen, Y. Li, D. Valenzuela, M. C. Fishman, and E. J. Neer. 1998. Effect of deletion of the major brain G-protein α subunit (αo) on coordination of G-protein subunits and on adenylyl cyclase activity. J. Neurosci. Res. 54:263-272. [DOI] [PubMed] [Google Scholar]
- 35.Metzenberg, R. L., and J. Grotelueschen. 1989. Restriction polymorphism maps of Neurospora crassa: update. Neurospora Newsl. 36:51-57. [Google Scholar]
- 36.Metzenberg, R. L., J. N. Stevens, E. U. Selker, and E. Morzycka-Wroblewska. 1985. Identification and chromosomal distribution of 5S rRNA genes in Neurospora crassa. Proc. Natl. Acad. Sci. USA 82:2067-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mixon, M. B., E. Lee, D. E. Coleman, A. M. Berghuis, A. G. Gilman, and S. R. Sprang. 1995. Tertiary and quaternary structural changes in Giα1 induced by GTP hydrolysis. Science 270:954-960. [DOI] [PubMed] [Google Scholar]
- 38.Neer, E. J., and T. F. Smith. 1996. G protein heterodimer: new structures propel new questions. Cell 84:175-178. [DOI] [PubMed] [Google Scholar]
- 39.Obara, T., M. Nakafuku, M. Yamamoto, and Y. Kaziro. 1991. Isolation and characterization of a gene encoding a G-protein α subunit from Schizosaccharomyces pombe: involvement in mating and sporulation pathways. Proc. Natl. Acad. Sci. USA 88:5877-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orbach, M. J., E. B. Porro, and C. Yanofsky. 1986. Cloning and characterization of the gene for β-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant marker. Mol. Cell. Biol. 6:2452-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orbach, M. J., M. S. Sachs, and C. Yanofsky. 1990. The Neurospora crassa arg-2 locus. Structure and expression of the gene encoding the small subunit of arginine-specific carbamoyl phosphate synthetase. J. Biol. Chem. 265:10981-10987. [PubMed] [Google Scholar]
- 42.Pall, M. L., and J. P. Brunelli. 1994. New plasmid and lambda/plasmid hybrid vectors and a Neurospora crassa genomic library containing the bar selectable marker and the Cre/lox site-specific recombination system for use in filamentous fungi. Fungal Genet. Newsl. 41:63-65. [Google Scholar]
- 43.Perkins, D. D., A. Radford, D. Newmeyer, and M. Bjorkman. 1982. Chromosomal loci of Neurospora crassa. Microbiol. Rev. 46:426-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Philley, M. L., and C. Staben. 1994. Functional analyses of the Neurospora crassa MT a-1 mating type polypeptide. Genetics 137:715-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plesofsky-Vig, N., D. Light, and R. Brambl. 1983. Paedogenetic conidiation in Neurospora crassa. Exp. Mycol. 7:283-286. [Google Scholar]
- 46.Raju, N. B. 1992. Genetic control of the sexual cycle in Neurospora. Mycol. Res. 96:241-262. [Google Scholar]
- 47.Roberts, A. N., V. Berlin, K. M. Hager, and C. Yanofsky. 1988. Molecular analysis of a Neurospora crassa gene expressed during conidiation. Mol. Cell. Biol. 8:2411-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosen, S., J. H. Yu, and T. H. Adams. 1999. The Aspergillus nidulans sfaD gene encodes a G protein beta subunit that is required for normal growth and repression of sporulation. EMBO J. 18:5592-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Springer, M. L. 1993. Genetic control of fungal differentiation: the three sporulation pathways in Neurospora crassa. Bioessays 15:365-374. [DOI] [PubMed] [Google Scholar]
- 50.Staben, C., B. Jensen, M. Singer, J. Pollock, M. Schechtman, J. Kinsey, and E. Selker. 1989. Use of a bacterial hygromycin B resistance gene as a dominant selectable marker in Neurospora crassa transformation. Fungal Genet. Newsl. 36:79-81. [Google Scholar]
- 51.Studier, F. W, A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 52.Terenzi, H. F., M. M. Flawia, and H. N. Torres. 1974. A Neurospora crassa morphological mutant showing reduced adenylate cyclase activity. Biochem. Biophys. Res. Commun. 58:990-996. [DOI] [PubMed] [Google Scholar]
- 53.That, T. C., and G. Turian. 1978. Ultrastructural study of microcyclic macroconidiation in Neurospora crassa. Arch. Microbiol. 116:279-288. [DOI] [PubMed] [Google Scholar]
- 54.Turner, G. E., and K. A. Borkovich. 1993. Identification of a G protein α subunit from Neurospora crassa that is a member of the Gi family. J. Biol. Chem. 268:14805-14811. [PubMed] [Google Scholar]
- 55.Vann, D. C. 1995. Electroporation-based transformation of freshly harvested conidia of Neurospora crassa. Fungal Genet. Newsl. 42A:53. [Google Scholar]
- 56.Vogel, H. J. 1964. Distribution of lysine pathways among fungi: evolutionary implications. Am. Nat. 98:435-446. [Google Scholar]
- 57.Wang, P., J. R. Perfect, and J. Heitman. 2000. The G-protein beta subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol. Cell. Biol. 20:352-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watson, A. J., A. Katz, and M. I. Simon. 1994. A fifth member of the mammalian G-protein β-subunit family. J. Biol. Chem. 269:22150-22156. [PubMed] [Google Scholar]
- 59.Westergaard, M., and H. K. Mitchell. 1947. Neurospora V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34:573-577. [Google Scholar]
- 60.Whiteway, M., L. Hougan, D. Dignard, D. Y. Thomas, L. Bell, G. C. Saari, F. J. Grant, P. O'Hara, and V. MacKay. 1989. The STE4 and STE18 genes of yeast encode potential β and γ subunits of the mating factor receptor-coupled G protein. Cell 56:476-477. [DOI] [PubMed] [Google Scholar]
- 61.Wu, L., R. Valkema, P. J. M. Van Haastert, and P. N. Devreotes. 1995. The G protein β subunit is essential for multiple responses to chemoattractants in Dictyostelium. J. Cell Biol. 129:1667-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang, Q., J. A. Bieszke, and K. A. Borkovich. 2000. Differential complementation of a Neurospora crassa Gαi mutation using mammalian Gα protein genes. Mol. Gen. Genet. 263:712-721. [DOI] [PubMed] [Google Scholar]
- 63.Yang, Q., and K. A. Borkovich. 1999. Mutational activation of a Gαi causes uncontrolled proliferation of aerial hyphae and increased sensitivity to heat and oxidative stress in Neurospora crassa. Genetics 151:107-117. [DOI] [PMC free article] [PubMed] [Google Scholar]