Abstract
We report a random survey of 1 to 2% of the somatic genome of the free-living ciliate Paramecium tetraurelia by single-run sequencing of the ends of plasmid inserts. As in all ciliates, the germ line genome of Paramecium (100 to 200 Mb) is reproducibly rearranged at each sexual cycle to produce a somatic genome of expressed or potentially expressed genes, stripped of repeated sequences, transposons, and AT-rich unique sequence elements limited to the germ line. We found the somatic genome to be compact (>68% coding, estimated from the sequence of several complete library inserts) and to feature uniformly small introns (18 to 35 nucleotides). This facilitated gene discovery: 722 open reading frames (ORFs) were identified by similarity with known proteins, and 119 novel ORFs were tentatively identified by internal comparison of the data set. We determined the phylogenetic position of Paramecium with respect to eukaryotes whose genomes have been sequenced by the distance matrix neighbor-joining method by using random combined protein data from the project. The unrooted tree obtained is very robust and in excellent agreement with accepted topology, providing strong support for the quality and consistency of the data set. Our study demonstrates that a random survey of the somatic genome of Paramecium is a good strategy for gene discovery in this organism.
Alongside an ever-growing number of prokaryotic genomes, several fungal, invertebrate, plant, and vertebrate genomes have now been largely or completely sequenced and released to the public, providing a wealth of information for functional and comparative studies. Given the variety of unicellular eukaryotes and the great evolutionary distances even within protist phyla, it is striking that few protists have been subjected to systematic genomic investigation. Notable exceptions are the cellular slime mold Dictyostelium discoidium and a few parasites of great medical importance such as Plasmodium spp. Ciliates are one of the major eukaryotic groups for which no large-scale genome project has been undertaken.
Ciliate models (Paramecium, Tetrahymena) have allowed major discoveries in biology such as variant nuclear genetic codes (13, 49), ribozymes (38), telomerase (33), and histone acetyltransferase as a transcription factor (10) and present fascinating epigenetic phenomena acting at DNA (14, 21, 42), RNA (51), and protein (6) levels. Unique among unicellular eukaryotes, ciliates separate germinal and somatic lines, in the form of nuclei (50). Somatic development involves programmed rearrangements of the entire germ line genome at each sexual generation, so that ciliates provide excellent experimental models for studying somatic DNA rearrangements similar to those that generate antibody diversity and malignant states in vertebrates.
The germ line micronucleus is diploid, is transcriptionally silent during vegetative growth, and intervenes during sexual processes. The somatic macronucleus is highly polyploid and responsible for transcriptional activity but is not transmitted across sexual generations. A new macronucleus is formed from the zygotic nucleus at each sexual generation. Although examples of pseudogenes are now documented in Paramecium primaurelia, they are highly underrepresented in the somatic genome (19). Thus, macronuclear genes can be considered to all be potentially expressed.
We carried out a pilot project of random sequencing of somatic DNA of the ciliate P. tetraurelia, the species of Paramecium that has been the most extensively studied by genetics (54). The genome of this organism is estimated to be 100 to 200 Mb in size, and most of the germ line sequences are present in the rearranged somatic genome which is amplified, fragmented, and stripped of noncoding sequences during development (60). The 10 to 15% of germ line DNA that is eliminated during somatic development may represent the heterochromatic fraction of the genome (44) and contains both intact and degenerate transposable elements (45).
Our survey of more than 3,000 single-run sequences (average length, ∼500 nucleotides [nt]) from the ends of inserts of a genomic library (36), corresponding to 1 to 2% of the genome, and of the sequences of several complete library inserts indicates that the Paramecium somatic genome is as compact as the yeast genome with >68% coding sequences and uniformly small introns (18 to 35 nt). We were able to identify 722 protein coding genes by similarity searches against databanks. In addition, internal comparison of the data set allowed identification of 108 families of protein coding genes, including 119 novel genes not identified by similarity search against databanks. Our study demonstrates that random sequencing of somatic DNA is an efficient strategy for gene discovery in Paramecium, especially since the sequences can be rapidly exploited for functional analysis by gene silencing (51) or RNA interference (29) even before the complete gene sequence is established. Since our data support a compact genome, with uniformly small introns, we believe that Paramecium would be an excellent choice for systematic sequencing in the near future.
MATERIALS AND METHODS
Sequencing.
The indexed library of macronuclear DNA from the 100% homozygous d4-2 strain, constructed to facilitate cloning genes by functional complementation, has been described (36). The library of >60,000 clones isolated in 384-well microtiter plates consists of 4- to 12-kb inserts in the pBluescript II KS(−) vector. The library was experimentally estimated to cover ∼3 times the genome. No evidence of contaminating mitochondrial, micronuclear, or bacterial DNA was obtained either in the initial characterization of the library (36) or in the genome survey analysis described here.
Single run sequencing of the ends of 1,800 clones from the indexed library was carried out at the Institute of Biochemistry and Biophysics of the Polish Academy of Sciences, Warsaw, Poland, or at the Core Molecular Biology Facility, York University, Toronto, Ontario, Canada. Plasmid DNA was prepared by standard alkaline lysis procedures on early-stationary-phase cultures, and DNA sequencing was performed by dye termination cycle sequencing and analyzed by using ABI fluorescence automated sequencers.
Data analysis.
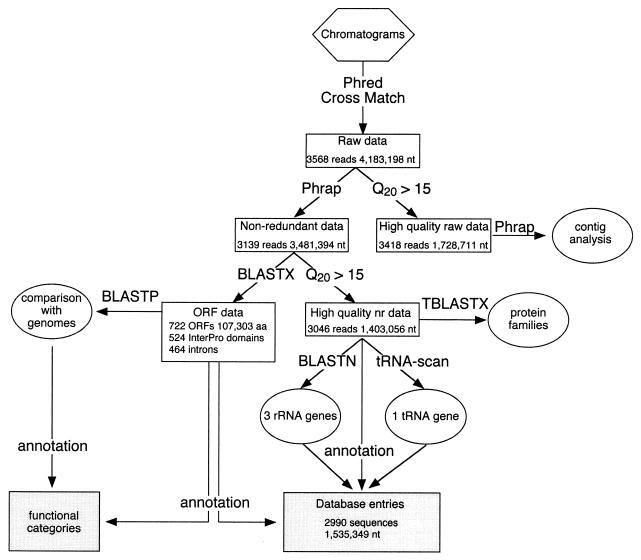
Data analysis was mainly performed at the INFOBIOGEN (French Bioinformatics Resource Centre) computing facility on a SunOS 5.6 Unix platform (see http://www.infobiogen.fr). All stages of the data analysis involved customized Perl scripts which we wrote to control data flow, extract information, or present the results (e.g., in HTML) in conjunction with programs given in Fig. 1 and Table 1.
FIG. 1.
Flow chart of the project. The square boxes represent data sets of nucleotide or protein sequences, and the programs used to generate the data sets are indicated over the arrows. Shaded boxes indicate annotated data submitted to the public nucleotide database or available at http://paramecium.cgm.cnrs-gif.fr. Ovals indicate aspects of the analysis detailed in the present article. Phrap, Phred, tRNA-SCAN, and BLAST programs are publicly available (see Materials and Methods). We wrote Perl scripts to cut the sequences according to Phrap quality values (Q20 > 15) and to annotate the data (tables of functional categories; database submission).
TABLE 1.
Similarity searchesa
| Target | Method | Query (GSS data set) | Result of query | |||
|---|---|---|---|---|---|---|
| Database or sequence | Date posted | No. of sequences | No. of letters | |||
| GenBank Alveolata | 8 August 2000 | 36,628 | 29,939,175 | BLASTN 2.0.14 | hq | 3 rRNA genes; 6 protein coding genes |
| Swall | BLASTX 2.0.12 | nr | 722 ORFs | |||
| Swissprot | 7 July 2000 | 86,593 | 31,411,157 | |||
| Swissprotnew | 8 July 2000 | 2,822 | 1,280,954 | |||
| Sptrembl | 28 June 2000 | 297,973 | 93,374,136 | |||
| Sptrnew | 8 July 2000 | 71,201 | 22,367,698 | |||
| Pfam | 21 September 2000 | 111,934 | 38,089,901 | BLASTP 2.0.14 | ORF | 524 PfamA domains |
| Proteome | BLASTP 2.0.14 | ORF | ||||
| E. coli | 4 January 2001 | 4,581 | 1,435,304 | 101 hitsb | ||
| S. cerevisiae | 15 November 2000 | 6,358 | 2,991,939 | 505 hitsb | ||
| C. elegans | 15 November 2000 | 19,704 | 8,596,400 | 586 hitsb | ||
| D. melanogaster | 15 November 2000 | 14,080 | 6,850,524 | 599 hitsb | ||
| A. thaliana | 6 January 2001 | 25,458 | 11,049,032 | 588 hitsb | ||
| H. sapiens | 15 November 2000 | 31,919 | 13,433,624 | 616 hitsb | ||
| hq | 2 November 2000 | 3,046 | 1,403,056 | TBLASTX 2.0.14 | hq | 108 families |
The genome survey sequence (GSS) data set used for each query is as follows: nr, nonredundant data (3,139 reads; 3,481,394 nt); hq, high-quality nonredundant data (3,046 reads, 1,403,056 nt); and ORF, set of 722 ORF fragments identified by similarity (107,303 aa).
Threshold, E < 10−4.
The Phred/Phrap package of programs (24, 25) was used to call bases from the chromatograms, screen vector, and construct contigs. The quality files generated by Phred were saved and used to automatically cut the sequences as required for subsequent analyses (see Results). For this project, we define “high quality” as the region of a sequence for which the probability of error in a nucleotide is less than 1 in 30 (Phred quality >15 over a 20-nt window; Q20 > 15).
The resulting set of unique sequences was used to search for similarity with known DNA and protein sequences, protein domains, and protein signatures in the public databases listed in Table 1, with appropriate implementations of the BLAST algorithm (2, 3). For BLASTX and TBLASTX analyses, the ciliate genetic code, the seg filter, and the blosum62 matrix were used.
Expert validation.
Although we tried to develop automatic data analysis that would also be suitable for a much larger project, some expert validation was necessary. Open reading frames (ORFs) were identified only on the basis of similarity with known sequences. BLASTX matches against the protein databases, initially retained with a very high expectation value (E < 10−2), were subjected to expert validation to decide the following. (i) Is the match significant? For example, heptad repeats indicative of coiled-coil domains, despite relatively low E-values, were not retained unless the match contained other indications of homology with the target sequence. In contrast, short but near-perfect matches with high E-values were retained. (ii) Does the sequence contain introns? The latter can change the reading frame of the match, which appears as multiple segments. When in phase, the introns appear as 6 to 11 unmatched amino acids, producing a gap in the alignment, sometimes containing a stop codon.
The set of 108 unique Paramecium introns (20 to 34 nt in length) from the invertebrate division of the EMBL/GenBank/DDBJ database, many of them identified on the basis of experimental evidence, were used to determine an intron profile by using a hidden Markov model (HMM). A list of potential introns, with scores, was then generated for the genome survey sequence (GSS) set. The majority of introns identified by expert validation were also identified by HMM and had high scores. However, the inverse was not true: the majority of high scoring potential introns identified by HMM profile analysis were not introns according to the expert validation. The HMM approach is thus promising, but more information is clearly required for automatic identification of Paramecium introns, a crucial step in the development of a Gene Finder.
Phylogenetic analysis.
CLUSTALW (v. 1.81) was used to align orthologous amino acid sequences, and the alignments were automatically trimmed to eliminate regions with insertions or deletions and concatenated using custom Perl scripts. Neighbor-joining trees were built by using pairwise distances between the amino acid sequences by using the programs PROTDIST with a Dayhoff scoring matrix and NEIGHBOR of the Phylip 3.53 package. Confidence levels were evaluated by using 100 bootstrap replicates. Unrooted trees were drawn with software available by ftp from pbil.univ-lyon1.fr (48). The data sets used and the alignments are available at http://paramecium.cgm.cnrs-gif.fr/phylogeny.
Nucleotide sequence accession numbers.
The final set of 2,990 nt sequences was deposited in the GSS division of the EMBL database (release date 2 November 2000; updates 8 January 2001 and 13 July 2001). The accession numbers are AL446043 to AL449029 and AL512551 to AL512553. The entries include annotation of potential protein coding sequences, domains, and signatures. The sequence numbers for the 7.4- and 11.1-kb complete library inserts used to estimate gene density are EMBL/GenBank/DDBJ accession numbers AJ437480 and AF487913, resepectively.
RESULTS
A brief history of the Paramecium genome survey pilot project, along with a preliminary description of the data now available, was published previously (17). We present here the data analysis and biological implications of the project.
General strategy.
Data collection involved distribution of 1,800 library clones to the two sequencing labs which, after DNA preparation and sequencing, yielded 3,568 chromatograms containing 4,183,198 bases of vector-screened raw data. Three nucleotide data sets, as well as a set of ORF fragments identified by similarity search, and the final annotated data set that was submitted to the public nucleotide database were generated as indicated in the flow chart in Fig. 1. The Phrap program was used to assemble the raw vector-screened data, yielding the nonredundant nucleotide data set that was used to search the protein databases by using the BLASTX program. For similarity searches with the BLASTN and TBLASTX programs and all other nucleotide analyses, the sequences in the nonredundant data set were cut at Q20 > 15 (Phrap quality value of 15 over a window of 20 nt; see Material and Methods) to yield the high-quality nonredundant data set. Table 1 recapitulates the database searches that were carried out in the course of the project and the data set used for each of the queries.
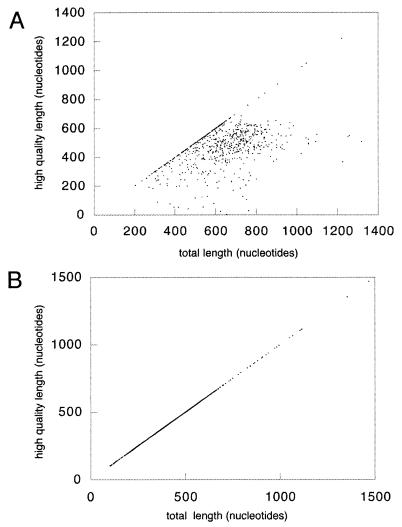
Since our primary objective was gene discovery, we used all of the sequence information available for similarity searches to identify protein coding genes, including low-quality sequence (i.e., Q20 < 15) at the ends of the runs. In this way we took advantage of the fact that genes can usually be identified well before every base in a sequence is known (8), at the risk of assembling chimeric contigs. Although this would not have been a good strategy for a larger project with longer sequence reads, it was highly successful for analysis of our data set of relatively short reads corresponding to only 1 to 2% of the genome. Scatter plots of length versus quality for each of the sequences in the final annotated data set show that, had we cut the sequences before database searching, we would have identified many fewer genes (Fig. 2A). Sequences with no significant BLASTX match were cut at Q20 > 15 (Fig. 2B) for database submission.
FIG. 2.
Quality versus length of GSS sequences. The plots represent high quality (Q20 > 15) length versus total length of the GSS sequences submitted to the public nucleotide database. (A) Sequences containing ORFs according to similarity search, annotated in the database entries using an mRNA feature. (B) Sequences with no mRNA feature were automatically cut at Q20 > 15 for database submission.
Characterization of the data set.
To evaluate the quality of the library used for random sequencing, we cut the vector-screened raw data (Q20 > 15) and then used Phrap to generate contigs. The average length of the reads input to Phrap was 505 nt; the output consisted of 2,924 singletons and 219 contigs. The library, indexed by handpicking colonies for the purpose of complementation cloning, contains duplicate clones within microtiter plates and occasionally in two consecutive microtiter plates (36). Examination of the 219 contigs proposed by Phrap revealed 162 duplicate reads and 45 bona fide two-member contigs. An additional 12 reads were exact duplicates but could represent two-member contigs since they were from nonconsecutive microtiter plates. No contigs contained more than two nonduplicate members. Contig analysis of these data, assuming a 100-Mb macronuclear genome, predicts 43 contigs, in good agreement with the value found of 45 to 57 contigs, suggesting that the library is nearly random. [That is, Ij = Ne−2c(1 − θ) (1 − e−c(1 − θ))j − 1 for j ≥ 1, where Ij is the expected number of j-member contigs or “islands,” N is the number of sequences, θ is the length of overlap required for detection of a match divided by the average sequence length (θ ≈ 50/500), and c is the relative genome coverage.] However, the actual size of the Paramecium somatic genome is not known precisely; if it is 150 Mb, then only 30 contigs are expected and the library used for sequencing may be less than perfectly random, although sufficiently so for the present analysis of a small fraction (1 to 2%) of the genome (39).
To assess the accuracy of the data set, we examined the alignments and chromatograms of the nine sequences identical to previously sequenced Paramecium tetraurelia genes revealed by BLASTN analysis of the high-quality data set, summarized in Table 2. Four sequences are 100% identical, and four sequences are ≥98.7% identical to the corresponding database sequence. The chromatogram of the one sequence that is 98% identical contains a small dye peak; we note that Phred base calls were not corrected manually. The other discrepancies are unambiguous on the chromatograms, and some could represent genetic polymorphisms since different strains were sequenced.
TABLE 2.
Accuracy of the data seta
| GSS ID (query) | nt identity (%)b | GenBank ID (target) | Gene | Stock |
|---|---|---|---|---|
| PT005A23R | 475/475 (100) | PTALPHA51 | Surface antigen | 51 |
| PT007O13R | 472/472 (100) | PTU19464 | Dynein beta heavy chain | 51 |
| PT009I01U | 323/323 (100) | PTE272425 | Eta-tubulin | d4-2 |
| PT011E03U | 621/624 (99) | AF149979 | X gene in rRNA spacer | 51 |
| PT012E03R | 366/367 (99.7) | PTSSRRN | 18S rRNA gene | 51 |
| PT021E13U | 490/500 (98) | PTY17649 | Calcium ATPase | 51 |
| PT012A14U | 485/486 (99.8) | AF149979 | 26S rRNA gene | 51 |
| PT014M07R | 453/453 (100) | PTU47117 | TMP4a secretory protein | d4-2 |
| PT014O15R | 150/152 (98.7) | PTG1IA1 | Surface antigen | 51 |
The database identification (ID) of the nine GSS sequences with BLASTN matches to previously characterized Paramecium genes are given with statistics concerning the matches. Examination of the chromatograms revealed a small dye peak in PT021E13U that accounts for the discrepancy with sequence PTY17649. No other obvious errors in base calling were found and, for at least some of the four other matches presenting with <100% identity, the differences may be due to genetic polymorphism since we have sequenced strain d4-2 derived from stock 51 and stock 29, whereas some Paramecium labs work with stock 51 (54).
That is, the number of matching nucleotides/total number of nucleotides.
Analysis of protein coding sequences: BLASTX analysis.
The principal objective of our project was gene discovery. The limited amount of Paramecium sequence data available at the outset of our project was not sufficient for development of a computational approach to finding genes, which in any event would not have been very useful given the relatively short length of the single-read genome survey sequences. Fortunately, Paramecium introns are uniformly small, facilitating identification of ORFs by sequence similarity with known proteins.
(i) ORF identification.
The nonredundant nucleotide data set of 3,139 sequences was compared to the Swissprot and the Sptrembl databases by using the BLASTX program with a very high threshold E-value of 10−2. Matches were found for 1,230 sequences. The data were then subjected to expert validation. The expert validation phase was necessary in order to retain short, near-perfect matches with high E-values, to eliminate matches with lower E-values owing to secondary structure (i.e., coiled-coil domains or cysteine repeats) in the absence of other indications of homology to the target sequence, and to identify introns. The latter either introduce frameshifts and output with multiple segments or, when in frame, introduce gaps in the alignment. Introns were validated if they had canonical GT…AG junctions and restored an uninterrupted alignment of the query and target sequences. In case of any ambiguity, an improved BLASTX score was also required. This procedure for identifying introns was first tested on high-quality sequences (Q20 > 15) in regions of unambiguous homology (> 40% amino acid identity) and permitted identification of 212 introns. The procedure was then extended to the entire data set and allowed identification of an additional 250 introns. Once the introns were identified, spliced sequences were generated automatically and used for another round of BLASTX similarity search against the same nonredundant protein database. The resulting output was used both for further validation of the ORFs and introns and for the final annotation of the genome survey sequences (GSS division of the EMBL/GenBank/DDBJ public nucleotide database; accession numbers AL446043 to AL449029 and AL512551 to AL512553), which includes an mRNA feature for each ORF identified, with information about the best BLASTX match.
Of the 1,230 initial matches, 722 ORF fragments were validated, and 348 of these partial ORFs contained one or more introns. A table of the 722 ORFs with links to the BLASTX output and the final annotated database entries is available at http://paramecium.cgm.cnrs-gif.fr/funcat_seq.
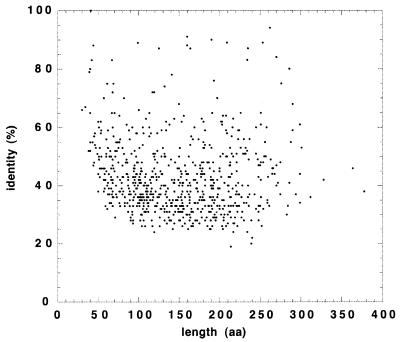
In order to evaluate a posteriori the expert validation, we examined the percent amino acid identity of the best match for each ORF as a function of the length of the match (Fig. 3). The validated BLASTX alignments range from <40 amino acids (aa) to >300 aa (average, 149 ± 63 aa). The fact that the average length is close to the average high-quality nucleotide length of the data set argues for the quality of the sequences (few insertion or deletion errors). The fact that the length and the identity are not correlated except for the very shortest matches (<40 aa) argues for the consistency of the expert validation.
FIG. 3.
Partial ORFs identified by similarity search. Each point represents the percent amino acid identity as a function of the length of the match for the best BLASTX match to the nonredundant protein database (see Table 1), after removal of introns from the coding regions. For the whole set of partial ORFs, the average amino acid identity with the best match is 42.9% ± 12.9%, and the average match length is 149 ± 63 aa.
(ii) Protein domains.
Conserved protein domains in the set of ORF fragments were identified by using the BLASTP program (threshold E-value of 10−4) to search the Pfam protein domain database (55). The output of the search was validated automatically by verifying the extent of overlap between the Pfam domain and the query sequence, with a minimum overlap of 15 aa required. After validation, 498 ORF fragments contained one PfamA domain and 26 contained two domains. The 524 PfamA domains were linked to the InterPro database (4), which facilitated comparison of the frequency of the domains in the Paramecium genome survey ORFs with the frequency of the same domains in complete eukaryotic genomes (table available at http://paramecium.cgm.cnrs-gif.fr/interpro.html). The surprising result of this analysis is that an amazingly high proportion of the Paramecium ORFs and protein domains identified by similarity search are protein kinases (119 eukaryotic protein kinase domains, 22.7% of total identified domains). Protein kinases are usually the most abundant proteins and domains in eukaryotic genomes; however, the percentage of InterPro domains that are eukaryotic protein kinase domains is at most 4.3% (Arabidopsis thaliana), 3% (Saccharomyces cerevisiae), or 2.8% (Caenorhabditis elegans) in the available proteomes (ftp.ebi.ac.uk/pub/databases/SPproteomes).
There are two possible explanations for this seeming overrepresentation of kinase domains in the GSS data set, which are not mutually exclusive. One explanation is some bias in the data or in our method for identifying ORFs. The bias could arise from the BLASTX analysis, either because kinases are very conserved or because many protozoan, ciliate, or Paramecium kinases are available in the protein databases. The other possibility is that Paramecium really does have an enormous number of kinases. We observed a high frequency of G-protein WD-40 repeats, ras family domains, dual-specificity protein phosphatase domains, cyclic nucleotide-binding domains, cyclic nucleotide-gated K+ ion channel trans-membrane domains, and phosphatidylinositol-4-phosphate 5-kinase domains, further suggesting that Paramecium does devote a large part of its coding capacity to signal transduction.
Other well-represented domains are serine carboxypeptidase domains and glycosyl hydrolases family 31 domains. These domains probably belong to lysosomal enzymes involved in digesting bacteria and other microorganisms, the main food source of Paramecium.
(iii) Introns.
Previous studies of Paramecium genes revealed unusually small introns (23, 52). Many of the Paramecium introns in the invertebrate division of GenBank (“INV”) were validated experimentally by comparison of gene and cDNA sequences, and all are 20 to 34 nt in length. These introns contain canonical 5′ and 3′ exon-intron junctions (5′-GT…AG-3′) and are assumed to be recognized by a Paramecium spliceosome. Other organisms, including hypotrich ciliates (35, 40, 59), have some small introns, but they also have larger introns. The only exceptions are the nucleomorph genomes of chromophyte and chlorarachnean algae, organisms which originally gained photosynthetic functions by engulfing eukaryotic red and green algae in a process known as secondary endosymbiosis (18). The nucleomorph is the remnant of the nuclear genome of the algal endosymbiont. Nucleomorph introns are uniformally small: 42 to 52 nt in the chromophyte Guillardia theta (18) and 18 to 20 nt in the chlorarachnion CCMP621 (31).
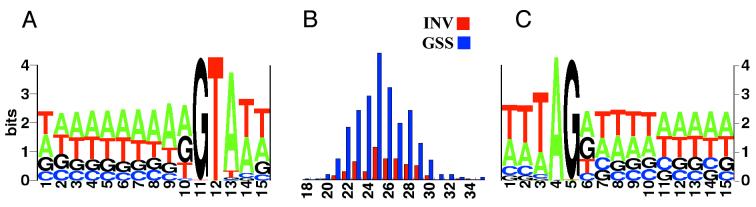
The 462 Paramecium introns identified in the course of our BLASTX analysis were compared to 108 introns extracted from a nonredundant set of Paramecium sequences from the invertebrate division of GenBank. As shown in Fig. 4, the introns identified in our GSS data set are similar to the previously characterized Paramecium introns in both sequence and length. The histogram shows similar size distributions for the two sets of introns (18 to 35 nt and 20 to 34 nt for GSS and INV, respectively). We note that if another class of much larger introns (>100 nt) exists in the Paramecium genome, it would not have been detected by the present study.
FIG. 4.
Characterization of Paramecium introns. The GSS introns were identified by inspection of gaps and phase shifts in the BLASTX alignments. The middle part of the figure (B) shows the histogram of intron length for the 462 GSS introns compared to 108 Paramecium introns from the INV division of GenBank. We note that only two of the GSS introns are shorter than 20 nt and consider that experimental validation will be necessary to assert that Paramecium introns can be as small as 18 nt. On the left (A) is shown the consensus sequence at the 5′ exon-intron junction for the GSS introns, and on the right of the histogram (C) is shown the consensus sequence at the 3′ intron-exon junction for the GSS introns. Consensus sequences were calculated at http://www.bio.cam.ac.uk/seqlogo/logo.cgi.
Comparison of the sequences from −10 to +5 of the 5′ junction and from −5 to +10 of the 3′ junction for the whole set of GSS introns is also shown in Fig. 4. The consensus sequence Purine|GTA…AG|Purine is found, although only the GT…AG at the intron boundaries are absolutely required. The preference for a purine (G or A) just before and just after the intron was observed by Russell et al. (52), who characterized a set of 50 introns from phosphatase, kinase, and small GTP-binding protein genes; this feature is typical of introns recognized by the spliceosome in other species. The surrounding exon sequences are roughly 65% AT; however, the intron sequences (with the exception of the G at each junction), are even more AT rich (83% AT for the GSS set, excluding the conserved GT and AG at the boundaries) and contain very few G residues.
(iv) Codon usage.
The GSS coding sequences corresponding to each ORF fragment identified by the BLASTX analysis were cut at Q20 > 15 for calculation of codon usage. For comparison with the codon usage of previously characterized Paramecium genes, a set of 114 nonredundant sequences were extracted from the invertebrate (INV) division of GenBank. The codon usage is not very different between the two data sets. Usage appears slightly less biased for GSS (71,818 codons) than for INV (33,183 codons); in particular, codons with G in the third position are slightly more frequent in the GSS data set, perhaps because the random genome survey sequences include more poorly expressed genes than the INV data set. Furthermore, there are many surface antigen genes in the INV set, and it is known that surface antigen codon usage is very biased. Tables of codon usage and amino acid composition for both data sets are available at http://paramecium.cgm.cnrs-gif.fr/codon_usage.html.
Comparison of the ORF set with proteomes.
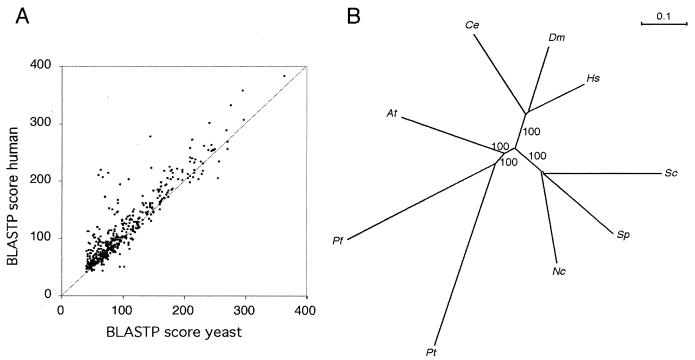
We compared the 722 ORF fragments identified by the BLASTX analysis with the available, annotated proteomes of Escherichia coli, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, Arabidopsis thaliana, and the partial proteome of Homo sapiens by using the BLASTP program (see Table 1). Using a threshold E-value of 10−4, we found that 447 of the Paramecium ORF fragments have homologues in each of the eukaryotic genomes and that, on average, the scores against human proteins are the highest and against yeast proteins are the lowest, suggesting that the Paramecium proteins are most similar to human proteins and least similar to yeast proteins (Fig. 5A). However, this does not necessarily reflect the phylogenetic relationships among these species. In order to determine the phylogenetic position of Paramecium, it is necessary to establish a distance matrix between homologous proteins in the genomes under comparison.
FIG. 5.
Comparative analysis. (A) BLASTP scores for the 447 conserved partial ORFs are shown as a scatter plot, with the best score against the yeast proteome on the abcissa and the best score against the human proteome on the ordinate axis. (B) Molecular phylogeny by using combined protein data. Unrooted distance matrix neighbor-joining tree obtained for nine species with a set of 38 partial proteins comprising 4,037 informative amino acids. The bootstrap values for 100 replicates were 100 at all nodes except for the two very closely spaced nodes connecting the three fungal species. At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Hs, Homo sapiens; Nc, Neurospora crassa; Pf, Plasmodium falciparum; Pt, Paramecium tetraurelia; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe.
Phylogenetic analysis with random, combined protein data.
We used the results of the comparison to proteomes to guide us in extracting a set of combined partial protein sequence data (9) to construct a phylogenetic tree by the neighbor distance method (53) by using the PHYLIP package (26). In order to obtain a set of homologous sequences with maximal probability of being orthologous, we compared the 447 Paramecium ORF fragments with each other and excluded all sequences belonging to multigene families. We also removed mitochondrial proteins to try to avoid genes acquired by horizontal transfer that do not reflect the evolutionary history of the species. We were left with 41 partial ORFs, which were compared to human, plant, fly, worm, budding yeast, and fission yeast proteomes (extracted from the Swissprot and Sptrembl protein databases [September 2001]) by using the BLASTP program and to the complete genome of Neurospora crassa (http://www-genome.wi.mit.edu/annotation/fungi/neurospora/) and finished sequences and updated contigs of Plasmodium falciparum (http://plasmodb.org) by using the TBLASTN program. For each sequence, the best match to each species was retained (threshold E-value of 10−20), and the eight “orthologues” were aligned with the corresponding Paramecium ORF fragment by using the CLUSTALW program. If there was no match satisfying the threshold criterion for one or more of the target species, the protein was not included in the data set used for tree construction. After visual inspection, trimming of the alignments, and concatenation, 4,037 informative amino acids from 38 proteins were used to construct a tree by neighbor-joining (see Materials and Methods). This unrooted tree, shown in Fig. 5B, is supported by highly significant bootstrap values and shares the topology found with small subunit rRNA or combined protein data (5, 58). The metazoans and fungi are monophyletic and are grouped together (opisthokontes). Paramecium and Plasmodium are also grouped together as expected for two alveolates.
The topology of the tree is very robust. However, the lengths of the branches of Paramecium and of Plasmodium are longer than the lengths of all other branches of the tree. Two nonexclusive explanations of the branch lengths seem possible. First, the proteomes of both Plasmodium and Paramecium may have evolved very rapidly. Second, these long branches may be a methodological artifact owing to the asymmetric role of the Paramecium partial ORF fragments, which were used to interrogate the database. This may have led to the use of paralogues rather than true orthologues. In order to try to discriminate between these hypotheses, the same method was applied to a subset of the GSS ORFs involved in translation, for which orthology is not a problem. A less-resolved but congruent tree was obtained, and the same trend was observed: both Paramecium and Plasmodium have long branches. This suggests that these alveolate proteomes have indeed evolved rapidly.
Our results, in particular the robust topology of the unrooted tree, favor the usefulness of unbiased, randomly generated sets of combined protein data for phylogenetic analyses as proposed by Brown et al. (9) and discussed by Olsen (46). Most important in the context of the present study, the approach provides strong support for the consistency of the Paramecium GSS data.
Functional categories.
The comparison of the Paramecium partial ORFs with the complete proteomes was also useful in establishing a tentative classification into functional categories. We used the MIPS yeast functional catalogue scheme as a reference (http://mips.gsf.de/proj/yeast/catalogues/funcat/), and for all ORFs with an unambiguous yeast homologue supported by the matches with the other proteomes the annotation was generated automatically. For ORFs with no clear yeast homologue, we consulted the available databases concerning the best matches to other proteomes or the best BLASTX matches for the sequence. It was relatively straightforward to assign categories to metabolic enzymes, transporters, ion channels, or cytoskeletal and motor proteins such as actin, tubulins, or kinesins. However, for most of the numerous kinases, we considered the function unknown unless there was substantial evidence that the Paramecium ORF fragment was the homologue of a particular, well-characterized kinase, for example, MEKK. There were also many clear homologues of genes of unknown function. Nonetheless, our tentative catalogue is probably the most immediately useful result of the project for Paramecium biology, and several genes identified by the genome survey have been or are currently under investigation, for example, delta-tubulin (30) and the membrane fusion protein NSF (28). The functional catalogue is available at http://paramecium.cgm.cnrs-gif.fr/funcat_des.
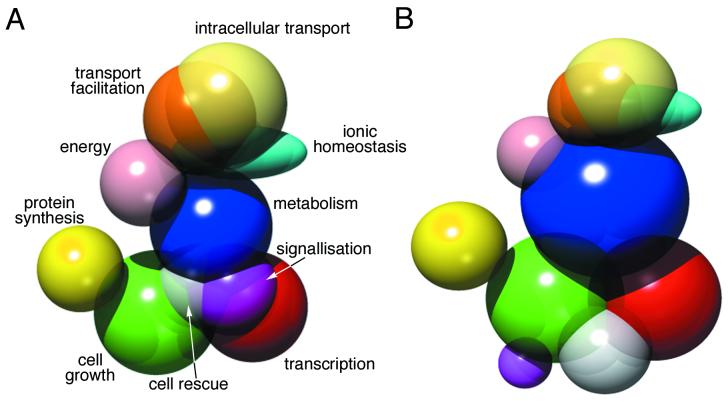
We attempted to represent the topology of the classification for the Paramecium ORFs compared to the entire yeast functional catalogue (Fig. 6). Since many genes belong to more than one functional category, we considered that a classic pie chart could not accurately represent the data. Instead, we have drawn intersecting solids representing the major categories and the major intersections between categories. The volumes of the solids are calculated to be directly related to the number of ORFs in the category; however, the arrangement of the solids is just one possible way to interpret the major relationships.
FIG. 6.
Functional categories. Three-dimensional drawing of the major functional categories, based on the yeast classification scheme (http://mips.gsf.de/proj/yeast/catalogues/funcat/), and of the major overlaps between categories. (A) Paramecium GSS partial ORFs. (B) Yeast genes. The drawing was made by using the POV-ray program, available at http://www.povray.org. Categories for which research is active in Paramecium (cellular organization, signaling, ion channels, cell growth and division) are probably overrepresented because the availability of Paramecium sequences in the database facilitated identification of the ORFs, while energy and protein synthesis are also well represented, probably because of high conservation of many mitochondrial and ribosomal proteins. Protein fate, which overlaps significantly with almost every other category, is not represented.
Identification of protein-coding genes with no database match.
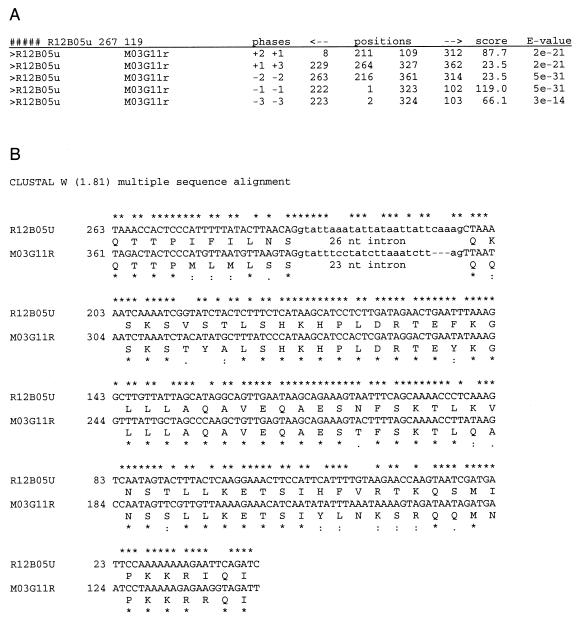
It is possible to detect protein-coding genes, even in the absence of a database match. If genes belong to a multigene family and at least two paralogues are present in the data set, they can be detected by using the TBLASTX program to compare the sequences to each other at the protein level in the 36 possible combinations of reading frames. We used the high-quality nonredundant nucleotide data set (cf. Fig. 1) in conjunction with the TBLASTX program to search for multigene families. Numerous significant alignments were obtained (E-value threshold of 10−12), indicating the occurrence of >100 families of protein coding genes within the data set. Each group of related sequences gave alignments in several different reading frames, with different BLAST scores. In all cases, one alignment had a higher score than the others. If the pressure for sequence conservation is at the protein level, then the true coding frame should be the one that gives the highest score. This was indeed the case for all of the families for which the coding frame was known from the BLASTX analysis. We therefore assumed that all of the novel genes identified also encode the protein sequences that give the best alignments. Figure 7 shows a family of two genes with no database homologues; moreover, in this case an intron interrupts the protein alignments.
FIG. 7.
Novel genes identified by TBLASTX internal comparison. An example of TBLASTX data (A) and of nucleotide and translated amino acid alignment (B) of a two-member family of novel genes is shown. The TBLASTX table shows that the best E-value was obtained for two segments in reading frames −1,−1 (score, 119) and −2,−2 (score, 23.5). Nucleotide alignment reveals the presence of an intron between the two coding segments. Neither of these putative genes has any homologue in the protein databases.
Altogether, 108 families, containing 252 ORF fragments, were identified: 95 families with two members, 7 families with three members, and 6 families with more than three members. A total of 46 families are composed of genes with homologues in the databases, 55 are families of novel genes, and 7 families have one known and one unknown member (the BLASTX match had not been validated for one of the sequences). In summary, we found 133 genes that had already been identified by similarity search and 119 novel genes. Among the 722 ORF fragments validated by the BLASTX analysis, 589 have no paralogues in the data set (“unique” genes). If we extrapolate from the proportion of genes with no database homologue in the families identified by internal comparison, we can predict that 527 more unique genes should be present in the genome survey data set, giving the estimation of 1,368 genes in all.
Gene density. (i) Complete inserts.
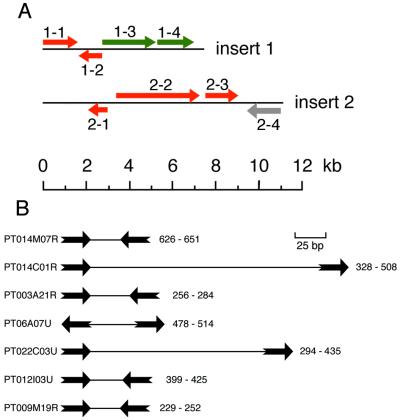
In order to independently evaluate the coding proportion of the somatic genome, we analyzed the sequences of two library inserts of 7.4 and 11.1 kb, respectively. As shown in Fig. 8A, the first insert, which was sequenced because it complements the nd2 mutant defective for secretory granule exocytosis, contains four identified genes. The first two ORFs, an aminopeptidase and a pseudouridine synthase, were validated by a BLASTX similarity search of the protein databases. The third ORF complements the nd2 mutation, and the fourth ORF represents a novel gene. The third and fourth ORFs were validated experimentally by homology-dependent gene silencing, a phenomenon related to RNAi which can be used to obtain the equivalent of a somatic knockout for a given gene (51). Secretory granule release is compromised by silencing the third ORF as expected, and the fourth ORF encodes an essential gene (M. Froissard, unpublished data). Thus, 87% of this insert corresponds to gene coding sequences (ORFs plus introns) and only 13% corresponds to intergenic sequences. The second insert was chosen randomly among large library inserts and sequenced for the purpose of estimating gene density. This insert harbors three protein-coding genes according to similarity searches and a possible fourth ORF with no database homologue, which we did not validate. Thus, 53.3% of this insert corresponds to validated gene coding sequences. We analyzed an additional 37 kb of unfinished sequences distributed among seven other library inserts (C. Klotz, F. Koll, A Krzywicka-Racka, and F. Ruiz, unpublished data; M. Froissard, unpublished data). ORFs were identified by similarity search and by inspection of the sequences. Potential ORFs with no database homologue were not validated. We found that for the ensemble of the nine contigs, corresponding to 55.5 kb that 68% of the DNA consists of validated coding sequences. This represents a minimal estimate of gene density in this sampling of library inserts, since there appear to be several novel ORFs that will require experimental validation. Although caution is required in extrapolating to the entire somatic genome, this estimation of gene density is consistent with the large number of ORF fragments found in the GSS data set.
FIG. 8.
Gene density. (A) ORF distribution of two complete library inserts. Colors: red, ORFs validated by significant BLASTX matches; green, ORFs validated by homology-dependent gene silencing; gray, unvalidated potential ORF. 1-1, aminopeptidase (best score, 140); 1-2, pseudouridine synthase (best score, 112); 1-3, ND2 gene required for regulated exocytosis; 1-4, novel essential gene; 2-1, translation initiation factor EIF-2B subunit (best score, 102); 2-2, DNA repair protein RAD5 homolog (best score, 124) with a SNF2 family helicase domain; 2-3, slime mold interaptin homolog (score, 73); 2-4 potential ORF. (B) Intergenic regions separating ORFs for the seven GSS sequences which contain two putative ORFs, as revealed by BLASTX similarity searches. The sequence identification is given to the left of the map, and the first and last nucleotides of the intergenic region are shown to the right of the map. ORFs are pictured as thick arrows, and the intergenic regions are pictured as thin lines. The ORFs are as follows: PT014M07R, TMP4a secretory granule protein and PRP8 splicing factor; PT014C01R, homolog of unknown Drosophila gene and RAC protein kinase; PT003A21R, unknown kinase and MEK kinase; PT006A07U, hypothetical protein and PRP19 splicing factor; PT022C03U, ribosomal protein L33 and YPT1-like GTP-binding protein; PT012I03U, ribosomal protein L36 and CTP phosphocholine cytidylyltransferase; and PT009 M19R, small GTP-binding protein and ring zinc-finger protein.
(ii) Intergenic regions.
We examined the intergenic regions of the seven genome survey sequences which contain two ORF fragments according to the BLASTX analysis (Fig. 8B). In five of the examples, we found fewer than 40 nt separating divergent or convergent genes, suggesting the existence of bidirectional and/or overlapping promoters and terminators and indicating that regulatory elements can be extremely small in Paramecium. In one case, the genes sequenced in the course of the genome survey project were already under study (PT014 M07R, containing the 3′ regions of the TMP-4a secretory protein gene and of the PRP8 splicing-factor gene). Experimental analysis of the transcripts by 3′ race experiments does support very short, overlapping 3′ terminator regions (D. J. Kobric and R. E. Pearlman, unpublished data). For the moment, we do not know whether these diminutive intergenic regions are the general case in Paramecium, or if we have selected unusually closely spaced genes by examining short DNA segments containing two validated ORFs. The average distance between genes in yeast is ca. 200 nt for convergent genes and ca. 400 nt for divergent genes (32). Among eukaryotes, only nucleomorph genomes (18) contain genes as closely spaced as the examples in Fig. 8B.
DISCUSSION
We have surveyed 1 to 2% of the somatic genome of the ciliate P. tetraurelia by random end sequencing of a plasmid library. The 2,990 sequences deposited in the GSS division of the public nucleotide database include 1 tRNA gene identified by tRNA-SCAN, 3 partial rRNA genes identical to previously established rRNA sequences, and 722 partial protein coding genes identified by similarity searches, only 6 of which were previously known. An additional 119 novel protein-coding genes were tentatively identified by internal comparison of the data set. High gene density, suggested by this rich harvest of ORF fragments, was further supported by examination of complete library inserts, giving a minimal estimate that 68% of somatic DNA is coding.
Gene discovery.
Random sequencing of genomic DNA has been used to generate sequence tags for genetic mapping as a means of rapid genome characterization prior to systematic sequencing (1, 11, 15, 16) and, in the case of hemiascomycete yeasts, as the basis for an in-depth study of genome evolution among closely related species by using the complete Saccharomyces cerevisiae genome as a reference (reference 56 and references therein). However, gene discovery has been generally achieved in eukaryotes by sequencing appropriately normalized cDNA libraries. This is because the genomes of most of the model organisms that have been studied, like the human genome, contain far more noncoding than coding DNA; moreover, the two types of sequences are interspersed since most genes contain introns, and the introns are at least as large as the coding regions they interrupt. The computational problem of identifying genes remains largely unresolved for such genomes, and EST data are crucial for their annotation and exploitation.
Unicells such as yeast have more compact genomes and few introns; annotation of the yeast genome did not require cDNA sequencing. We note, however, that cDNA sequencing is under way for the Tetrahymena genome (27). Although it is not yet possible to estimate gene density in this ciliate, Tetrahymena protein-coding genes contain much larger introns than Paramecium genes (range, 51 to 979 nt; average, 213 nt; for the set of 59 Tetrahymena thermophila introns available in GenBank [February 2002]). The rationale behind our pilot project was that, although many Paramecium genes do contain introns, their uniformly small size, confirmed by the project, would make genomic sequencing nearly as efficient for gene discovery as cDNA sequencing, without the problems of library normalization and/or redundant sequencing of highly expressed genes. The drawback of the approach is that only genes conserved among other species are readily identified by similarity searches. As demonstrated by the Génolevure's hemiascomycete project (41), ca. 40% of most organism's genes are species or phylum-specific “maverick” genes. This figure is consistent with our own data: 47% of the protein-coding gene fragments in the families identified by TBLASTX internal comparison had no database homolog.
Since all of the previously characterized Paramecium protein-coding genes fall into ca. 30 different families (e.g., surface antigens, cyclins, K+ ion channels, heat shock proteins, guanylyl cyclases, centrins, tubulins) or unique genes (e.g., calmodulin, ND7), the project has significantly multiplied the known Paramecium genes. This is particularly striking for genes involved in metabolism and energy production. Even for functions that have been well studied in Paramecium, many useful genes have been identified. For example, the project contains a number of genes involved in membrane trafficking (clathrin heavy chain, adaptor complex subunits, SEC61 ER translocator, coatamer alpha subunit, NSF, and possibly synaptobrevins), providing a direct argument that conserved eukaryotic trafficking mechanisms are used by Paramecium. The project has allowed identification of a number of helicases potentially involved in DNA recombination and repair; such genes could play key roles in the programmed DNA rearrangements involved in the development of the somatic nucleus. The project also contains genes that could open new avenues of investigation; for example, numerous histidine kinase homologues suggest the existence of two-component phosphorelay signal transduction pathways in Paramecium (57). In the case of gene families already under study (e.g., surface antigens, tubulins, trichocyst matrix proteins, small GTP-binding proteins, and K+ ion channels), the project has revealed new members, indicating that these gene families are larger than previously imagined.
Toward a genome project.
The long-term goal of our random survey is to promote a Paramecium genome project. Ciliates are an important eukaryotic group, and even ciliate species considered closely related are separated by vast evolutionary distances (100 MY separate the two oligohymenophoron models Paramecium and Tetrahymena) and profound biological differences. Of particular significance in elaborating a genome project, the precise relationship between germ line and somatic DNA is not the same in different ciliates (50).
During somatic development, Tetrahymena chromosomes are fragmented at a precise 15-bp sequence known as cbs (“chromosome breakage sequence”) (61) so that somatic DNA is a unique set of molecules that has been compared to a restriction digest (47). Paramecium does not have a unique somatic genome. During somatic development, chromosomes are fragmented not at precise sites but in regions leading to chromosome polymorphism even within a single cell (12). Internal eliminated sequences, unique germ line sequence elements that are removed by a precise DNA splicing mechanism (7), can have alternative junctions (20, 34). Finally, homology-dependent epigenetic controls allow the old somatic nucleus, which is discarded at each sexual generation, to influence the pattern of somatic genome rearrangements during development (21, 22, 42, 43), so that somatic lines successful under given environmental conditions can be maintained across sexual generations in the absence of any modification of the germ line genome.
Sequencing the macronuclear genome, as shown here, provides a direct approach to finding genes. However, the ultimate goal of a genome project is to establish the sequence of the germ line, micronuclear genome. Although technically more challenging because the 10 to 15% of the genome eliminated during macronuclear development is rich in repeated sequences and transposable elements (45) which are probably heterochromatic, it is the micronuclear genome that can tell us the most about the development and the evolution of the organism. Knowledge of the germ line is essential to full understanding of the developmental DNA rearrangements. Most intriguing, the germ line is marked by the evolutionary history of the organism, and may reveal how transposable elements invaded ciliates, were tamed and then exploited (37, 45) to give this unicell unique developmental and adaptive potentials. We believe that in Paramecium, sequencing somatic DNA will provide information about the expressed part of the genome essential for annotation and exploitation, thus fulfilling much the same role as cDNA sequencing in genome projects of higher eukaryotes.
Acknowledgments
We are indebted to all members of the Paramecium Genomics Consortium, who made the pilot project possible. We thank Janine Beisson, Mireille Bétermier, and Eric Meyer for critical reading of the manuscript and Catherine Klotz, France Koll, Anna Krzywicka-Racka, and Françoise Ruiz for allowing us to use their unpublished data. We are grateful to André Adoutte and Nicolas Lartillot for many useful discussions about interpreting molecular phylogenies.
Footnotes
Dedicated to the memory of André Adoutte.
REFERENCES
- 1.Aguero, F., R. E. Verdun, A. C. Frasch, and D. O. Sanchez. 2000. A random sequencing approach for the analysis of the Trypanosoma cruzi genome: general structure, large gene and repetitive DNA families, and gene discovery. Genome Res. 10:1996-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apweiler, R., T. K. Attwood, A. Bairoch, A. Bateman, E. Birney, M. Biswas, P. Bucher, L. Cerutti, F. Corpet, M. D. Croning, R. Durbin, L. Falquet, W. Fleischmann, J. Gouzy, H. Hermjakob, N. Hulo, I. Jonassen, D. Kahn, A. Kanapin, Y. Karavidopoulou, R. Lopez, B. Marx, N. J. Mulder, T. M. Oinn, M. Pagni, and F. Servant. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29:37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldauf, S. L., A. J. Roger, I. Wenk-Siefert, and W. F. Doolittle. 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290:972-977. [DOI] [PubMed] [Google Scholar]
- 6.Beisson, J., and T. M. Sonneborn. 1965. Cytoplasmic inheritance of the organization of the cell cortex in Paramecium aurelia. Proc. Natl. Acad. Sci. USA 53:275-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betermier, M., S. Duharcourt, H. Seitz, and E. Meyer. 2000. Timing of developmentally programmed excision and circularization of Paramecium internal eliminated sequences. Mol. Cell. Biol. 20:1553-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouck, J., W. Miller, J. H. Gorrell, D. Muzny, and R. A. Gibbs. 1998. Analysis of the quality and utility of random shotgun sequencing at low redundancies. Genome Res. 8:1074-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, J. R., C. J. Douady, M. J. Italia, W. E. Marshall, and M. J. Stanhope. 2001. Universal trees based on large combined protein sequence data sets. Nat. Genet. 28:281-285. [DOI] [PubMed] [Google Scholar]
- 10.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 11.Cameron, R. A., G. Mahairas, J. P. Rast, P. Martinez, T. R. Biondi, S. Swartzell, J. C. Wallace, A. J. Poustka, B. T. Livingston, G. A. Wray, C. A. Ettensohn, H. Lehrach, R. J. Britten, E. H. Davidson, and L. Hood. 2000. A sea urchin genome project: sequence scan, virtual map, and additional resources. Proc. Natl. Acad. Sci. USA 97:9514-9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caron, F. 1992. A high degree of macronuclear chromosome polymorphism is generated by variable DNA rearrangements in Paramecium primaurelia during macronuclear differentiation. J. Mol. Biol. 225:661-678. [DOI] [PubMed] [Google Scholar]
- 13.Caron, F., and E. Meyer. 1985. Does Paramecium primaurelia use a different genetic code in its macronucleus? Nature 314:185-188. [DOI] [PubMed] [Google Scholar]
- 14.Chalker, D. L., and M. C. Yao. 1996. Non-Mendelian, heritable blocks to DNA rearrangement are induced by loading the somatic nucleus of Tetrahymena thermophila with germ line-limited DNA. Mol. Cell. Biol. 16:3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crollius, H. R., O. Jaillon, C. Dasilva, C. Ozouf-Costaz, C. Fizames, C. Fischer, L. Bouneau, A. Billault, F. Quetier, W. Saurin, A. Bernot, and J. Weissenbach. 2000. Characterization and repeat analysis of the compact genome of the freshwater pufferfish Tetraodon nigroviridis. Genome Res. 10:939-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Degrave, W. M., S. Melville, A. Ivens, and M. Aslett. 2001. Parasite genome initiatives. Int. J. Parasitol. 31:531-535. [DOI] [PubMed] [Google Scholar]
- 17.Dessen, P., M. Zagulski, R. Gromadka, H. Plattner, R. Kissmehl, E. Meyer, M. Betermier, J. E. Schultz, J. U. Linder, R. E. Pearlman, C. Kung, J. Forney, B. H. Satir, J. L. Van Houten, A. Keller, M. Froissard, L. Sperling, and J. Cohen. 2001. Paramecium genome survey: a pilot project. Trends Genet. 17:306-308. [DOI] [PubMed] [Google Scholar]
- 18.Douglas, S., S. Zauner, M. Fraunholz, M. Beaton, S. Penny, L. T. Deng, X. Wu, M. Reith, T. Cavalier-Smith, and U. G. Maier. 2001. The highly reduced genome of an enslaved algal nucleus. Nature 410:1091-1096. [DOI] [PubMed] [Google Scholar]
- 19.Dubrana, K., and L. Amar. 2000. Programmed DNA under-amplification in Paramecium primaurelia. Chromosoma 109:460-466. [DOI] [PubMed] [Google Scholar]
- 20.Dubrana, K., A. Le Mouel, and L. Amar. 1997. Deletion endpoint allele-specificity in the developmentally regulated elimination of an internal sequence (IES) in Paramecium. Nucleic Acids Res. 25:2448-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duharcourt, S., A. Butler, and E. Meyer. 1995. Epigenetic self-regulation of developmental excision of an internal eliminated sequence on Paramecium tetraurelia. Genes Dev. 9:2065-2077. [DOI] [PubMed] [Google Scholar]
- 22.Duharcourt, S., A. M. Keller, and E. Meyer. 1998. Homology-dependent maternal inhibition of developmental excision of internal eliminated sequences in Paramecium tetraurelia. Mol. Cell. Biol. 18:7075-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dupuis, P. 1992. The beta-tubulin genes of Paramecium are interrupted by two 27-bp introns. EMBO J. 11:3713-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 25.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 26.Felsensetin, J. 1989. PHYLIP: phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 27.Fillingham, J. S., N. D. Chilcoat, A. P. Turkewitz, E. Orias, M. Reith, and R. E. Pearlman. Analysis of expressed sequence tags (ESTs) in the ciliated protozoan Tetrahymena thermophila. J. Eukarot. Microbiol., in press. [DOI] [PubMed]
- 28.Froissard, M., R. Kissmehl, J. C. Dedieu, T. Gulik-Krzywicki, H. Plattner, and J. Cohen. NSF is required to organize functional exocytotic microdomains in Paramecium. Genetics, in press. [DOI] [PMC free article] [PubMed]
- 29.Galvani, A., and L. Sperling. 2002. RNA interference by feeding in Paramecium. Trends Genet. 18:11-12. [DOI] [PubMed] [Google Scholar]
- 30.Garreau De Loubresse, N., F. Ruiz, J. Beisson, and C. Klotz. 2001. Role of delta-tubulin and the C-tubule in assembly of Paramecium basal bodies. BMC Cell Biol. 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilson, P. R., and G. I. McFadden. 1996. The miniaturized nuclear genome of eukaryotic endosymbiont contains genes that overlap, genes that are cotranscribed, and the smallest known spliceosomal introns. Proc. Natl. Acad. Sci. USA 93:7737-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goffeau, E. A. 1997. The yeast genome directory. Nature 387:1-105. [PubMed] [Google Scholar]
- 33.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405-413. [DOI] [PubMed] [Google Scholar]
- 34.Haynes, W. J., K. Y. Ling, R. R. Preston, Y. Saimi, and C. Kung. 2000. The cloning and molecular analysis of pawn-B in Paramecium tetraurelia. Genetics 155:1105-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaufmann, J., V. Florian, and A. Klein. 1992. TGA cysteine codons and intron sequences in conserved and nonconserved positions are found in macronuclear RNA polymerase genes of Euplotes octocarinatus. Nucleic Acids Res. 20:5985-5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keller, A. M., and J. Cohen. 2000. An indexed genomic library for Paramecium complementation cloning. J. Eukaryot. Microbiol. 47:1-6. [DOI] [PubMed] [Google Scholar]
- 37.Klobutcher, L. A., and G. Herrick. 1997. Developmental genome reorganization in ciliated protozoa: the transposon link. Prog. Nucleic Acid Res. Mol. Biol. 56:1-62. [DOI] [PubMed] [Google Scholar]
- 38.Kruger, K., P. J. Grabowski, A. J. Zaug, J. Sands, D. E. Gottschling, and T. R. Cech. 1982. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31:147-157. [DOI] [PubMed] [Google Scholar]
- 39.Lander, E. S., and M. S. Waterman. 1988. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics 2:231-239. [DOI] [PubMed] [Google Scholar]
- 40.Liang, A., and K. Heckmann. 1993. The macronuclear gamma-tubulin-encoding gene of Euplotes octocarinatus contains two introns and an in-frame TGA. Gene 136:319-322. [DOI] [PubMed] [Google Scholar]
- 41.Malpertuy, A., F. Tekaia, S. Casaregola, M. Aigle, F. Artiguenave, G. Blandin, M. Bolotin-Fukuhara, E. Bon, P. Brottier, J. de Montigny, P. Durrens, C. Gaillardin, A. Lepingle, B. Llorente, C. Neuveglise, O. Ozier-Kalogeropoulos, S. Potier, W. Saurin, C. Toffano-Nioche, M. Wesolowski-Louvel, P. Wincker, J. Weissenbach, J. Souciet, and B. Dujon. 2000. Genomic exploration of the hemiascomycetous yeasts: 19. Ascomycetes-specific genes. FEBS Lett. 487:113-121. [DOI] [PubMed] [Google Scholar]
- 42.Meyer, E. 1992. Induction of specific macronuclear developmental mutations by microinjection of a cloned telomeric gene in Paramecium primaurelia. Genes Dev. 6:211-222. [DOI] [PubMed] [Google Scholar]
- 43.Meyer, E., A. Butler, K. Dubrana, S. Duharcourt, and F. Caron. 1997. Sequence-specific epigenetic effects of the maternal somatic genome on developmental rearrangements of the zygotic genome in Paramecium primaurelia. Mol. Cell. Biol. 17:3589-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyer, E., and S. Duharcourt. 1996. Epigenetic regulation of programmed genomic rearrangements in Paramecium aurelia. J. Eukaryot. Microbiol. 43:453-461. [DOI] [PubMed] [Google Scholar]
- 45.Meyer, E., and O. Garnier. 2002. Non-mendelian inheritance and homology-dependent effects in ciliates. Adv. Genet. 46:305-338. [DOI] [PubMed] [Google Scholar]
- 46.Olsen, G. J. 2001. The history of life. Nat. Genet. 28:197-198. [DOI] [PubMed] [Google Scholar]
- 47.Orias, E. 1998. Mapping the germ-line and somatic genomes of a ciliated protozoan, Tetrahymena thermophila. Genome Res. 8:91-99. [DOI] [PubMed] [Google Scholar]
- 48.Perriere, G., and M. Gouy. 1996. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 49.Preer, J. R., Jr., L. B. Preer, B. M. Rudman, and A. J. Barnett. 1985. Deviation from the universal code shown by the gene for surface protein 51A in Paramecium. Nature 314:188-190. [DOI] [PubMed] [Google Scholar]
- 50.Prescott, D. M. 1994. The DNA of ciliated protozoa. Microbiol. Rev. 58:233-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruiz, F., L. Vayssié, C. Klotz, L. Sperling, and L. Madeddu. 1998. Homology-dependent gene silencing in Paramecium. Mol. Biol. Cell 9:931-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russell, C. B., D. Fraga, and R. D. Hinrichsen. 1994. Extremely short 20-33 nucleotide introns are the standard length in Paramecium tetraurelia. Nucleic Acids Res. 22:1221-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic tree. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 54.Sonneborn, T. M. 1974. Paramecium aurelia, p. 469-594. In R. King (ed.), Handbook of genetics. Plenum Publishing Corp., New York, N.Y.
- 55.Sonnhammer, E. L., S. R. Eddy, E. Birney, A. Bateman, and R. Durbin. 1998. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 26:320-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Souciet, J., M. Aigle, F. Artiguenave, G. Blandin, M. Bolotin-Fukuhara, E. Bon, P. Brottier, S. Casaregola, J. de Montigny, B. Dujon, P. Durrens, C. Gaillardin, A. Lepingle, B. Llorente, A. Malpertuy, C. Neuveglise, O. Ozier-Kalogeropoulos, S. Potier, W. Saurin, F. Tekaia, C. Toffano-Nioche, M. Wesolowski-Louvel, P. Wincker, and J. Weissenbach. 2000. Genomic exploration of the hemiascomycetous yeasts. 1. A set of yeast species for molecular evolution studies. FEBS Lett. 487:3-12. [DOI] [PubMed] [Google Scholar]
- 57.Thomason, P., and R. Kay. 2000. Eukaryotic signal transduction via histidine-aspartate phosphorelay. J. Cell Sci. 113:3141-3150. [DOI] [PubMed] [Google Scholar]
- 58.Wainright, P. O., G. Hinkle, M. L. Sogin, and S. K. Stickel. 1993. Monophyletic origins of the metazoa: an evolutionary link with fungi. Science 260:340-342. [DOI] [PubMed] [Google Scholar]
- 59.Wang, W., R. Skopp, M. Scofield, and C. Price. 1992. Euplotes crassus has genes encoding telomere-binding proteins and telomere-binding protein homologs. Nucleic Acids Res. 20:6621-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao, M. C., S. Duharcourt, and D. L. Chalker. 2002. Genome-wide rearrangements of DNA in ciliates, p. 730-758. In R. C. N. Craig, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 61.Yao, M. C., C. H. Yao, and B. Monks. 1990. The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell 63:763-772. [DOI] [PubMed] [Google Scholar]