Abstract
The Cdc42p GTPase controls polarized growth and cell cycle progression in eukaryotes from yeasts to mammals, and its precise subcellular localization is essential for its function. To examine the cell cycle-specific targeting of Cdc42p in living yeast cells, a green fluorescent protein (GFP)-Cdc42 fusion protein was used. In contrast to previous immunolocalization data, GFP-Cdc42p was found at the plasma membrane around the entire cell periphery and at internal vacuolar and nuclear membranes throughout the cell cycle, and it accumulated or clustered at polarized growth sites, including incipient bud sites and mother-bud neck regions. These studies also showed that C-terminal CAAX and polylysine domains were sufficient for membrane localization but not for clustering. Time-lapse fluorescence microscopy showed that GFP-Cdc42p clustered at the incipient bud site prior to bud emergence and at the mother-bud neck region postanaphase as a diffuse, single band and persisted as two distinct bands on mother and daughter cells following cytokinesis and cell separation. Initial clustering occurred immediately prior to actomyosin ring contraction and persisted postcontraction. These results suggest that Cdc42p targeting occurs through a novel mechanism of membrane localization followed by cell cycle-specific clustering at polarized growth sites.
Protein localization to sites of polarized growth is an integral part of coordinating the temporal and spatial regulation of cell growth and cytokinesis during the cell cycle. The highly conserved Rho family of GTPases, including Cdc42p, are essential for regulating multiple aspects of cell polarity in eukaryotic cells. In mammalian cells, Cdc42p has been implicated in actin rearrangements, asymmetric cell division, polarized growth, vesicular transport, endocytosis, and cytokinesis (10, 13, 17, 39). In Saccharomyces cerevisiae, Cdc42p is essential for proper selection of polar bud sites, rearrangement of the actin cytoskeleton during bud emergence, and directing actin-dependent secretion into an enlarging bud (14).
Proper targeting of Cdc42p is a vital component of these processes and is essential for cell viability. In previous immunolocalization experiments, endogenous S. cerevisiae Cdc42p was observed at incipient bud sites and the tips and sides of enlarging buds (43), and at the mother-bud neck region in cells overexpressing green fluorescent protein (GFP)-tagged Cdc42p (33). Mutational analyses also showed that the Cdc42p C-terminal 188CTIL CAAX box and 183KKSKK polylysine domain were involved in localization (8, 43). In Schizosaccharomyces pombe, GFP-Cdc42p was observed around the entire cell periphery, at internal membranes, and clustered at the medial region at the site of cell division (23), while mammalian Cdc42p has been observed at internal membranes and sites of polarized growth (14). However, the cell cycle-specific Cdc42p targeting and timing mechanisms remain unclear.
To examine Cdc42p cell cycle-specific targeting, a functional single-copy GFP-tagged Cdc42p was observed in S. cerevisiae. In contrast to previous immunolocalization data, GFP-Cdc42p localized to the entire cell periphery and to internal membranes and clustered at polarized growth sites, including the incipient bud site, during G1 phase and the mother-bud neck region during and after cytokinesis and cell separation. The observed localization patterns are consistent with a novel model in which Cdc42p is targeted to membranes and then clustered at different cellular locations during the cell cycle.
MATERIALS AND METHODS
Reagents, media, and strains.
Enzymes, PCR kits, and other reagents were obtained from standard commercial sources and used as specified by the suppliers. Oligonucleotide primers for sequencing and PCR were obtained from Genosys (The Woodlands, Tex.) and are available upon request. Growth media, maintenance of bacterial and yeast strains, and yeast transformations have been described previously (34, 36). Selection of transformants was done on synthetic complete drop-out medium lacking one or more specified amino acids and containing 2% glucose as a carbon source.
The yeast strains used are listed in Table 1. To create strain TRY100, the integrating plasmid pRS306(GFP-CDC42), which was linearized within the URA3 gene, was transformed into CDC42/Δcdc42::TRP1 heterozygous diploid DJD6-11; stable Ura+ transformants had GFP-CDC42 under the control of the endogenous CDC42 promoter integrated at the ura3 locus, which was confirmed by PCR and DNA-DNA hybridization analysis (data not shown). Complementation of the cdc42-1ts mutant allele by GFP-CDC42 in strain DJTD2-16A was determined by using plasmids p415MET25 (27), pGAL1-CDC42, and pRS415MET25(GFP-CDC42) (32). Transformants were selected on plates of SC medium lacking Leu (SC−Leu plates) at 23°C, and individual transformants were streaked to SC plates lacking Leu and Met (SC−Leu−Met plates) containing 2% galactose and incubated at 23°C or 37°C.
TABLE 1.
Yeast strains
| Strain | Genotype | Reference |
|---|---|---|
| C276 | MATa/MATα gal2/gal2 | 41 |
| DJTD2-16A | MATacdc42-1 his4 leu2 trp1 ura3 | 15 |
| DJD6-11 | MATa/MATα Δcdc42::TRP1/+ his3Δ200/+ his4/+ leu2/+ can1/+ lys2-801/lys2-801 trp1-Δ1/trp1-Δ101 ade2-101/+ ura3-52/ura3-52 | 25 |
| CSO-1B | MATacdc12-6 leu2 ura3 | B. Haarer |
| TRY11-7D | MATα leu2 ura3 trp1 his | 33 |
| W303 | MATa/MATα ade2-101/ade2-101 his3-11,5/his3-11,5 leu2-3,112/leu2-3,112 trp1-Δ1/trp1-Δ1 ura3-1/ura3-1 can1-100/can1-100 | R. Rothstein |
| TRY100 | MATa/MATα Δcdc42::TRP1/+ his3Δ200/+ his4/+ leu2/+ can1/+ lys2-801/lys2-801 trp1-Δ1/trp1-Δ101 ade2-101/+ ura3-52/ura3-52:pRS306(p42-GFP-CDC42):URA3 | This study |
| YEF2125 | MATa/MATα his3/his3 leu2/leu2 lys2/lys2 trp1/trp1 ura3/ura3 HOF1:HA::HIS3/HOF1:HA::HIS3 myo1::HIS3/myo1::HIS3 | 38 |
| YEF2126 | MATa/MATα his3/his3 leu2/leu2 lys2/lys2 trp1/trp1 ura3/ura3 HOF1:HA::HIS3/HOF1:HA::HIS3 bni1::HIS3/bnil::HIS3 | 38 |
DNA and protein analysis.
Recombinant DNA manipulations (34) and plasmid isolation from Escherichia coli (5) were performed as described previously (8). Automated DNA sequencing at the Vermont Cancer Center DNA Sequencing Facility was used to sequence all gene constructs. Site-directed mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.).
The plasmids pRS315(cdc42C188S) (42), p415MET(GFP) (37), p415MET(GFP-CDC42), p416MET(GFP-CDC42) (32), and pLP08(MYO1-GFP) (35) were described previously. p415MET(GFP-cdc42C188S) was constructed by PCR of cdc42C188S from template pRS315(cdc42C188S). The resulting product was cut with NotI plus XhoI and inserted into p415MET(GFP) cut with NotI plus XhoI. p415MET(GFP-KKSKKCTIL) was constructed by cutting pGAL(GFP-KKSKKCTIL) (kindly provided by Johanna Whitacre) with BamHI plus XhoI and inserting the resulting fragment into p415MET cut with BamHI plus XhoI. p415MET(GFP-CTIL) was constructed by PCR of GFP by using a 3′ primer that included CTIL sequences. The resulting PCR product was inserted into p415MET cut with SpeI plus HindIII.
p415MET(GFP-cdc42G12V) and p415MET(GFP-cdc42D118A) were created by using the QuikChange kit with p415MET(GFP-CDC42) as the DNA template. The cycling parameters for the mutagenesis were 12 cycles of 95°C for 30 s, 55°C for 30 s, and 68°C for 17 min. p415MET(CFP-CDC42) was created by incorporating the F64L, S65T, Y66W, N146I, M153T, and V163A mutations into GFP by using the QuikChange kit in a two-step mutagenesis scheme with p415MET(GFP-CDC42) as the original DNA template. pLP08(MYO1-YFP) and p415MET(YFP-CDC24) were created by incorporating the S65G, V68L, S72A, and T203Y mutations in GFP by using the QuikChange kit in a two-step mutagenesis scheme with pLP08(MYO1-GFP) and p415MET(GFP-CDC24) as the original DNA templates, respectively. The cycling parameters for these mutagenesis reactions were 18 cycles of 95°C for 30 s, 55°C for 30 s, and 68°C for 32 min.
Protein was isolated from wild-type strain C276, CDC42/Δcdc42 heterozygous stain DJD6-11, and TRY100. Immunoblot analysis was performed as previously described (43) with minor changes.
Photomicroscopy.
Cells were grown in the appropriate liquid medium to mid-log phase and were collected, sonicated, and examined morphologically. Methods for preparing and staining cells with FM4-64 have been described previously (29). Cells containing the various GFP-Cdc42p and Myo1p-GFP constructs were grown in SC−Ura−Met, SC−Leu−Met, and SC−Ura−Leu−Met medium as appropriate for expression from the methionine-repressible promoter.
Photomicroscopy and time-lapse microscopy with phase contrast optics and Omega XF100 optical filter cube, Chroma Yellow GFP (41028) optical filter cube, and Chroma Cyan GFP V2 (31044 V2) optical filter cube to visualize GFP, yellow fluorescent protein (YFP), and cyan fluorescent protein (CFP) fluorescence, respectively, were performed on an E400 Nikon microscope (Omega Optical, Brattleboro, Vt.). Digital cell images were obtained and analyzed as previously described (24).
For time-lapse microscopy, wild-type TRY11-7D cells containing one or two plasmids with various GFP, YFP, and/or CFP constructs were grown to mid-log phase, and an aliquot of cells was layered onto a microscope slide thin-layered with an appropriately selective SC−Met medium-agarose slab. The slide was then placed on the microscope, and cell division and fluorescence were monitored and captured in still-frame pictures at specified time intervals. Where indicated, cells from the same culture but different fields were assembled into collages by using Adobe Photoshop 5.0.
RESULTS
GFP-Cdc42p localized to the plasma membrane and internal membranes and clustered at the incipient bud site and mother-bud neck region.
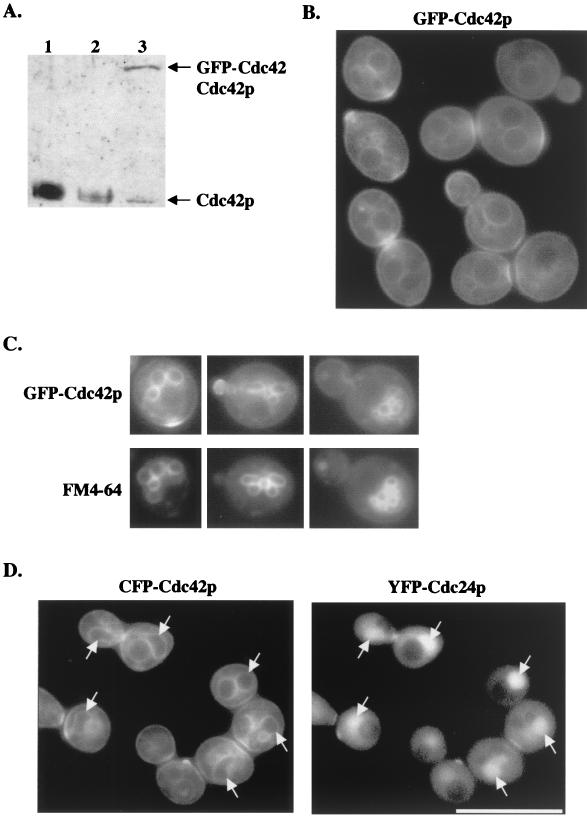
Preliminary characterization of an overexpressed GFP-Cdc42p construct in S. cerevisiae indicated that it was functional (i.e., able to complement a cdc42-1ts mutant) and was observed at sites of polarized growth, including the bud site and mother-bud neck region (32). To further examine these localization patterns, this GFP-Cdc42p construct was expressed under the CDC42 endogenous promoter and integrated into a CDC42/Δcdc42 heterozygous diploid at the URA3 locus. PCR, DNA-DNA hybridization, and immunoblot analysis confirmed that this strain (designated TRY100) had the GFP-CDC42 construct integrated at the correct locus (data not shown) and that GFP-Cdc42p was being expressed at levels comparable to endogenous Cdc42p (Fig. 1A). Under these conditions, GFP-Cdc42p still localized to polarized growth sites (see below) but was also seen around the entire cell periphery and at internal membranes (Fig. 1B) in cells at all stages of the cell cycle. Cdc42p localization at the plasma and internal membranes had not been observed previously in S. cerevisiae but was recently observed in S. pombe (23).
FIG. 1.
(A) GFP-Cdc42p levels were comparable to endogenous Cdc42p levels. Protein was isolated from wild-type strain C276 (lane 1), CDC42/Δcdc42 heterozygous strain DJD6-11 (lane 2), and TRY100 (lane 3), and immunoblots were probed with anti-Cdc42p antibody diluted 1:1,000. (B) GFP-Cdc42p clusters at polarized growth sites and localizes to internal membranes and the plasma membrane. TRY100 was grown in synthetic complete medium to mid-log phase and then observed for GFP-Cdc42p localization. (C) TRY100 were grown as in B, stained with FM4-64, and then viewed for colocalization of GFP-Cdc42p and vacuolar staining. (D) Colocalization of CFP-Cdc42p and YFP-Cdc24p. Plasmids p416MET(CFP-CDC42) and p415MET(YFP-CDC24) were transformed into wild-type strain TRY11-7D, and transformants were grown to mid-log phase in SC−Leu−Ura−Met medium. Arrows indicate CFP-Cdc42p localization to nuclear membranes and YFP-Cdc24p targeting to nuclei. Collages of images were assembled in Adobe Photoshop 5.0.
An enhanced GFP-Cdc42p signal was observed at polarized growth sites, including the presumptive bud site prior to bud emergence, at the tips and sides of enlarging buds, and at the mother-bud neck region in large-budded cells (Fig. 1B), suggesting that Cdc42p accumulates or clusters at these polarized growth sites. Identical targeting patterns were observed in a and α haploid cells and diploid cells (data not shown). GFP-Cdc42p localized to vacuolar membranes, as indicated by costaining with the vacuolar membrane stain FM4-64 (Fig. 1C). In addition, Cdc42p colocalized with its exchange factor Cdc24p at bud sites and the mother-bud neck region and was localized to the periphery of the nucleus, as delimited by Cdc24p internal nuclear localization (Fig. 1D) (Richman et al., unpublished data). These observations are consistent with a role for Cdc42p in intracellular membrane trafficking and with a mechanism for Cdc42p targeting involving membrane localization, followed by clustering at particular sites (see Discussion). The localization patterns of single-copy and plasmid-borne GFP-Cdc42p were identical, and thus plasmid-borne GFP-Cdc42p was used for all subsequent experiments.
C terminus of Cdc42p is sufficient for localization to membranes but not for clustering.
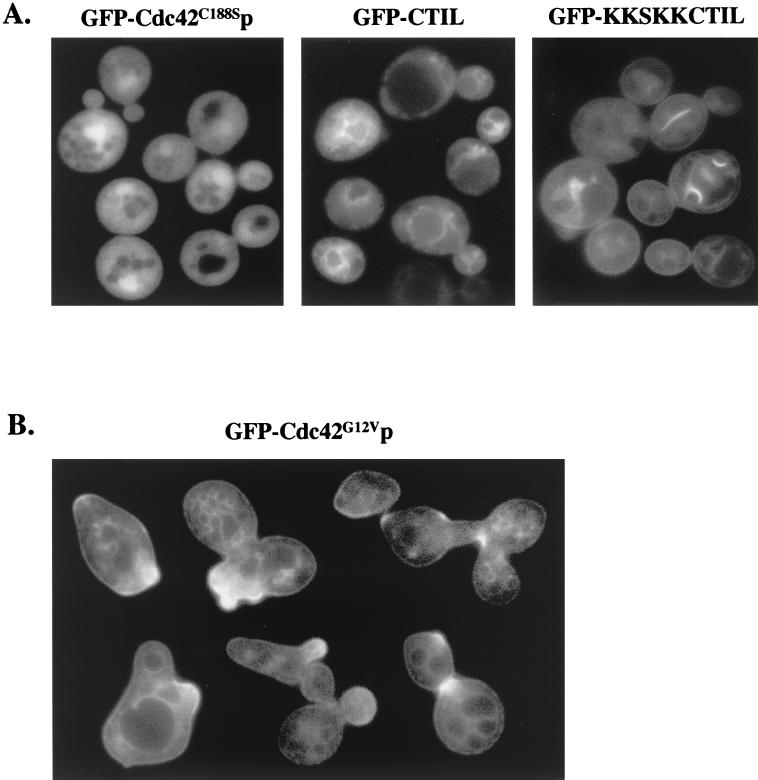
Mutational analysis of the Cdc42p C-terminal 188CTIL CAAX box and 183KKSKK polylysine domain have previously implicated these sequences in the localization and attachment of Cdc42p to membranes (8, 43). Geranylgeranylation of the Cys188 residue is necessary for Cdc42p attachment to membranes (43), and as expected, mutant GFP-Cdc42C188Sp no longer localized to the plasma membrane or internal membranes and did not cluster (Fig. 2A).
FIG. 2.
(A) Membrane localization required Cdc42p CTIL and polylysine residues. Plasmid p415MET(GFP-cdc42C188S) was transformed into wild-type strain TRY11-7D, and plasmids p415MET(GFP-CTIL) and p415MET(GFP-KKSKKCTIL) were transformed into wild-type strain W303. Transformants were grown to mid-log phase in SC−Leu−Met medium. (B) Constitutively active Cdc42G12Vp showed aberrant clustering. Plasmid p415MET(GFP-cdc42G12V) was transformed into wild-type TRY11-7D cells, and transformants were grown to mid-log phase in SC−Leu+Met medium. Images were assembled as in Fig. 1.
The conserved CTIL and polylysine residues were fused to GFP to test whether these sequences were sufficient for membrane targeting. A GFP-CTIL fusion protein localized to internal membranes but not plasma membranes or polarized growth sites, while a GFP-KKSKKCTIL fusion protein localized to both plasma and internal membranes but not to polarized growth sites (Fig. 2A). Mutant GFP-STIL and GFP-QQSQQSTIL proteins did not localize to plasma or internal membranes (data not shown). These results indicated that prenylation of the Cys residue was sufficient for internal membrane targeting, but the addition of the polylysine sequences was needed for targeting to the plasma membrane. These results also suggest that additional Cdc42p residues play a role in Cdc42p clustering at polarized growth sites.
To examine the role of other Cdc42p residues, previously characterized cdc42 mutations were tested for their effects on GFP-Cdc42p clustering. GTP-bound GFP-Cdc42G12Vp targeted normally but was clustered at multiple bud sites or bud tips simultaneously and was persistently polarized to tips of elongated buds (Fig. 2B), suggesting that the nucleotide-bound state of Cdc42p was important for timing and/or persistence of clustering. In contrast, dominant-negative GFP-Cdc42D118Ap showed no aberrant clustering (data not shown). GFP-Cdc42Y32Kp, containing the Y32K effector domain mutation that abolishes interactions with multiple effectors and regulators, including Cla4p, Skm1p, Gic1p, Gic2p, Iqg1p, and Bem3p but not Bni1p (32), showed normal clustering and targeting to membranes (data not shown), similar to that previously seen with the D38E and V44A effector domain mutations (32, 33). In addition, cla4Δ (33), gic1Δ, and gic2Δ mutants (data not shown) did not have defects in Cdc42p targeting or clustering. Although this does not completely rule out effector domain involvement in clustering, these results suggest that Cdc42p interactions with Cla4p, Skm1p, Gic1p, Gic2p, Iqg1p, and GTPase-activating protein (GAP) Bem3p were not required.
Time-lapse microscopy of Cdc42p localization during the cell cycle.
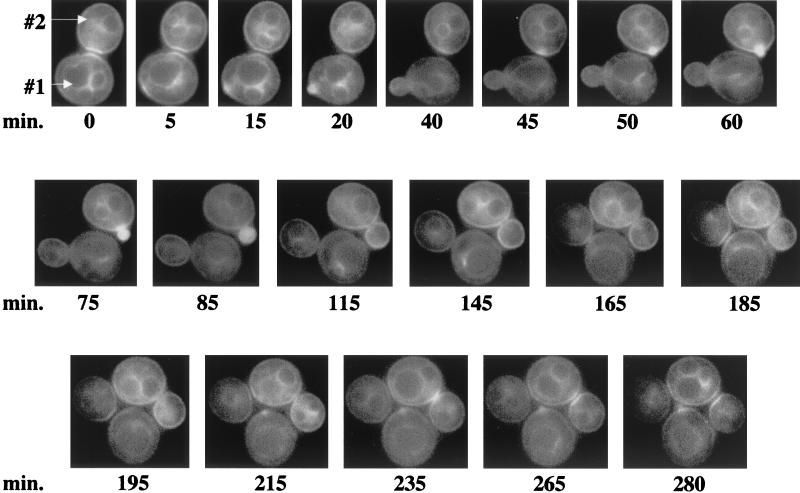
The cell cycle-specific timing of GFP-Cdc42p clustering was determined by time-lapse fluorescence microscopy (Fig. 3). At time zero, GFP-Cdc42p was clustered at the mother-bud neck, and 5 min later, GFP-Cdc42p clustered at a site on the mother cell (designated cell #1), where ≈10 min later, a small bud started to emerge. GFP-Cdc42p persisted at the original mother-bud neck region until after the new bud emerged (time = 20 min). As the cell #1 bud enlarged, an initial clustering event occurred on the daughter cell (#2), followed ≈10 min later by an emerging bud (time = 40 to 50 min). In cell #2, GFP-Cdc42p remained polarized to the growing bud until it was medium to large (time = 115 min), at which point GFP-Cdc42p appeared more diffusely along the tips and sides of the growing bud. GFP-Cdc42p clustered at the mother-bud neck region of cell #2 as a diffuse single band (time = 235 min) and then as bands on mother and daughter cells (probably following cytokinesis and cell separation; see below) that persisted for the remainder of the time course (time = 265 to 280 min). GFP-Cdc42p remained around the cell periphery and internal membranes of both mother and daughter cells throughout the time course.
FIG. 3.
Time-lapse photomicroscopy of GFP-Cdc42p localization during the cell cycle. Wild-type haploid strain TRY11-7D was transformed with p415MET(GFP-CDC42), and transformants were grown to mid-log phase in SC−Leu−Met medium. Cells were placed onto a thin-layered agar slab made with SC−Leu−Met medium and viewed by fluorescent microscopy. GFP-Cdc42p localization was captured at ≈5- to 15-min intervals. The cells designated #1 and #2 are described in the text. These cells are representative of at least 18 cells documented with similar localization patterns.
GFP-Cdc42p clustered to incipient bud sites on average ≈11 min prior to bud emergence (Table 2), which is comparable to the timing of actin and septin localization to this site (11, 16, 19). Furthermore, these experiments established that Cdc42p was indeed clustering at the site where a future bud would emerge. GFP-Cdc42p clustering at the mother-bud neck occurred postanaphase, as indicated by time-lapse microscopy of YFP-Cdc24p and CFP-Cdc42p colocalization, in which localization of both proteins occurred only after nuclei were clearly partitioned to mother and daughter cells (data not shown) (Richman et al., unpublished data). Upon clustering at the mother-bud neck region, the appearance of distinct Cdc42p bands on both mother and daughter cells took on average ≈10 min (Table 2). In general, GFP-Cdc42p persisted at the mother-bud neck region until a new budding cycle was initiated.
TABLE 2.
Timing of GFP-Cdc42p localization during the cell cyclea
| Localization pattern | No. of cells observed | Timing of event (min)
|
|
|---|---|---|---|
| Avg | Range | ||
| Incipient bud site | 19 | 11.4 ± 2.9 | High, 20; low, 7.5 |
| Mother-bud neck | 18 | 9.6 ± 3.4 | High, 16; low, 5 |
| bni1Δ | 6 | 24.8 ± 2.7 | High, 29; low, 21 |
For the incipient bud site, time was measured from GFP-Cdc42p's first clustering at the incipient bud site to the start of bud emergence in wild-type cells (TRY11-7D). Fluorescence at this time was captured approximately every 5 min. The timing of GFP-Cdc42p localization at the mother-bud neck region was measured from the first clustering of GFP-Cdc42p at the mother-bud neck region to the time it appeared as two distinct bands on mother and daughter cells in wild-type cells (TRY11-7D). Time was measured from the first clustering of GFP-Cdc42p at the mother-bud neck region to the time it appeared as two distinct bands on mother and daughter cells in the bni1Δ mutant (YEF5126).
Cdc42p clusters at the mother-bud neck region immediately prior to myosin ring contraction.
To determine whether cells with mother-bud neck clustering had completed cytokinesis and cell separation, fixed cells expressing GFP-Cdc42 were treated with the cell wall-digesting enzyme glusalase. Pre-glusalase treatment, ≈69% of cells were budded and the appearance of clustered Cdc42p on both mother and daughter cells was seen in ≈58% of cells exhibiting GFP-Cdc42p at the mother-bud neck. After glusalase treatment, only ≈50% were budded, suggesting that ≈19% of the budded cells had undergone cytokinesis and cell separation, and the appearance of clustered Cdc42p on both mother and daughter cells also decreased to ≈32%. These results suggest a correlation between the completion of cytokinesis and cell separation and GFP-Cdc42p clustering on both mother and daughter cells.
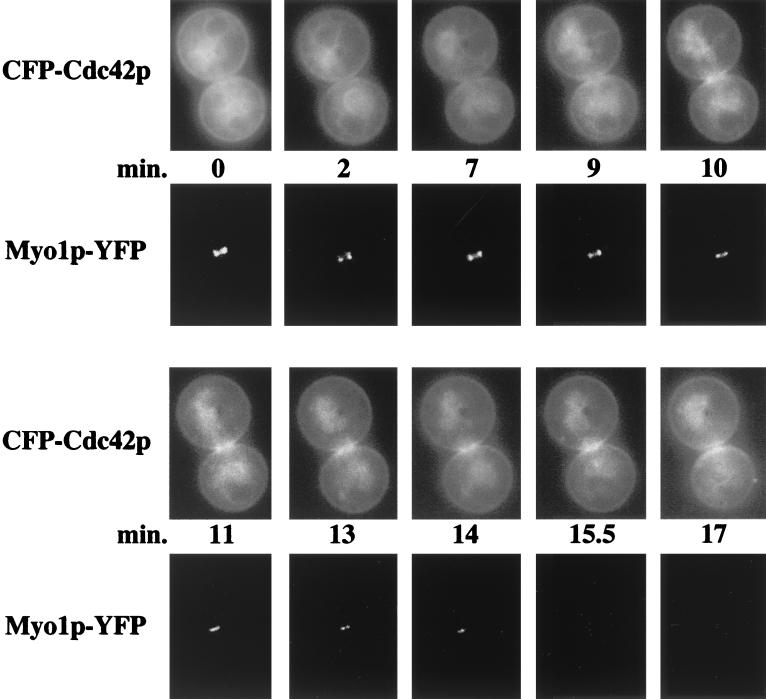
To pinpoint when Cdc42p clustering occurred in relation to actomyosin ring contraction, CFP-Cdc42p and Myo1p-YFP (the type II myosin component of the actomyosin ring) were observed together. Myo1p-YFP appeared at the mother-bud neck region in small-, medium-, and large-budded cells as previously described (4) (data not shown). Myo1p-YFP and CFP-Cdc42p both localized to the mother-bud neck, but cells showing CFP-Cdc42p clustered on both mother and daughter cells never concurrently showed a myosin ring, suggesting that the myosin ring had contracted by the time Cdc42p clustered on both mother and daughter cells.
Time-lapse microscopy confirmed this observation (Fig. 4; indicative of eight cells documented). At time zero, CFP-Cdc42p was not clustered and Myo1p-YFP localized to the mother-bud neck region in a double ring structure of diameter 1 μm. Within ≈9 min, CFP-Cdc42p had begun to cluster at the mother-bud neck region and the Myo1p-YFP rings had fused into a single ring, with the diameter of the ring remaining unchanged, indicating that Cdc42p accumulates prior to actomyosin ring contraction. Within ≈1 min, CFP-Cdc42p clustering intensified until the Myo1p-YFP ring began to contract to a diameter of 0.74 μm (time = 11 min) and to 0.49 μm ≈2 min later, and then disappeared ≈2.5 min later, suggesting that myosin ring contraction was complete in a total of ≈5.5 min, which was in the range seen previously (4). Overall, these observations suggest that Cdc42p clusters at the mother-bud neck region prior to actomyosin ring contraction and persists on mother and daughter cells after cytokinesis.
FIG. 4.
Localization of CFP-Cdc42p and Myo1p-YFP during cytokinesis. Plasmids p416MET(CFP-CDC42) and pLP08 containing MYO1-YFP under the MYO1 endogenous promoter were transformed into wild-type strain TRY11-7D. The cells were then prepared and observed as in Fig. 3. Large-budded cells were observed at ≈1- to 2-min intervals through CFP-Cdc42p's clustering at the mother-bud neck and Myo1p-YFP's starting to contract and disappear.
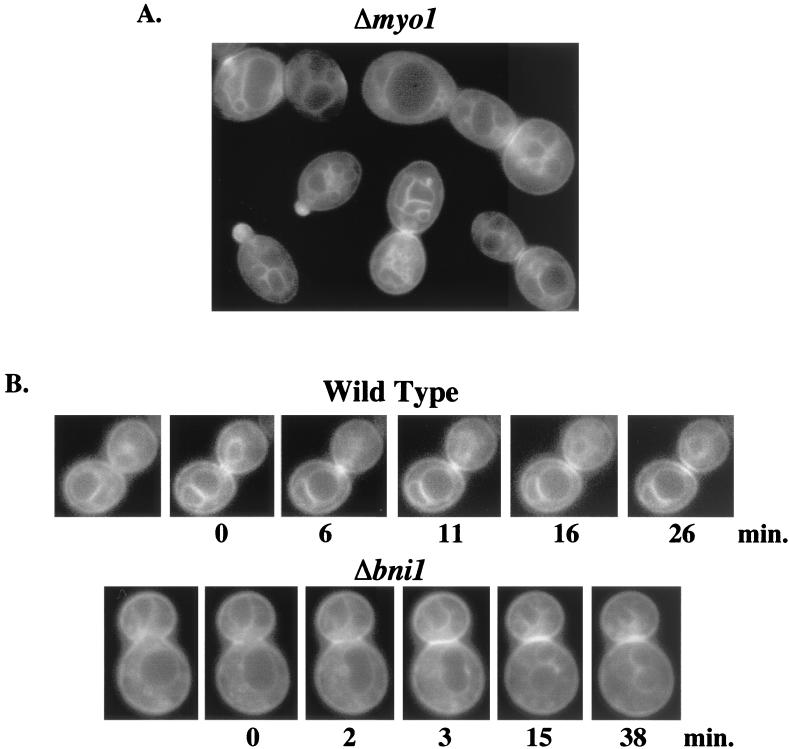
GFP-Cdc42p clustering at the mother-bud neck region is altered in septin cdc12-6ts and bni1Δ mutants but not myo1Δ mutants.
To determine whether GFP-Cdc42p clustering at the mother-bud neck region was dependent on the actomyosin ring structure, GFP-Cdc42p was observed in septin, bni1Δ, and myo1Δ mutant backgrounds, which have actomyosin ring localization or contraction defects. In myo1Δ mutant cells, GFP-Cdc42p was still observed at the plasma and internal membranes and at the mother-bud neck region (Fig. 5A), indicating that an intact myosin ring was not required for Cdc42p clustering at the mother-bud neck region. In bni1Δ mutant cells, GFP-Cdc42p clustered normally, but single banded mother-bud neck clustering seemed to be more predominant compared to wild-type cells, suggesting that the appearance of clustered Cdc42p on both mother and daughter cells depended on Bni1p.
FIG. 5.
(A) Cdc42p localization in myo1Δ mutant. p415MET(GFP-CDC42) was transformed into myo1Δ strain YEF2125, and transformants were grown to mid-log phase in SC−Leu−Met medium. Images were assembled as in Fig. 1. (B) Time-lapse microscopy of GFP-Cdc42p clustering at the mother-bud neck region in wild-type and bni1Δ cells. p415MET(GFP-CDC42) was transformed into wild-type (TRY11-7D) and bni1Δ (YEF2126) cells, and transformants were grown to mid-log phase in SC−Leu−Met medium and observed as in Fig. 3. Fluorescence pictures were captured at ≈1- to 2-min intervals, with time zero representing the time at which GFP-Cdc42p was first observed at the mother-bud neck region. The cells shown are representative of at least 18 wild-type (upper panels) and 6 bni1Δ (lower panels) cells documented.
Previously, bni1Δ mutants were shown to have a delay in myosin ring contraction and defects in cell separation (38). By using time-lapse microscopy, the appearance of clustered Cdc42p on both mother and daughter cells averaged ≈25 min in bni1Δ cells (Table 2), ≈2.5-fold longer than was seen in wild-type cells. Furthermore, true separation between the two bands, as was typically seen within 10 to 15 min of GFP-Cdc42p clustering in wild-type cells (Fig. 5B; time = 11 to 16 min), never occurred distinctly in bni1Δ cells (Fig. 5B; time = 38 min). These results reinforce the role of Bni1p in cytokinesis and cell separation and indicate that the appearance of clustered Cdc42p on both mother and daughter cells is a good reporter for the completion of cytokinesis and cell separation.
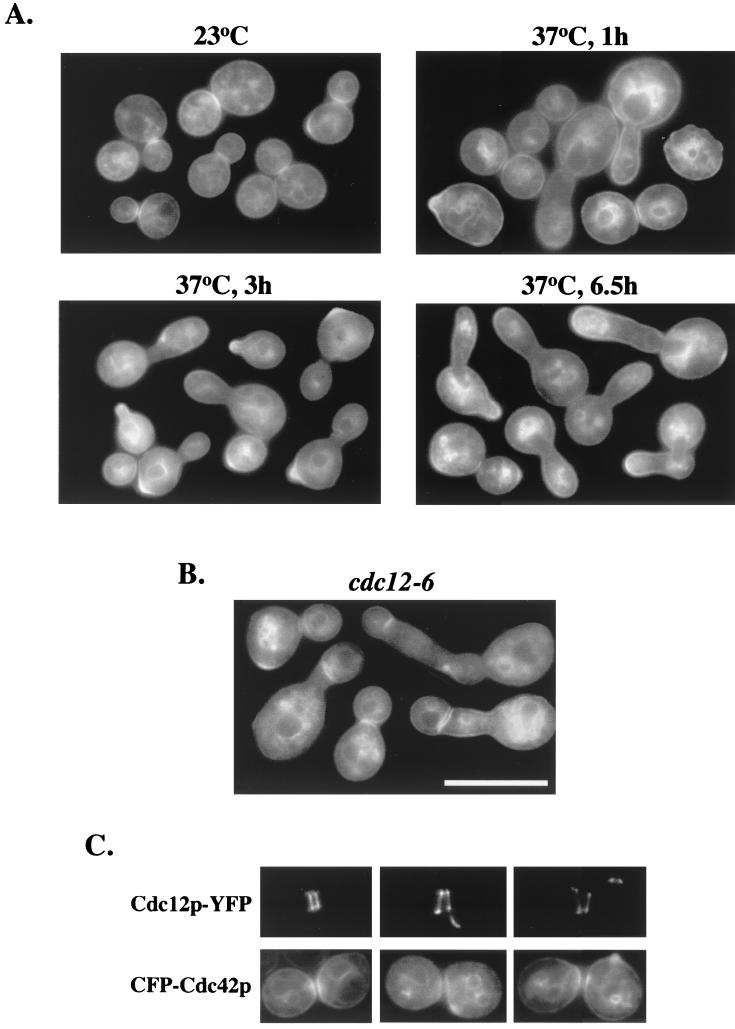
In a cdc12-6ts septin mutant at permissive temperatures, GFP-Cdc42p clustered normally at the mother-bud neck region (Fig. 6A, upper left panel). Upon shifting to restrictive temperatures for 1 h, clustering was still observed in normally budded cells (Fig. 6A, upper right panel), but elongated budded cells did not have GFP-Cdc42p at the mother-bud neck region (Fig. 6A, upper right panel). Over time at restrictive temperatures, some elongated cells showed GFP-Cdc42p clustering at new bud sites, with no GFP-Cdc42p clustering at the mother-bud neck region (Fig. 6A, lower left panel), indicating either that cells reset the budding cycle without going through cytokinesis, bypassing the time at which Cdc42p clustered at the mother-bud neck region, or that GFP-Cdc42p was not clustering at the mother-bud neck region due to the loss of the septin ring.
FIG. 6.
Cdc42p localization in septin cdc12-6ts mutant. Plasmid p415MET(GFP-CDC42) was transformed into septin mutant cdc12-6ts cells (CSO-1B), and transformants were grown to early to mid-log phase in SC−Leu−Met medium at 23°C. Half of the culture was then shifted to the restrictive temperature of 37°C. Both 23°C and 37°C cultures were observed for fluorescence. Upper left panel shows transformants grown at 23°C; upper right, lower left, and lower right panels show transformants shifted to 37°C for 1 h, 3 h, and 6.5 h, respectively. (B) Transformed cdc12-6 cells from A were shifted to the restrictive temperature of 37°C for 2.5 h, shifted back to the permissive temperature of 23°C for 1.5 h, and then shifted back to 37°C for 1 h. (C) Colocalization of CFP-Cdc42p and Cdc12p-YFP. Plasmids p416(CFP-CDC42) and pRS315 containing CDC12-YFP under the control of the CDC12 promoter (32) were transformed into wild-type strain TRY11-7D, and transformants were grown to mid-log phase in SC−Leu−Ura−Met medium.
By 6.5 h post-temperature shift, GFP-Cdc42p persistently polarized to the tips of elongated buds and also at some incipient bud sites, but mother-bud neck region clustering was never observed (Fig. 6A, lower right panel). These results raise the possibility that Cdc42p clustering to the mother-bud neck region required an intact septin ring (see below). Interestingly, internal membrane-bound GFP-Cdc42p was coalesced in various regions of these cells (Fig. 6A, lower right panel), which was also observed in S. pombe upon overexpression of the vacuolar component Nrf1p/Vtc1p (28), reinforcing a possible link between Cdc42p, septins, and vacuolar fusion events (9, 26).
Septin ring defects seen in a cdc12-6ts mutant trigger the Swe1p morphogenetic checkpoint that delays progression through the G2/M transition and the apical-isotropic switch (3, 33), as evidenced by GFP-Cdc42p remaining persistently polarized to the tips of elongated buds. Thus, the absence of clustered GFP-Cdc42p at the mother-bud neck region could be due to the cell cycle delay, the septin ring defect, or both.
To distinguish between these possibilities, cdc12-6ts cells were shifted to the restrictive temperature of 37°C for 2.5 h, shifted back to the permissive temperature of 23°C for 1.5 h (until cells started to progress through the G2/M transition), and then shifted back to 37°C for 1 h. These cells showed GFP-Cdc42p clustering at the mother-bud neck, although the intensity of clustering was decreased and some clustered GFP-Cdc42p bands appeared abnormally positioned (Fig. 6B).
Since the septin ring is lost ≈30 min after a shift to restrictive temperatures (12), these results suggested that Cdc42p is able to cluster at the neck region independently of the septin ring, but its positioning depends directly or indirectly on an intact septin ring. Accordingly, coexpressed septin Cdc12p-YFP and CFP-Cdc42p both localized to the incipient bud site and the mother-bud neck region, and both persisted at the old mother-bud neck region after a new bud began emerging (Fig. 6C), suggesting a possible functional relationship between Cdc42p and the septin ring at the incipient bud site and the mother-bud neck region.
DISCUSSION
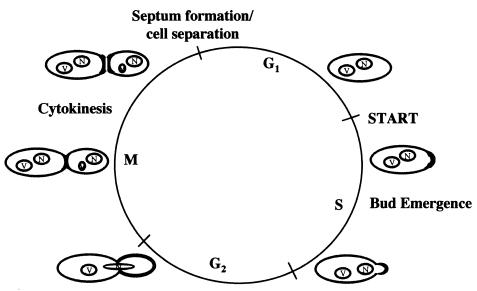
Proper targeting of Cdc42p is essential for its function and the function of a number of its effectors (7, 18). The results reported herein indicate that Cdc42p is targeted to the plasma and internal membranes and clusters at polarized growth sites during the cell cycle (Fig. 7). The presence of GFP-Cdc42p at plasma and internal membranes was a function of the C-terminal CAAX and polylysine domain, but the ability of Cdc42p to cluster at polarized growth sites was independent of this domain.
FIG. 7.
Model of Cdc42p localization and targeting. Boldfaced regions represent sites of Cdc42p clustering. See text for details.
In immunolocalization experiments, we previously had shown that Cdc42p was not observed around the plasma or internal membranes and was only variably seen at the mother-bud neck region (43). The disparity between these patterns and GFP-Cdc42p targeting patterns described herein may be due to the nature of the anti-Cdc42p antibody used. The antibody was raised against a peptide sequence containing amino acids 165 to 181, a region adjacent to the membrane-targeting domain (42) and likely to be in close proximity to the plasma membrane, raising the possibility that steric hindrance interfered with efficient binding. An underestimation of Cdc42p membrane targeting may also be due to the immunolocalization protocol used, including the use of cell wall-digesting enzymes and sodium dodecyl sulfate, required for efficient Cdc42p visualization. These possibilities were supported by the observation that the GFP-Cdc42p immunolocalization pattern with anti-Cdc42p antibody was similar to that seen previously with untagged endogenous Cdc42p (data not shown). Taken together, these data suggest that the GFP-Cdc42p localization pattern represents the bona fide Cdc42p pattern.
The presence of GFP-Cdc42p at the plasma and internal membranes throughout the cell cycle suggested that the mechanism by which Cdc42p clustered to sites of polarized growth was more complex than targeting solely through the action of a single protein or the secretory pathway. Upon proper signaling, Cdc42p may laterally diffuse through the membrane to concentrate at sites of polarized growth. However, a combination of specific targeting proteins, localized secretion, and lateral diffusion may all be involved in Cdc42p clustering at polarized growth sites. Some abnormal or decreased GFP-Cdc42p clustering was observed in sec1ts and sec6ts late secretion mutants (unpublished results), although it was difficult to discern whether these effects were specific to GFP-Cdc42p targeting, general defects in the secretory pathway, or cell death.
The function of Cdc42p at either nuclear or vacuolar membranes is not entirely clear, but various studies now link Cdc42p to roles in endocytosis and vacuolar fusion (9, 26, 29, 40). Persistence of GFP-Cdc42p at internal membranes throughout the cell cycle suggests that its function at these membranes occurs throughout the duration of the cell cycle. Interestingly, Cdc42p has recently been implicated in a specific docking stage of vacuolar fusion (9, 26). In addition, the localization of Cdc24p to nuclei and Cdc42p to the periphery of the nucleus suggested that these proteins may either have nuclear functions or interact prior to colocalization at the plasma membrane.
Time-lapse experiments indicated that Cdc42p clustered at an incipient bud site ≈11 min prior to bud emergence, which is in the range of actin and septin localization to the incipient bud site. Loss of Cdc42p function led to depolarized actin (1) and delocalized septins (21, 33), and Cdc42p clustered independently of actin at the incipient bud site (2). Together, these data are consistent with a model in which Cdc42p localization is required for actin and septin polarization to the incipient bud site.
Cdc42p clusters postanaphase at the mother-bud neck region immediately prior to actomyosin ring contraction and then persists on mother and daughter cells ≈10 min later following cytokinesis and cell separation. Localization studies with S. pombe Cdc42p also suggested a role in mediating ring contraction and/or septum formation (23). One can envision multiple potential roles for Cdc42p at this site, including signaling changes in septin ring structure at the mother-bud neck region immediately prior to actomyosin ring contraction (20), inducing actomyosin ring contraction after the split of the septin ring into two rings (20), triggering the ring-to-plate structure transition of Bni1p and Cbk1p (30, 31), and mediating actin-dependent septum formation and cell separation.
Over 40 S. cerevisiae proteins have been shown to target to the mother-bud neck region late in the cell cycle. They can be grouped into several distinct localization patterns, including double rings often associated with the septin double ring, single rings associated with the actomyosin ring, single or double rings that have not yet been correlated to a specific structural ring, and single rings specifically on the mother side of the mother-bud neck region. The Cdc42p banding patterns comprise a subset of proteins that include Cdc24p, Gic1p, Gic2p (6, 7), chitinase regulator protein kinase Cbk1p (31), Bni1p (30), and Syp1p (22), a multicopy suppressor of the actin-binding protein profilin. Many of these proteins have also been implicated in regulating bud emergence, suggesting that similar Cdc42p-associated complexes may function in the establishment of bud emergence and in reestablishing cell polarity during cytokinesis and cell separation.
These analyses provide new insights into Cdc42p targeting and localization patterns and the timing of its clustering at sites of polarized growth, especially at the mother-bud neck region. Given the highly conserved structure and function of Cdc42p in all eukaryotes examined to date, the establishment of these temporal and spatial patterns should provide an important context in which to analyze the timing and localization of other cell signaling proteins with respect to Cdc42p clustering. Future experiments building on the timing of Cdc42p clustering will hopefully lead to a more defined hierarchy of protein localization during the eukaryotic cell cycle.
Acknowledgments
We thank Rong Li, Erfei Bi, and Johanna Whitacre for sharing valuable reagents, Kurt Toenjes for technical assistance, and members of the Johnson laboratory for valuable discussions and critical comments on this manuscript.
This work was supported by NSF grants MCB-0076826 and MCB-0110138.
REFERENCES
- 1.Adams, A. E. M., D. I. Johnson, R. M. Longnecker, B. F. Sloat, and J. R. Pringle. 1990. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 111:131-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayscough, K. R., J. Stryker, N. Pokala, M. Sanders, P. Crews, and D. G. Drubin. 1997. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed by using the actin inhibitor latrunculin-A. J. Cell Biol. 137:399-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barral, Y., M. Parra, S. Bidlingmaier, and M. Snyder. 1999. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 13:176-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi, E., P. Maddox, D. J. Lew, E. D. Salmon, J. N. McMillan, E. Yeh, and J. R. Pringle. 1998. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142:1301-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, J. L., M. Jaquenoud, M.-P. Gulli, J. Chant, and M. Peter. 1997. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 11:2972-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, G.-C., Y.-J. Kim, and C. S. M. Chan. 1997. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 11:2958-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, C. R., T. J. Richman, S. B. Deliduka, J. O. Blaisdell, C. C. Collins, and D. I. Johnson. 1998. Analysis of the mechanisms of action of the Saccharomyces cerevisiae dominant lethal cdc42G12V and dominant negative cdc42D118A mutations. J. Biol. Chem. 273:849-858. [DOI] [PubMed] [Google Scholar]
- 9.Eitzen, G., N. Thorngren, and W. Wickner. 2001. Rho1p and Cdc42p act after Ypt7p to regulate vacuole docking. EMBO J. 20:5650-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson, J. W., and R. A. Cerione. 2001. Multiple roles for Cdc42 in cell regulation. Curr. Opin. Cell Biol. 13:153-157. [DOI] [PubMed] [Google Scholar]
- 11.Ford, S. K., and J. R. Pringle. 1991. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC11 gene product and the timing of events at the budding site. Dev. Genet. 12:281-292. [DOI] [PubMed] [Google Scholar]
- 12.Haarer, B. K., and J. R. Pringle. 1987. Immunofluorescence localization of the Saccharomyces cerevisiae CDC12 gene product to the vicinity of the 10-nm filaments in the mother-bud neck. Mol. Cell. Biol. 7:3678-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joberty, G., C. Petersen, L. Gao, and I. G. Macara. 2000. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2:531-539. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, D. I. 1999. Cdc42: an essential rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63:54-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, D. I., and J. R. Pringle. 1990. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J. Cell Biol. 111:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, H. B., B. K. Haarer, and J. R. Pringle. 1991. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J. Cell Biol. 112:535-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kroschewski, R., A. Hall, and I. Mellman. 1999. Cdc42 controls secretory and endocytic transport to the basolateral plasma membrane of MDCK cells. Nat. Cell Biol. 1:8-13. [DOI] [PubMed] [Google Scholar]
- 18.Leberer, E., C. Wu, T. Leeuw, A. Fourest-Lieuvin, J. E. Segall, and D. Y. Thomas. 1997. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 16:83-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lew, D. J., and S. I. Reed. 1993. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120:1305-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lippincott, J., K. B. Shannon, W. Y. Shou, J. Deshaies, and R. Li. 2001. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J. Cell Sci. 114:1379-1386. [DOI] [PubMed] [Google Scholar]
- 21.Longtine, M. S., D. J. DeMarini, M. L. Valencik, O. S. Al-Awar, H. Fares, C. DeVirgilio, and J. R. Pringle. 1996. The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol. 8:106-119. [DOI] [PubMed] [Google Scholar]
- 22.Marcoux, N., S. Cloutier, E. Zakrzewska, P. M. Charest, Y. Bourbonnais, and D. Pallotta. 2000. Suppression of the profilin-deficient phenotype by the RHO2 signaling pathway in Saccharomyces cerevisiae. Genetics 156:579-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merla, A., and D. I. Johnson. 2000. The Cdc42p GTPase is targeted to the site of cell division in the fission yeast Schizosaccharomyces pombe. Eur. J. Cell Biol. 79:469-477. [DOI] [PubMed] [Google Scholar]
- 24.Merla, A., and D. I. Johnson. 2001. The Schizosaccharomyces pombe Cdc42p GTPase signals through Pak2p and the Mkh1p-Pek1p-Spm1p MAP kinase pathway. Curr. Genet. 39:205-209. [DOI] [PubMed] [Google Scholar]
- 25.Miller, P. J., and D. I. Johnson. 1997. Characterization of the S. cerevisiae cdc42-1ts allele and new temperature-conditional-lethal cdc42 alleles. Yeast 13:561-572. [DOI] [PubMed] [Google Scholar]
- 26.Müller, O., D. I. Johnson, and A. Mayer. 2001. Cdc42p functions at the docking stage of yeast vacuole membrane fusion. EMBO J. 20:5657-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mumberg, D., R. Müller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray, J. M., and D. I. Johnson. 2000. Isolation and characterization of Nrf1p, a novel negative regulator of the Cdc42p GTPase in Schizosaccharomyces pombe. Genetics 154:155-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray, J. M., and D. I. Johnson. 2001. The Cdc42p GTPase and its regulators Nrf1p and Scd1p are involved in endocytic trafficking in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 276:3004-3009. [DOI] [PubMed] [Google Scholar]
- 30.Ozaki-Kuroda, K., Y. Yamamoto, H. Nohara, M. Kinoshita, T. Fujiwara, K. Irie, and Y. Takai. 2001. Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:827-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Racki, W. J., A. M. Becam, F. Nasr, and C. J. Herbert. 2000. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J. 19:4524-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richman, T. J., and D. I. Johnson. 2000. Saccharomyces cerevisiae Cdc42p GTPase is involved in preventing the recurrence of bud emergence during the cell cycle. Mol. Cell. Biol. 20:8548-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richman, T. J., M. M. Sawyer, and D. I. Johnson. 1999. The Cdc42p GTPase is involved in a G2/M morphogenetic checkpoint regulating the apical-isotropic switch and nuclear division in yeast. J. Biol. Chem. 274:16861-16870. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Shannon, K. B., and R. Li. 1999. The multiple roles of Cyk1p in the assembly and function of the actomyosin ring in budding yeast. Mol. Biol. Cell 10:283-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics: laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Toenjes, K. A., M. M. Sawyer, and D. I. Johnson. 1999. The guanine-nucleotide-exchange factor Cdc24p is targeted to the nucleus and polarized growth sites. Curr. Biol. 9:1183-1186. [DOI] [PubMed] [Google Scholar]
- 38.Vallen, E. A., J. Caviston, and E. Bi. 2000. Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell 11:593-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Aelst, L., and C. D'Souza-Schorey. 1997. Rho GTPases and signaling networks. Genes Dev. 11:2295-2322. [DOI] [PubMed] [Google Scholar]
- 40.White, W. H., and D. I. Johnson. 1997. Characterization of synthetic-lethal mutants reveals a role for the Saccharomyces cerevisiae guanine-nucleotide exchange factor Cdc24p in vacuole function and Na+ tolerance. Genetics 147:43-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilkinson, L. E., and J. R. Pringle. 1974. Transient G1 arrest of S. cerevisiae cells of mating type α by a factor produced by cells of mating type a. Exp. Cell Res. 89:175-187. [DOI] [PubMed] [Google Scholar]
- 42.Ziman, M., J. M. O'Brien, L. A. Ouellette, W. R. Church, and D. I. Johnson. 1991. Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol. Cell. Biol. 11:3537-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ziman, M., D. Preuss, J. Mulholland, J. M. O'Brien, D. Botstein, and D. I. Johnson. 1993. Subcellular localization of Cdc42p, a Saccharomyces cerevisiae GTP-binding protein involved in the control of cell polarity. Mol. Biol. Cell 4:1307-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]