Abstract
The trisubstituted pyrrole 4-[2-(4-fluorophenyl)-5-(1-methylpiperidine-4-yl)-1H-pyrrol-3-yl]pyridine (compound 1) has in vivo activity against the apicomplexan parasites Toxoplasma gondii and Eimeria tenella in animal models. The presumptive molecular target of this compound in E. tenella is cyclic GMP-dependent protein kinase (PKG). Native PKG purified from T. gondii has kinetic and pharmacologic properties similar to those of the E. tenella homologue, and both have been functionally expressed as recombinant proteins in T. gondii. Computer modeling of parasite PKG was used to predict catalytic site amino acid residues that interact with compound 1. The recombinant laboratory-generated mutants T. gondii PKG T761Q or T761M and the analogous E. tenella T770 alleles have reduced binding affinity for, and are not inhibited by, compound 1. By all other criteria, PKG with this class of catalytic site substitution is indistinguishable from wild-type enzyme. A genetic disruption of T. gondii PKG can only be achieved if a complementing copy of PKG is provided in trans, arguing that PKG is an essential protein. Strains of T. gondii, disrupted at the genomic PKG locus and dependent upon the T. gondii T761-substituted PKGs, are as virulent as wild type in mice. However, unlike mice infected with wild-type T. gondii that are cured by compound 1, mice infected with the laboratory-generated strains of T. gondii do not respond to treatment. We conclude that PKG represents the primary molecular target responsible for the antiparasitic efficacy of compound 1.
The apicomplexan parasite Toxoplasma gondii is an opportunistic human pathogen and the causative agent of toxoplasmosis, a disease whose clinical impact is largely restricted to immunocompromised patients (15, 23-25). A notable clinical exception follows from transplacental transmission of the parasite and in utero damage to the developing fetus (32). The primary therapeutic choice for the former patient population is the combination of pyrimethamine and sulfa drugs. While efficacious in many cases, this regimen is not optimal due to toxicity in some patients (10, 14, 19). The identification of alternative therapies for toxoplasmosis is urgent.
In a recent publication we reported on the trisubstituted pyrrole 4-[2-(4-fluorophenyl)-5-(1-methylpiperidine-4-yl)-1H-pyrrol-3-yl]pyridine (compound 1) and its potent activity against several apicomplexan parasites, including Eimeria tenella and T. gondii, both in cell-based assays and in animal models (13, 26). Using a [3H]compound 1 ligand-binding assay, a cyclic GMP (cGMP)-dependent protein kinase (PKG) was biochemically purified from E. tenella (13). The trisubstituted pyrrole is a potent (50% inhibitory concentration [IC50] of 1 nM) ATP-competitive inhibitor of E. tenella PKG (Et-PKG) activity. Remarkably, compound 1 is a much less potent inhibitor of both mammalian and avian PKG activities (13) and therefore represents an opportunity for selective intervention. Structural differences in the parasite PKG catalytic site that account for differences in inhibitor sensitivity could reflect an overall divergence of function between the parasite and animal enzymes. Vertebrate PKGs are known to play a role in smooth muscle physiology and fluid homeostasis in the gut (9, 29, 30, 35), processes for which there are no obvious counterparts in the parasite. A second characteristic that distinguishes apicomplexan parasite PKGs from invertebrate, avian, and mammalian homologues is the presence of a larger activation domain encompassing a putative third allosteric site (13). The additional third nucleotide binding site, revealed by analysis of the deduced protein coding regions of cloned cDNAs, represents a striking departure from all PKG proteins that have been reported to date. Site-directed mutagenesis studies of T. gondii and E. tenella PKGs demonstrate that the third nucleotide binding site is functionally important and suggest that it is responsible for the unique cGMP-dependent activation properties of the Eimeria enzyme (3, 13, 35a).
Protein kinases play key roles in eukaryotic signal transduction pathways and for that reason have been extensively studied as targets for therapeutic intervention in many different disease indications (11, 16, 39). In this report, we provide data to demonstrate that cGMP-dependent protein kinase is an essential gene product for T. gondii and as such represents an attractive therapeutic target. Like the majority of kinase inhibitors, compound 1 is an ATP-competitive inhibitor of parasite PKG. We have used molecular modeling to predict amino acid residues important for the association of compound 1 within the ATP-binding site of PKG. A site-directed mutation at one of these residues (T761 in T. gondii PKG or T770 in E. tenella PKG) renders recombinant PKG insensitive to compound 1 but equivalent to wild-type enzyme by all other criteria. Conservation at the ATP-binding site among protein kinases predicts that inhibitor selectivity will represent a substantial obstacle, but our studies with this site-directed mutation of PKG suggest otherwise. We have constructed a strain of T. gondii disrupted at the genomic PKG locus and dependent upon the compound 1-insensitive form of PKG. This modified strain of T. gondii is as virulent as wild type in a mouse model. Where compound 1 cures mice challenged with the wild-type strain (26), mice infected with the mutant strain do not respond to therapy. These results lead us to conclude that PKG represents the primary molecular target responsible for the antiparasitic efficacy of compound 1. Moreover, parasite PKG represents the first protein kinase target to be validated in an animal model of an infectious disease agent.
MATERIALS AND METHODS
Host cells and parasite cultures.
Cell culture media and reagents were obtained from Life Technologies, Inc. T. gondii tachyzoites of the RH strain and the derived HXGPRT knockout strain (RHΔHXGPRT) were provided by the NIH AIDS Research and Reference Reagent Program (Bethesda, Md. [http:www.niaid.nih.gov/reagent]). Parasites were maintained by serial passage in primary human foreskin fibroblast (HFF) cultures. Host cells were grown in Dulbecco modified Eagle medium with 10% heat-inactivated newborn bovine serum. This culture medium was replaced with modified Eagle medium containing 2% dialyzed fetal bovine serum immediately prior to parasite infection (34).
PKG cloning, plasmid construction, and mutagenesis.
DNA manipulations, hybridizations, etc., were performed according to standard procedures (36). Plasmid DNA was purified by using the Qiagen Maxi kit (Qiagen), sequenced with an ABI Prism sequencer and fluorescent sequencing reagents (Perkin-Elmer), and analyzed by using Vector NTI Suite software (InforMax). The complete sequence of the T. gondii PKG genomic locus and cDNA copies corresponding to full-length E. tenella and T. gondii PKGs have been deposited in GenBank under accession numbers AF448496, AF411961, and AF413570.
Toxoplasma PKG λ clones were screened from T. gondii cDNA and genomic libraries in λ ZAP-II and λ DASH-II vectors, respectively (vectors from Stratagene; libraries from NIH AIDS Reference and Reagent Repository, Bethesda, Md., catalog numbers 1896 and 2862). The cloning of Eimeria and Toxoplasma full-length cDNAs, which encode 1,003 and 994 amino acid polypeptides, respectively, has been described elsewhere (13). Parasite cDNAs encoding PKG open reading frames were modified prior to subcloning by appending N-terminal and C-terminal FLAG epitopes (Kodak) via PCR amplification (Pfu polymerase; Stratagene). DNA fragments encoding FLAG epitope-tagged PKGs were subcloned into plasmid pGEM-T Easy (Promega) for further modification by site-directed mutagenesis or placed under the control of a Toxoplasma α-tubulin promoter for expression in Toxoplasma (3). PKG mutations were generated utilizing either the Chameleon or QuickChange site-directed mutagenesis procedures (Stratagene) according to the manufacturers' protocols. Gel-purified grade mutagenic oligonucleotides bearing modified codons in the middle to introduce desired amino acid substitutions were obtained from Integrated DNA Technologies, Inc. Mutations were confirmed by DNA sequencing of the entire PKG minigene (∼3 kb for Eimeria and Toxoplasma PKGs).
A combination of restriction enzyme mapping and DNA sequence analysis was used to generate a map of the ∼20-kb Toxoplasma PKG genomic locus. Genomic clones used to generate gene targeting vectors and subclones for DNA sequencing were constructed by using restriction fragments isolated from overlapping λ clones. DNA sequence analysis of the locus, performed by using information from mostly one strand only, was completed with the use of 42-oligonucleotide primers. Gene targeting vectors were made by combining genomic PKG fragments in plasmids carrying the HXGPRT marker (4). Knockout vector gPKG-KO combines a 3.6-kb XbaI-SalI fragment and distal 3.9-kb SalI-SacII fragment, creating an internal 1.6-kb deletion. Similarly, the hit-and-run vectors gPKG-h&r#1 and gPKG-h&r#2 were created by splicing together genomic fragments spanning the 5′ and 3′ untranslated regions (UTRs) of the PKG locus.
Selection of transgenic parasites.
General procedures for the transient or stable transformation of T. gondii by electroporation have been described (17, 34, 38). The T. gondii HXGPRT gene, the Escherichia coli chloramphenicol acetyltransferase (CAT) gene, and drug-resistant Tg-PKG and Et-PKG genes were used as selectable markers. Clones were obtained following selections by limiting dilution in 96-well plates. HXGPRT+ parasites were selected in 25 μg of mycophenolic acid/ml supplemented with 50 μg of xanthine/ml (4, 27). Selection for loss of the HXGPRT marker from PKG genomic locus pseudodiploids, during the resolution step of hit-and-run mutagenesis, was performed in 320 μg of 6-thioxanthine (6-TX)/ml as described previously (5, 27). Because gene-targeting vectors carry the HXGPRT marker, knockout experiments with transgenic parasites expressing recombinant PKGs were done with lines in which PKG transgenes had been previously selected by using chloramphenicol and the CAT marker (1). Alternatively, knockout experiments with parasites expressing drug-resistant Eimeria and Toxoplamsa PKGs (Tg-PKG T761Q, Tg-PKG T761M, Et-PKG T770Q, and Et-PKG T770M mutants) were done with lines selected directly by serial passage (three or four times) in 1 μg of compound 1/ml. Southern blot analysis of several independent parasite clones showed that Tg-PKG T761Q, Tg-PKG T761M, Et-PKG T770Q, or Et-PKG T770M transgenes selected with compound 1 exhibited the same copy number as transgenes selected with the HXGPRT marker (one to three copies [4]).
Purification of native and recombinant PKG.
Parasites from 10 T-175 cultures were filtered through 3-μm (pore-size) polycarbonate membranes (Millipore) to remove host cell debris, spun over 50% Percoll, and washed in phosphate-buffered saline (PBS). The resulting pellet was resuspended in HBS lysis buffer (50 mM HEPES [pH 7.4], 1 mM EDTA, 1% Nonidet P-40, 10 mM NaF, 0.1 mM sodium orthovanadate, 1 mM dithiothreitol) supplemented with a cocktail of protease inhibitors (Sigma). After sonication with a Branson Sonifier Microtip (three 15-s blasts with intermittent cooling), the extract was centrifuged successively at 4°C for 10 min at 3,000 × g and for 30 min at 100,000 × g to remove insoluble debris. Glycerol was added to a final concentration of 10% (vol/vol). After the extract was filtered through a 0.2-μm (pore-size) filter unit (Millipore), 500 μl was loaded onto a SMART system FPLC 100-μl MonoQ anion-exchange column (Pharmacia). A linear NaCl gradient was applied (0 to 500 mM in 3 ml at 100 μl/min), and 100-μl fractions were collected and assayed for PKG and compound 1 binding activity. Compound 1 binding was performed by using 96-well gel filtration blocks (Edge Biosystems). Then, 15 μl of each fraction was incubated in a total volume of 100 μl of binding assay buffer (20 mM HEPES [pH 7.4], 1 mM MgCl2, 20 nM 3H-labeled compound 1 [specific activity, 35 mCi/mg]) for 60 min on ice. Then, 50 μl of the reaction was loaded onto the filtration block, which was spun at 1,500 rpm for 4 min. Scintillation cocktail (190 μl) was added to the filtrate in a collection plate, which was sealed and counted. FLAG epitope-tagged PKGs were purified from Toxoplasma crude extracts with anti-FLAG M2-agarose affinity gel (Sigma). Recombinant enzyme was eluted with 0.5 mg of FLAG peptide/ml, after washing of the matrix with lysis buffer, and concentrated in an Ultrafree Biomax-30K filter device (Millipore). Coomassie brilliant blue R-250 (Sigma) staining of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)-8% Tris-glycine gels (Novex) revealed a single band of ∼110 kDa for N-terminally FLAG epitope-tagged PKGs purified in this way. Silver staining was done with a kit (Daiichi, Tokyo, Japan). Protein was measured with a micro-BCA reagent (Pierce) by using a TCA-sodium deoxycholate step (2) to remove residual FLAG peptide prior to quantitation. Approximately 20 to 30 μg of purified recombinant FLAG PKG was obtained from pooled cultures of transgenic parasites (T-175). Affinity purification of recombinant and native PKGs was done as described previously (13) by using 8-AET-cGMP-agarose (Biolog A019).
Immunological reagents.
Antisera against PKG-specific peptide epitopes (TR4, GNDAEDQLEIFRD; TR7, DEDDTIVLEDEYDWDKDF) were generated by Covance Research Products. Antisera to the FLAG epitope was purchased from Sigma. Chemiluminescent reagents (Amersham Pharmacia) were used to detect primary antisera in Western blot experiments.
Enzyme assays and steady-state kinetics.
PKG activity was assayed essentially as described previously (13) by using 96-well multiscreen phosphocellulose plates (MAPH-NOB; Millipore) to capture 33P-phosphorylated kemptide peptide substrate. Kinase reactions were carried out in 40-μl volumes containing 25 mM HEPES (pH 7.4), 10 mM MgCl2, 20 mM β-glycerolphosphate, 2 mM dithiothreitol, 10 μM cGMP, 250 μM kemptide, 0.1 mg of bovine serum albumin/ml, 2 to 50 μM ATP, and 0.01 μM [γ-33P]ATP (3,000 Ci/mmol; NEN/DuPont). The reactions were initiated by the addition of 0.2 to 2 nM PKG (final concentration) and incubated at room temperature for 60 min. Dilutions of enzyme were used that ensured that reactions were linear over a time course of up to 2 h. Reactions were terminated by the addition of phosphoric acid (2.5 mM final concentration) before the samples were applied to a phosphocellulose plate mounted on a vacuum manifold. Wells were washed in 75 mM phosphoric acid and then dried, and radiolabeled peptide was detected by scintillation counting on a Microbeta 1450 scintillation counter (Perkin-Elmer Wallac). Kinetic constants were determined from nonlinear regression (curve fit) analysis (GraphPad Prism Software, Inc., San Diego, Calif.).
Computer modeling.
The sequence of the catalytic domain of Et-PKG was aligned with the sequence of PKA from the crystal structure (18) with the aid of the homology modeling software LOOK v3.5 (Molecular Applications Group, Palo Alto, Calif.). The alignment was adjusted further by using the automated homology modeling and energy minimization procedures SEGMOD (20) and ENCAD (21), implemented in LOOK. The resulting homology model was used to generate binding models of inhibitors of the trisubstituted imidazole class in the ATP binding site, based on the orientation of related compounds in the crystal structure of the p38 mitogen-activated protein (MAP) kinase (40).
Metabolic labeling of tachyzoites.
Labeling of tachyzoites with [3H]uracil (28) was done by using infected HFF cell monolayers growing in Cytostar-T scintillating 96-well microplates (Amerhsam Pharmacia). Plates were counted in a Microbeta 1450 scintillation counter (Perkin-Elmer Wallac) 48 h after the addition of 2 × 104 parasites and 1 μCi of [3H]uracil (33 Ci/mmol; Dupont/NEN) to each well.
In vivo efficacy studies.
Tachyzoites of T. gondii collected from freshly lysed HFF host cell monolayers were counted in a hemocytometer and diluted in minimal essential medium to a concentration of 1 × 103 to 3 × 103 T. gondii tachyzoites per 500 μl for intraperitoneal injection into C57BL/6 strain mice (Jackson Laboratories, Bar Harbor, Maine). Parasite titers were confirmed by plaque assay (34). Compound 1 was dissolved in phosphate-buffered saline and administered in 100-μl doses by intraperitoneal injections given twice daily starting 24 h after parasite inoculation. Mice were monitored twice daily for clinical evidence of toxoplasmosis and mortality throughout the experimental period. Moribund mice, displaying severe malaise or lethargy, were euthanized by using CO2 gas. All procedures were performed in accordance with the highest standards for the humane handling, care, and treatment of research animals and were approved by the Merck Institutional Animal Care and Use Committee. Procedures for the care and use of research animals at Merck meet or exceed all applicable local, national, and international laws and regulations.
RESULTS
Characterization of native PKG from T. gondii.
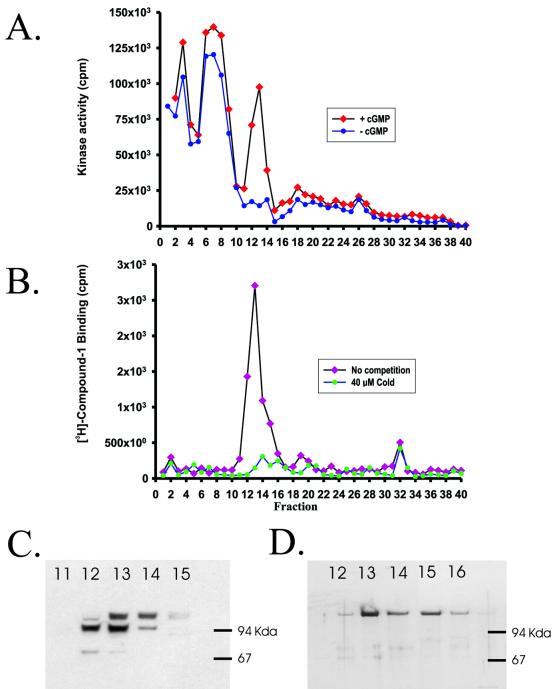
Fractionation of a T. gondii tachyzoite detergent lysate by MonoQ anion-exchange chromatography resulted in the detection of a single peak of cGMP-dependent kinase activity (Fig. 1A). This catalytic activity was coincident with the sole peak of 3H-labeled compound 1 ligand binding activity (Fig. 1B). Nucleotide-independent kinase activities eluted earlier in the gradient, but these fractions did not bind radiolabeled compound 1. Western blot analysis with antisera directed against a conserved peptide epitope from within the E. tenella and T. gondii PKG catalytic domains (13) detected two immunoreactive polypeptides at 100 and 110 kDa in the MonoQ fraction which contained the highest catalytic and ligand-binding activities (Fig. 1C). Titration experiments demonstrated that ligand binding was competitive with ATP but was not competitive with peptide substrate (data not shown). These data suggest that compound 1 interacts with Toxoplasma PKG at the ATP-binding site. Biochemical, kinetic, and pharmacological properties of partially purified native Toxoplasma PKG (pool of fractions 12 to 14, Fig. 1C) are shown in Table 1.
FIG. 1.
Correlation between PKG activity and compound 1 binding in fractionated Toxoplasma extracts. (A) A crude detergent (1% NP-40) extract of T. gondii tachyzoites was passed over a MonoQ column, and proteins were eluted with a NaCl gradient. Fractions were assayed for kinase activity in the presence (⧫) or absence (•) of 10 μM cGMP as indicated, with kemptide as the peptide substrate. (B) Ligand-binding activity was assayed by incubating fractions with 3H-labeled compound 1, filtering samples through a gel filtration plate, and scintillation counting to quantitate protein-bound radiolabel. Specific binding was determined by incubation in the presence (•) or absence (⧫) of 40 μM cold compound 1. (C) Western blot of fractions containing compound 1 binding activity with anti-peptide antisera (TR4) specific to an epitope shared by Eimeria and Toxoplasma PKG enzymes. (D) Recombinant FLAGTg-PKG, recovered from a lysate of transgenic parasites in a single-step FLAG-immunoaffinity procedure (see Materials and Methods) was subjected to MonoQ chromatography (as in panels A and B). The fractions of peak activity were electrophoretically resolved and silver stained.
TABLE 1.
Biochemical properties of native and recombinant expressed PKGa
| Enzyme | Kemptide Km (μM) ± SD | ATP Km (μM) ± SD | cGMP Ka (μM) ± SD | Hill constant (H) ± SD | kcat (min−1) ± SD | kcat/Km(ATP) (μM−1 min−1) | Compound 1 IC50 (nM) ± SD |
|---|---|---|---|---|---|---|---|
| Native (Tg) | 25.3 ± 3.8 | 7.3 ± 0.1 | 1.7 ± 1.0 | 1.9 ± 0.4 | 1.0 ± 0.5 | ||
| FLAGTg-PKG | 27.7 ± 0.1 | 6.5 ± 0.4 | 1.0 ± 0.2 | 2.3 ± 0.1 | 5.5 ± 0.2 | 0.85 | 2.3 ± 1.0 |
| FLAGTg-PKG T761M | 8.4 ± 0.3 | 2.9 ± 0.3 | 0.5 ± 0.2 | 2.2 ± 0.1 | 3.5 ± 0.5 | 1.20 | 950 ± 90 |
| FLAGTg-PKG T761Q | 17.3 ± 6.6 | 8.5 ± 2.3 | 0.9 ± 0.1 | 2.2 ± 0.2 | 10.3 ± 0.5 | 1.21 | 21,000 ± 200 |
| FLAGEt-PKG | 78.7 ± 1.9 | 5.4 ± 1.0 | 0.9 ± 0.1 | 2.1 ± 0.2 | 2.5 ± 0.4 | 0.46 | 2.4 ± 0.3 |
| FLAGEt-PKG T770M | 11.0 ± 1.0 | 2.7 ± 0.3 | 0.4 ± 0.1 | 2.3 ± 0.4 | 1.2 ± 0.2 | 0.44 | 2,500 ± 750 |
| FLAGEt-PKG T770Q | 55.0 ± 3.0 | 5.4 ± 0.4 | 0.9 ± 0.1 | 2.3 ± 0.4 | 1.1 ± 0.3 | 0.20 | 42,000 ± 400 |
Recombinant parasite PKG enzymes were purified from representative transgenic Toxoplasma strains by immunoaffinity chromatography. Partially purified native enzyme was prepared as described in Fig. 1C (pool of fractions 12 to 14). Steady-state rate constants were calculated by using nonlinear regression (curve fit) analysis (Graphpad Prism 3.0). The errors shown represent standard deviations of samples assayed in duplicate.
Expression of recombinant parasite PKGs in Toxoplasma.
We have recently reported the expression in Toxoplasma of two isoforms of both T. gondii PKG (Tg-PKG) and E. tenella PKG (Et-PKG) from constructs bearing a FLAG epitope appended to the C-terminal end of the cDNA clones (PKGFLAG). The two recombinant isoforms, expressed from Toxoplasma and Eimeria PKG cDNAs, correspond in size by SDS-PAGE to polypeptides observed in preparations of native enzyme from Toxoplasma and Eimeria (3) spp. The larger parasite PKG polypeptide (isoform I) is modified by N-terminal acylation and is membrane associated; the smaller soluble polypeptide (isoform II) appears to result from translational initiation at a methionine codon distal to the primary initiator methionine. We obtained improved yields of recombinant PKG from transgenic parasites expressing N-terminally FLAG-tagged PKGs. Placement of a FLAG epitope at the N terminus of PKG (FLAGPKG) prevents dual acylation of isoform I by myristate and palmitate (3) and facilitates the recovery of active soluble recombinant PKG isoform I with >95% purity from crude parasite lysates after a single FLAG epitope immunoaffinity chromatography step (see Materials and Methods). Since the isoform II enzyme expressed from the FLAGPKG transgene lacks a portion of the PKG amino terminus, it is not labeled with the FLAG epitope and is therefore not recovered. An additional MonoQ anion-exchange chromatography step results in purification of fully active enzyme, essentially to homogeneity (Fig. 1D). In a side-by-side comparison with purified native T. gondii PKG, the recombinant parasite proteins have similar catalytic site substrate binding affinities (Km, kemptide; Km, ATP), allosteric site cooperativity (cGMP dependent [Ka] and H values) and sensitivity to compound 1(IC50, summarized in Table 1).
Identification of catalytic site amino acid residues in PKG that are critical for compound 1 efficacy.
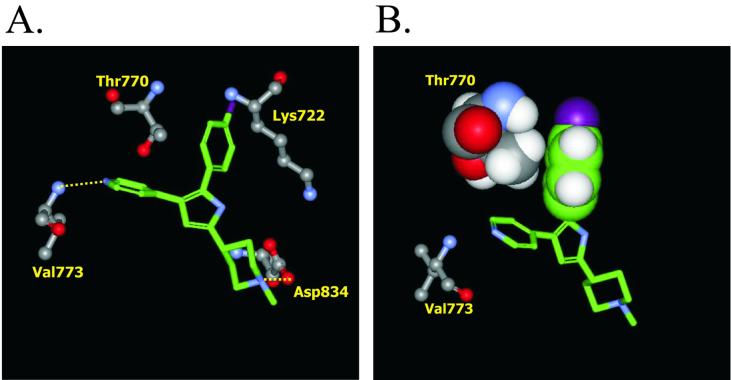
To explore the molecular basis for PKG inhibitor specificity, we generated homology models of the active site of parasite (Eimeria and Toxoplasma) PKGs based on the crystal structure data from cAMP-dependent protein kinase (PKA) and p38 MAP kinase (18, 40). PKG active-site residues within 5 Å of the docked inhibitors were analyzed to identify amino acids potentially important for ligand binding. Trisubstituted imidazole inhibitors of kinases, which are structurally related to compound 1, are known to exploit a hydrophobic pocket adjacent to the catalytic lysine in the active site (18, 40). A key threonine residue in this pocket (T106 of human p38 kinase) has been shown to interact with halophenyl substituents from this structural class of compounds. Substitution of this threonine residue with amino acids carrying bulky side chains such as glutamine or methionine (T106Q, T106M) prevented inhibitor binding and resulted in an enzyme that was insensitive to this class of compounds (12, 22). Our modeling of parasite PKGs implicated the analogous residues T770 of Et-PKG and T761 of Tg-PKG as potentially crucial for the stabilization of the fluorophenyl moiety of compound 1 (Fig. 2). Consequently, we introduced methionine and glutamine substitutions at T761 of Tg-PKG and T770 of Et-PKG and selected transgenic parasites expressing these alleles (T761M, T761Q, T770M, and T770Q) in Toxoplasma.
FIG. 2.
Model of compound 1 docked and minimized in the ATP-binding site of Et-PKG. (A) Four conserved active site residues in close proximity to compound 1, which is shown in green with nitrogens in blue and fluorine in purple. Oxygen atoms in PKG are shown in red. Dotted lines denote predicted interactions: a hydrogen bond between the amide nitrogen of Val773 and the compound 1 pyridine nitrogen, a salt bridge between the carboxyl group of the essential catalytic Asp834, and the basic amine of the compound 1 piperidine moiety. (B) Space-filling view of Et-PKG residue T770 and the fluorophenyl moiety of compound 1. Substitution of T770 for residues with bulky side chains would be predicted to disrupt this hydrophobic interaction.
The consequences of the substitutions on recombinant enzyme activity in vitro are shown in Table 1. Substitution of the targeted active-site threonine of Eimeria and Toxoplasma PKGs by methionine (Tg-PKG T761M, Et-PKG T770M) increases the IC50 for compound 1 by ca. 3 orders of magnitude. The glutamine substitution (Tg-PKG T761Q, Et-PKG T770Q) has an even a more pronounced effect, shifting the sensitivity of both Eimeria and Toxoplasma PKGs by ca. 4 orders of magnitude. In contrast, the steady-state kinetic properties of the recombinant mutant enzymes are similar to the wild-type Tg-PKG and Et-PKG enzymes. The most noticeable difference, a two- to fivefold reduction in substrate Km and cGMP-dependent Ka values in the case of the Tg-PKG T761M and Et-PKG T770M substitutions, does not result in significant changes in catalytic efficiency (kcat/Km ATP) or cGMP-dependent cooperativity (Hill coefficient values).
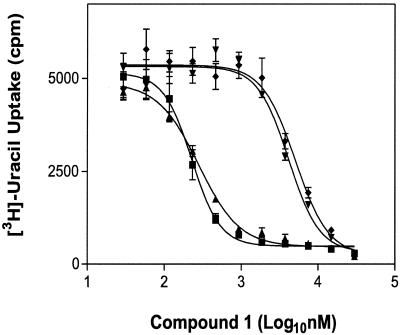
To assess the affect of PKG catalytic site mutations on drug sensitivity in Toxoplasma, the metabolic activity of transgenic parasites growing in drug-treated HFF cell monolayer cultures was measured by determining [3H]uracil uptake (28). Parasites expressing C-terminally FLAG-tagged allelic forms of T. gondii and E. tenella PKGs were chosen for these experiments, since the level of expression and subcellular location of these recombinant enzymes better reflects the membrane-associated and cytosolic isoforms of corresponding native enzymes than the nonacylated soluble FLAGPKG variants (3). Dose-response titrations show that parasites expressing either the T761M or T761Q Tg-PKGFLAG proteins are substantially less sensitive to compound 1 than parasites expressing the T761 Tg-PKGFLAG enzyme or untransformed parasites expressing only native Tg-PKG (Fig. 3). As shown in Table 2, the calculated IC50 values for parasites expressing these Tg-PKGFLAG T761M and T761Q alleles are increased 19- and 25-fold, respectively, compared to untransformed wild-type parasites. Similarly, when the Et-PKGFLAG T770Q protein is expressed in Toxoplasma, a 15-fold increase in the IC50 for compound 1 was observed. Taken together, these results are consistent with the interpretation that the primary molecular target for the antiparasitic activity of compound 1 is PKG. The data also suggest that compound 1 acts at secondary targets, but ones that are considerably less sensitive.
FIG. 3.
Dose response for inhibition of parasite growth by compound 1. Dose-response curves indicating IC50 values (see Table 2) for in vitro-grown parasites are shown for a representative experiment as follows: ▪, RHΔHXGPRT wild-type strain; ▴, transgenic strain expressing Tg-PKGFLAG; ▾, transgenic Tg-PKGFLAG T761M; and ⧫, transgenic Tg-PKGFLAG T761Q. The metabolic activity of parasites growing in HFF cell monolayers was measured by determining [3H]uracil uptake. IC50 values were determined from plots of samples run in duplicate by using curve-fitting software from GraphPad Prism (v.3).
TABLE 2.
Sensitivity of untransformed and transgenic Toxoplasma to compound 1a
| Parasite strain | Compound 1 IC50 (nM) ± SD | Fold change |
|---|---|---|
| Wild-type (RHΔHXGPRT) | 200 ± 60 | |
| Tg-PKGFLAG | 300 ± 100 | 1.5 |
| Tg-PKGFLAG T761M | 3,800 ± 360 | 19 |
| Tg-PKGFLAG T761M-KO | 4,900 ± 600 | 25 |
| Tg-PKGFLAG T761Q | 5,000 ± 160 | 25 |
| Tg-PKGFLAG T761Q-KO | 7,932 ± 100 | 40 |
| Et-PKGFLAG | 300 ± 50 | 1.5 |
| Et-PKGFLAG T770Q | 2,900 ± 340 | 15 |
The IC50 values shown are the average of two to three experiments (the standard deviation is indicated). At least two independent clones derived from each construct were evaluated.
Gene targeting experiments at the Tg-PKG genomic locus.
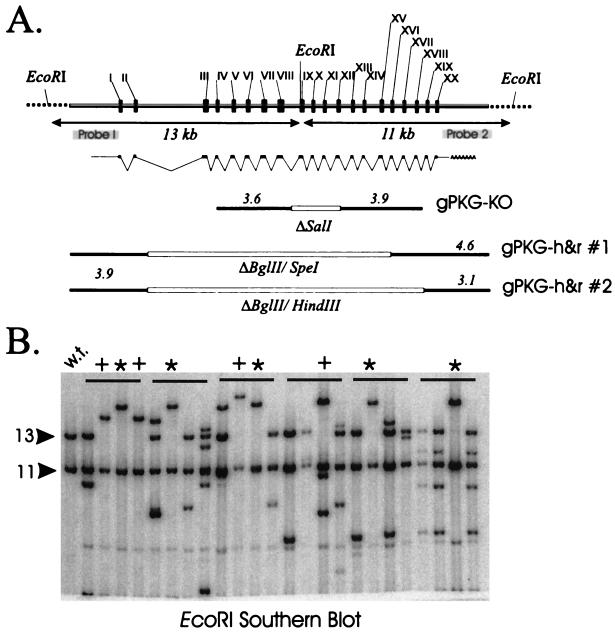
A map of the PKG genomic locus cloned from T. gondii is shown in Fig. 4A. A comparison of the PKG genomic DNA sequence with a full-length cDNA sequence encoding Tg-PKG (13; GenBank AF413570) revealed that the locus contains 20 exons and spans 15.1 kb. The cloned locus contains an additional 2.4 and 3.0 kb of 5′ and 3′ UTRs, respectively. Southern blotting indicated the presence of only a single Toxoplasma PKG gene (not shown).
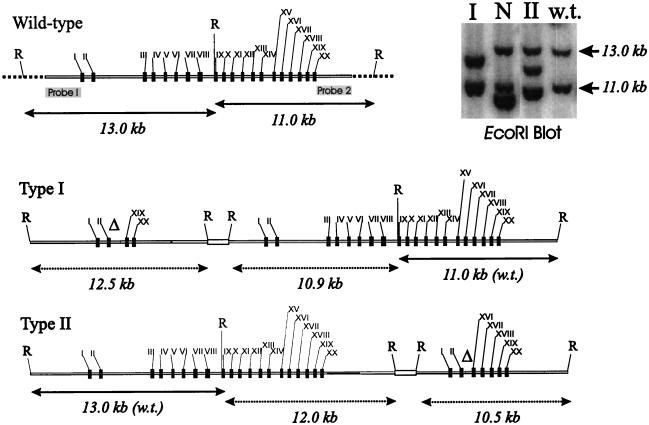
FIG. 4.
Toxoplasma PKG genomic locus and gene knockouts. (A) Schematic map of the PKG locus (cloned sequence, gray bar). The transcript splicing pattern and location of 20 exons (black boxes) was deduced from comparisons of genomic and cDNA sequence data. The locations of probes within 5′ and 3′ UTRs, used for Southern blot analysis of recombinants (in panel B), are indicated. Genomic sequences present in circular gene targeting vectors (of indicated kilobase sizes) are shown as linear molecules: gPKG-KO contains 3.6- and 3.9-kb fragments flanking a 1.6-kb deletion (ΔSalI); the gPKG-h&r vectors #1 and #2 (used to generate pseudodiploids, Fig. 6) contain genomic fragments spanning 5′ and 3′ UTRs that border internal deletions of 11.3 and 13.5 kb, respectively (restriction enzyme sites used are indicated). (B) Parasite clones obtained in a knockout experiment in which a transgenic line expressing the Tg-PKGFLAG transgene was transformed with gPKG-KO. A total of 24 clones (in groups of 4) derived from six independent electroporations were randomly screened by Southern blot analysis (EcoRI digests). Wild-type parasites (w.t.) show the 11- and 13-kb bands characteristic of the intact PKG locus (see panel A). Lanes marked with an asterisk indicate the shifted band (∼19 kb) expected of recombinants arising from a single reciprocal exchange between the proximal 3.6-kb vector fragment and the genomic locus. Other homologous recombinants (“+”) contain additional rearrangements or insertions.
Because the Toxoplasma tachyzoite is haploid, disruption of an essential gene is a lethal event. However, gene knockout experiments can be designed to provide indirect evidence to conclude that a gene is required for parasite growth. We employed two strategies to explore whether PKG is dispensable for parasite survival in culture. Both strategies utilize gene targeting vectors (illustrated in Fig. 4A), which aim to disrupt the Tg-PKG gene by a single reciprocal homologous recombination event. Circular vectors are employed, since they are efficient at producing homologous recombination in Toxoplasma (5, 6).
In the first method, we used a knockout vector (gPKG-KO) designed to create a null mutation as a result of genomic integration at the PKG locus. According to this strategy, the dispensability of the T. gondii PKG locus is tested by comparing the frequency of knockout events in wild-type parasites with the frequency of knockout events in transgenic parasites expressing functional recombinant PKG. If the PKG gene is dispensable, gene knockouts should be obtained in wild-type and transgenic parasites at similar frequencies. Conversely, if PKG is essential, only parasites expressing a complementing transgenic copy should yield recombinants. The knockout vector gPKG-KO was designed to contain a large portion of the Tg-PKG protein coding region but contains a 1.6-kb deletion (ΔSalI) of the gene which removes exons IX to XI and part of exon XII. The deletion results in a null mutation, since mRNAs expressed from this template after integration into the genomic locus would be expected to yield truncated polypeptides lacking most of the PKG catalytic domain. Southern blot analysis with EcoRI was used to detect recombinants resulting from homologous integration events between the knockout vector and the PKG genomic locus. Recombination events within the proximal 3.6-kb portion of the gPKG-KO vector (type I) would be predicted to yield recombinants that disrupt a 13-kb EcoRI fragment at the genomic locus. Recombination within the distal 3.9-kb portion of the knockout vector (type II) would modify the diagnostic 11-kb EcoRI fragment at the PKG genomic locus.
Figure 4B shows the results of a gene targeting experiment in which parasites expressing the Tg-PKGFLAG transgene were stably transformed with vector gPKG-KO. Of 21 independent clones, 8 are disrupted at the genomic PKG locus, and all of these are type I knockouts where recombination has occurred to disrupt the 13-kb EcoRI fragment. Three of these eight gPKG-KO vector insertions contain additional rearrangements at the PKG locus or insertions elsewhere in the genome as a result of nonhomologous integration events. None of the homologous recombination events were of the type II variety, suggesting a recombinational preference for the proximal of the two PKG fragments contained within the gPKG-KO vector (Fig. 4A). Although we have no explanation for this bias, recombination can occur within either proximal or distal ends of the PKG locus with hit-and-run targeting vectors (see Table 3).
TABLE 3.
Homologous recombination at the Toxoplasma PKG genomic locusa
| Targeting vector | Parasite strain | No. of homologous recombinants/total no. (%) | Description |
|---|---|---|---|
| Knockout gPKG-KO (ΔSal) | Wild-type (RHΔHXGPRT) | 0/68 (0)b | No recombinants obtained |
| ::Tg-PKGFLAG | 5/21 (24) | 5 knockouts, type I | |
| ::Tg-PKGFLAG T761M | 4/20 (20) | 4 knockouts, type I | |
| ::Tg-PKGFLAG T761Q | 1/20 (5) | 1 knockout, type I | |
| ::Et-PKGFLAG | 1/14 (7) | 1 knockout, type I | |
| ::FLAGEt-PKG | 2/20 (10) | 2 knockouts, type I | |
| Hit-and-run | |||
| gPKG-h&r#1 | Wild-type (RHΔHXGPRT) | 14/86 (16)b | 14 pseudodiploids, type II |
| gPKG-h&r#2 | Wild-type (RHΔHXGPRT) | 5/27 (18) | 5 pseudodiploids, type I |
Untransformed or transgenic parasites were transfected with circular targeting vectors, and stable transformants were selected with mycophenolic acid. Transgenic parasite strains used as recipients for knockout transformations were initially selected with either chloramphenicol (Tg-PKGFLAG, Et-PKGFLAG, and FLAGEt-PKG) or with compound 1 (Tg-PKGFLAGT761M, Tg-PKGFLAGT761Q [see Materials and Methods]). Randomly picked clones were screened by Southern blot analysis (Fig. 4B) to identify targeted disruptions that introduce null mutations (knockout vector) or that regenerate a genomic copy at the PKG locus (hit-and-run vectors). The frequency of the desired targeting event among the independent clones analyzed is indicated, as well as the class of recombinant obtained.
Sum of two experiments.
Similar experiments were performed with transgenic parasite lines expressing other recombinant PKG constructs, and the frequency of independent homologous recombination events detected was between 5 and 20% in these transgenic lines (Table 3). In conclusion, successful targeted disruption of the PKG locus is readily achieved in transgenic parasites expressing recombinant PKGs. By way of contrast, in parallel experiments with wild-type parasites in which at least 68 independent gPKG-KO transformants were examined, no evidence of homologous recombination at the PKG genomic locus was obtained. Although indirect, these data imply that knockout mutants are not recovered from wild-type parasites due to the essential nature of the PKG gene. Metabolic activity assays performed on each of the knockout strains that carry a recombinant PKG (including Tg-PKGFLAG, Tg-PKGFLAG T761M, Tg-PKGFLAG T761Q, Et-PKGFLAG, and FLAGEt-PKG) failed to show detectable differences in growth rate compared with wild-type parasites (data not shown). This suggests that recombinant Tg-PKG and Et-PKG enzymes are both fully functional in Toxoplasma strains disrupted at the genomic locus.
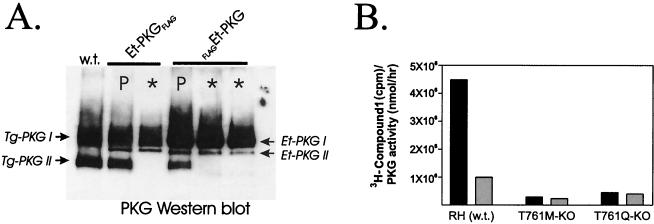
Confirmation that the knockout strategy successfully prevents expression of native PKG from the genomic PKG locus was obtained by Western blot analysis of knockout lines of parasites expressing recombinant Eimeria PKGs (Et-PKGFLAG and FLAGEt-PKG). Unlike the PKG isoform I polypeptides of Toxoplasma and Eimeria which fail to resolve on SDS-PAGE gels, the faster-moving isoform II polypeptides can be readily separated (3). Immunoreactive Toxoplasma PKG isoforms I and II are detected in wild-type parasites (Fig. 5A, lane w.t.) and in the parental T. gondii lines that were used to make the genomic knockout strains (Fig. 5A, lanes P). In contrast, genomic PKG knockout progeny from these parental lines (Fig. 5A, lanes ✽) are devoid of isoform Tg-PKG II. Lacking a Tg-PKG selective antisera, we infer that Tg-PKG isoform I (which comigrates with recombinant Et-PKG isoform I) is also lost as a result of the genomic integration event.
FIG. 5.
Knockout mutants lack native Toxoplasma PKG. (A) Native Tg-PKG isoform II (Tg-PKG II) is absent in knockout lines expressing the Et-PKGFLAG or FLAGEt-PKG transgenes (lanes with asterisk) but present in parental lines (lanes P) and in the wild-type strain (w.t.). Recombinant Et-PKG isoform II (Et-PKGII) was observed in the transgenic lines. Native and recombinant isoform I of Et-PKG and Tg-PKG comigrate and cannot be resolved by SDS-PAGE. PKG enzymes were purified from parasites by cGMP affinity chromatography and detected with antisera to a conserved C-terminal peptide epitope, TR7. (B) 3H-labeled compound 1 binding activity in knockout lines expressing Tg-PKGFLAG T761Q and Tg-PKGFLAG T761M alleles (T761 M-KO, T761Q-KO) compared with wild-type parasites (w.t.). Parasite lysates (obtained from ca. 5 × 109 tachyzoites) were fractionated by MonoQ chromatography as described in Fig. 1. Fractions with peak cGMP-dependent kinase activity were pooled, and 3H-labeled compound 1 binding activity was assayed in the absence (black bars) or presence (gray bars) of 40 μM unlabeled ligand. Activities presented are normalized for the total PKG activity of the pooled samples. The total PKG activity recovered from either knockout line (T761M-KO or T761Q-KO) was approximately fourfold higher than the total recovered from untransformed parasites of the parental strain (RHΔHXGPRT).
Ligand-binding experiments with knockout lines that express alleles of Tg-PKG that are not sensitive to compound 1 (Tables 2 and 3, T761M-KO and T761Q-KO) also support the conclusion that the expression of native enzyme is ablated as a result of the genomic integration event. Parasite lysates were prepared from the wild-type T. gondii strain and the Tg-PKGFLAG T761Q and Tg-PKGFLAG T761M transgenic strains disrupted at the PKG genomic locus. MonoQ ion-exchange chromatography was performed as described in Fig. 1, and fractions containing PKG activity were pooled and assayed for 3H-labeled compound 1 binding activity (Fig. 5B). Since the T761Q and T761M modified recombinant versions of TgPKG do not significantly bind compound 1 (Table 1), any ligand-binding activity would be contributed by the native enzyme. Indeed, significant levels of activity were detected in wild-type parasites, but no specific binding activity was detected in the knockout strains.
The second method used to address whether PKG is essential for T. gondii employs a two-step hit-and-run strategy (5). In this approach, an initial targeting event disrupts the PKG locus but regenerates a functional genomic copy at the site of integration. The resulting chimeric pseudodiploid gene can be resolved in a second recombination step to yield wild-type or knockout mutant segregants. The targeting vectors (gPKG-h&r#1 and gPKG-h&r#2) used in this protocol are shown in Fig. 4A. Both vectors contain DNA fragments from the 5′ and 3′ UTRs of the PKG genomic locus and bookend internal deletions that constitute most of the PKG coding region. In the first step of the procedure, circular hit-and-run vectors recombine with the targeted locus to create a pseudodiploid, in which a chimeric, functional copy of the gene is flanked by a nonfunctional copy bearing the targeted mutation (5, 6). As with the knockout vector gPKG-KO, recombination between hit-and-run vectors and the PKG locus should occur either within the proximal or distal genomic PKG fragments of the vector to generate two types of pseudodiploid, termed type I or II (5). Of the parasite transformants obtained with vector gPKG-h&r#1, 14 of 86 independent clones examined (16%) were determined to be pseudodiploids by Southern blot analysis (Fig. 6 and Table 3). The second hit-and-run vector, gPKG-h&r#2, resulted in 27 independent transformants, and 5 of these (18%) were pseudodiploids. Interestingly, pseudodiploids derived from vector gPKG-h&r#1 were exclusively of the type II variety, whereas pseudodiploids generated from vector gPKG-h&r#2 were exclusively type I (Table 3).
FIG. 6.
Identification of pseudodiploids at the genomic PKG locus. Circular hit-and-run vectors (gPKG-h&r, Fig. 4) integrate at the PKG genomic locus to form two types of pseudodiploids: type I, which results from recombination within the proximal 3.9-kb vector-borne genomic fragment; and type II, which results from recombination between the distal 4.6-kb (or 3.1 kb) vector fragments. The EcoRI Southern blot (inset) shows the banding pattern observed for “I,” a representative type I recombinant obtained with vector gPKG-h&r#2; “N,” an example of a nonhomologous recombinant resulting from vector integration outside of the PKG genomic locus; “II,” a representative type II recombinant obtained with gPKG-h&r#1; and “w.t.,” wild type. The accompanying illustrations show the banding patterns predicted for wild-type parasites and type I and type II pseudodiploids. Included are EcoRI sites (R), introduced deletions (Δ), and vector sequences including pKS (narrow white bar), and marker HXGPRT (thick white bar).
If Tg-PKG is not an essential gene, then resolution of the pseudodiploids by intrachromosomal homologous recombination should generate both wild-type and knockout mutant segregants. To complete the mutagenesis procedure, four clones representative of each of the two types of pseudodiploid were subjected to 6-TX treatment to select for loss of plasmid vector sequences integrated at the PKG locus. Five 6-TX-resistant clones from each selected population were obtained by limiting dilution and subjected to Southern blot analysis (a total of 40 clones). None of the clones displayed a banding pattern consistent with a successful gene replacement. Instead, the majority displayed the 11- and 13-kb PKG EcoRI fragments characteristic of wild-type segregants (90 and 50% of the clones derived from type I and II pseudodiploids, respectively). The remainder of the clones exhibited the same banding pattern as the parental pseudodiploid, suggesting that resistance to 6-TX in these cases was acquired as a result of inactivation of the HXGPRT-selectable marker during the counterselection procedure or by epigenetic mechanisms.
PKG as the primary determinant of antiparasite efficacy of compound 1 in the murine Toxoplasma model.
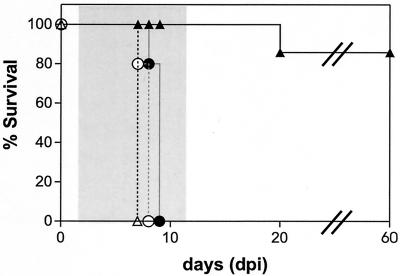
The absence of native enzyme in PKG knockout lines (Fig. 5) and the inferred dependence of such strains on complementing recombinant PKGs for parasite growth (Table 3) allowed us to further test whether this enzyme represents the primary target for the antiparasitic activity of compound 1. To this end, T. gondii strains disrupted at the genomic PKG locus and dependent upon recombinant Tg-PKGs refractory to compound 1 binding (Table 2, T761M-KO and T761Q-KO) were evaluated in a murine toxoplasmosis survival model (Fig. 7). Mice infected with either wild-type parasites or the Tg-PKG T761M-KO or Tg-PKG T761Q-KO strains (103 parasites each) succumbed to acute toxoplasma infections within 8 days. This result demonstrates that the T761M and T761Q Tg-PKG alleles do not compromise parasite virulence in the mouse. As observed previously (26), compound 1 effectively controls an acute toxoplasma infection in 90% of the mice infected with the RH strain or the virulent RHΔHXGPRT variant used in these studies. In contrast, compound 1 is incapable of protecting mice infected with the T761M-KO and T761Q-KO strains; treated animals succumb within 24 h of the untreated control groups infected with the parental strain of Toxoplasma. If the therapeutic efficacy of compound 1 were attributed to an essential biochemical target distinct from PKG, one would predict that the T761M-KO and T761Q-KO strains would be controlled by this treatment regimen. These data provide further support for the conclusion that PKG is the primary molecular target for the antiparasitic activity of compound 1 in T. gondii.
FIG. 7.
Lack of compound 1 efficacy in mice infected with T. gondii strains dependent upon the Tg-T761Q and Tg-T761M alleles. Groups of 10 C57BL/6 mice were infected with 103 tachyzoites of T. gondii and either treated with a twice-daily intraperitoneal dose of 50 mg of compound 1/kg or with vehicle for 10 consecutive days (shaded area). Vehicle-treated mice infected with parental RHΔHXGPRT parasites (▵, dashed line) or with the T761Q-KO strain (○, dashed line) died within 8 days. Mice infected with untransformed parasites and treated with compound 1 survived acute toxoplasmosis (▴), whereas animals infected with the T761Q-KO strain (•) did not respond to compound 1 treatment. Similar results were obtained in a second trial that also included the T761M-KO parasite line (not shown).
DISCUSSION
In a related study (13) we reported that the trisubstituted pyrrole, 4-[2-(4-fluorophenyl)-5-(1-methylpiperidine-4-yl)-1H-pyrrol-3-yl]pyridine (compound 1) has antiparasitic activity against several apicomplexan parasites, including T. gondii. Biochemical analysis and molecular cloning indicate that the chemotherapeutic target of compound 1 is the cGMP-dependent protein kinase (PKG) in E. tenella. Because of the technical limitations faced when working with the Eimeria parasite, we conducted molecular characterization and target validation studies of PKG in the related coccidian pathogen, T. gondii. We expressed Toxoplasma and Eimeria PKG enzymes in Toxoplasma and show here that the recombinant enzymes resemble each other, as well as the respective native enzymes, by all biochemical and kinetic criteria.
In an attempt to understand the molecular basis of drug specificity and drug binding, we used a combination of computer modeling and site-directed mutagenesis approaches to predict and then to elucidate key interactions between compound 1 and residues in the ATP-binding domain of parasite PKG. Guided by the results of previous investigations of the p38 MAP kinase (12, 22), an enzyme that is inhibited by trisubstituted imidazoles that are structurally related to compound 1, we targeted a conserved catalytic-site threonine in PKG from both T. gondii and E. tenella. Our predictions were fulfilled by the observations that amino acid residues T761 of Tg-PKG and T770 of Et-PKG are critical for the binding of compound 1 (Table 1). Like the analogous substitutions at T106 of p38 kinase, T761M and T761Q substitutions within Tg-PKG appear to prevent the binding of compound 1 by blocking access of the fluorophenyl substituent of compound 1 to an active-site hydrophobic pocket. Similar results were demonstrated in the T770M and T770Q substitutions within Et-PKG. The T761 and T770 substitutions within Tg-PKG and Et-PKG, respectively, do not profoundly affect enzyme activity, suggesting that this binding pocket is not important for the binding of ATP or peptide substrates (Table 1). Not only do the substitutions have little affect on the activity of the recombinant enzymes in vitro, but the mutant PKG alleles also have apparently no effect on PKG function in Toxoplasma tachyzoites cultivated in vitro or in vivo. Specifically, the mutant enzymes can complement a PKG genomic disruption that prevents expression of the native Toxoplasma enzyme without a discernible effect on parasite growth or virulence (Fig. 5B and 7).
In this report we have shown that transgenic parasites expressing laboratory-generated alleles of Tg-PKG are far less sensitive to compound 1 in both in vitro and in vivo models. T. gondii strains expressing recombinant Tg-PKG T761M or T761Q mutants exhibit a 19- or 25-fold increase in IC50 compared to wild-type parasites. In the background of a PKG genomic knockout, these values are increased to 25-fold for T761M and 40-fold for the T761Q mutant, respectively (Table 2). The magnitude of the shift in sensitivity to compound 1 is perhaps surprising, given the much more modest changes observed in other apicomplexan drug resistance models (7, 31, 33, 41, 42). In the Toxoplasma dihydrofolate thymidylate synthase (DHFR-TS) model (7, 33), combinations of pyrimethamine resistance alleles of T. gondii DHFR-TS that mirror clinically relevant P. falciparum variants confer only a twofold increase in IC50 in cell-based assays. In the malaria P-glycoprotein (Pgh1) model, resistance to quinine, mefloquine, and halofantrine is modulated over a two- to fourfold range as a result of swapping resistance and/or sensitivity determinants in different field isolates of Plasmodium (31). In the context of such studies, the magnitude in the IC50 shift conferred by the mutant PKG alleles expressed in Toxoplasma reflects a high degree of selectivity of compound 1 for the PKG enzyme relative to other parasite kinases that might also be important for parasite growth. In conclusion, the ability of engineered variants of PKG to confer resistance to compound 1 in transgenic Toxoplasma cultivated in vitro, combined with its lack of efficacy against such strains in the animal model, unequivocally demonstrates that PKG is the primary determinant for parasite sensitivity to the compound 1.
Since the therapeutic use of compound 1 to cure murine toxoplasmosis is not accompanied by signs of toxicity (26), drug selectivity with respect to off-target interactions in the vertebrate host also appears to be favorable. Animal PKG and p38 kinases are the most obvious candidates as enzymes most likely to exhibit off-target interactions with compound 1. PKG is the most closely related host enzyme based on amino acid sequence similarity (13), and p38 enzymes are inhibited by compounds structurally related to compound 1 (12, 22). Primary amino acid sequence alignments of assorted PKG proteins reveal that animal homologues contain a methionine in the positions corresponding to Tg-PKG T761 or Et-PKG T770 (13). From the mutagenesis studies described here for PKG and those published for p38 (12, 22), we know that a bulky side chain in this position will interfere with binding of the compound. Indeed, PKG partially purified from chicken displays at least a 1,000-fold lower affinity for compound 1 than parasite PKG (13). In the case of host p38 enzymes, molecular modeling predicts that binding of compound 1 is suboptimal compared with more potent related compounds (S. Singh, unpublished data).
Aside from validating PKG as the molecular target of compound 1 in Toxoplasma, Tg-PKG and Et-PKG alleles that do not bind this ligand constitute useful dominant selectable markers for the stable transformation of Toxoplasma or other apicomplexan parasites. Transgenic parasites expressing Tg-PKG T761Q, Tg-PKG T761M, Et-PKG T770Q, or Et-PKG T770M alleles were successfully selected by serial passage in 1 μg/ml of compound 1. Transgenic T. gondii lines obtained in this way, such as the Tg-PKG T761Q or T761M strains used for the construction of knockouts (Tables 2 and 3), show no evidence of gene amplification or enhanced resistance compared with analogous transgenic lines obtained by conventional selection with the HXGPRT marker (not shown).
Knockout experiments with targeting vectors provide compelling evidence that the single-copy PKG genomic locus is essential for parasite growth. Two indirect strategies were used to demonstrate the indispensability of the locus, employing gene knockout and “hit-and-run” vectors, respectively. High frequencies of homologous recombination were detected at the genomic locus with either class of vector, but only in parasites expressing a complementing Tg-PKG or Et-PKG cDNA copy (Table 3, knockout vector) or in recombinants in which a functional genomic copy was regenerated at the locus (Table 3, hit-and-run vectors). Attempts to generate strains of T. gondii disrupted at the PKG locus by homologous recombination and which do not express recombinant PKG were never successful and apparently are not viable. The strategy which employed hit-and-run vectors was able to generate several strains with a pseudodiploid PKG genomic locus. Each attempt to resolve these pseudodiploids yielded wild-type segregants but no vector-borne null alleles. Taken together, these data allow us to conclude that PKG is essential for the survival of T. gondii tachyzoites in cell culture, an important criterion for a drug target in a pathogenic organism.
In addition to showing that the Toxoplasma PKG gene is essential, gene knockout experiments have also demonstrated the close functional similarity of the PKG enzymes from Toxoplasma and Eimeria. Toxoplasma PKG knockout lines expressing either acylated (Et-PKGFLAG) or nonacylated (FLAGEt-PKG) Eimeria PKG (Fig. 5A) appear to grow normally, suggesting that acylation per se may not be essential for parasite PKG function, at least in Toxoplasma tachyzoites. The ability of Eimeria PKG to complement a knockout of the Toxoplasma PKG gene (Table 3 and Fig. 5A) also underscores the utility of the Toxoplasma model system to investigate biochemical targets from related parasites. Indeed, because the analogous site-directed mutation in Eimeria PKG confers a similar phenotype when expressed in transgenic Toxoplasma parasites, we can infer that PKG is likely to be a key determinant for compound 1 sensitivity in Eimeria as well. Furthermore, the spectrum of efficacy of compound 1, combined with the amino acid sequence conservation observed among apicomplexan parasite PKG enzymes compared with homologues from their vertebrate hosts (13), makes it likely that PKG catalytic site inhibitors will be useful against other apicomplexan parasites.
Numerous efforts to identify small molecule inhibitors of cellular protein kinases have been aggressively pursued in several therapeutic areas. The most recent and obvious success story in this regard has been the use of the kinase inhibitor STI-571 (Gleevec) to treat chronic myeloid leukemia (8, 37). Unfortunately, there have been no parallel successes in the area of infectious diseases, even insofar as the identification and validation of a protein kinase target. The cGMP-dependent protein kinases of apicomplexan parasites represent the first entry into this arena. Validation of this novel enzyme target for chemotherapeutic intervention provides impetus for future discovery efforts and establishes a precedent for the use of kinase inhibitors to treat infections of pathogenic microorganisms.
Acknowledgments
We thank Tesfaye Biftu for providing compound 1 for our studies. We also thank the University of Pennsylvania (David Roos) and Stanford University (John Boothroyd) for licenses to use Toxoplasma reagents.
REFERENCES
- 1.Black, M., F. Seeber, D. Soldati, K. Kim, and J. C. Boothroyd. 1995. Restriction enzyme-mediated integration elevates transformation frequency and enables cotransfection of Toxoplasma gondii. Mol. Biochem. Parasitol. 74:55-63. [DOI] [PubMed] [Google Scholar]
- 2.Brown, R. E., K. L. Jarvis, and K. J. Hyland. 1989. Protein measurement using bicinchoninic acid: elimination of interfering substances. Anal. Biochem. 180:136-139. [DOI] [PubMed] [Google Scholar]
- 3.Donald, R. G. K., and P. A. Liberator. 2002. Molecular characterization of a coccidian parasite cGMP-dependent protein kinase. Mol. Biochem. Parasitol. 120:165-175. [DOI] [PubMed] [Google Scholar]
- 4.Donald, R. G. K., D. Carter, B. Ullman, and D. S. Roos. 1996. Insertional tagging, cloning and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. J. Biol. Chem. 271:14010-14019. [DOI] [PubMed] [Google Scholar]
- 5.Donald, R. G. K., and D. S. Roos. 1998. Gene knock-outs and allelic replacements in Toxoplasma gondii: HXGPRT as a selectable marker for hit-and-run mutagenesis. Mol. Biochem. Parasitol. 91:295-305. [DOI] [PubMed] [Google Scholar]
- 6.Donald, R. G. K., and D. S. Roos. 1994. Homologous recombination and gene replacement at the dihydrofolate reductase-thymidylate synthase locus in Toxoplasma gondii. Mol. Biochem. Parasitol. 63:243-253. [DOI] [PubMed] [Google Scholar]
- 7.Donald, R. G. K., and D. S. Roos. 1993. Stable molecular transformation of Toxoplasma gondii: a selectable dihydrofolate reductase-thymidylate synthase marker based on drug-resistance mutations in malaria. Proc. Natl. Acad. Sci. USA 90:11703-11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Druker, B. J., C. L. Sawyers, H. Kantarjian, D. J. Resta, S. F. Reese, J. M. Ford, R. Capdeville, and M. Talpaz. 2001. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N. Engl. J. Med. 344:1038-1042. [DOI] [PubMed] [Google Scholar]
- 9.Francis, S. H., and J. D. Corbin. 1999. Cyclic nucleotide-dependent protein kinases: intracellular receptors for cAMP and cGMP action. Crit. Rev. Clin. Lab. Sci. 36:275-328. [DOI] [PubMed] [Google Scholar]
- 10.Fung, H. B., and H. L. Kirschchenbaum. 1996. Treatment regimens for patients with toxoplasmic encephalitis. Clin. Ther. 18:1037-1056. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Echeverria, C., P. Traxler, and D. B. Evans. 2000. ATP site-directed competitive and irreversible inhibitors of protein kinases. Med. Res. Rev. 20:28-57. [DOI] [PubMed] [Google Scholar]
- 12.Gum, R. J., M. M. McLaughlin, S. Kumar, Z. Wang, M. J. Bower, J. C. Lee, J. L. Adams, G. P. Livi, E. J. Goldsmith, and P. R. Young. 1998. Acquisition of sensitivity of stress-activated protein kinases to the p38 inhibitor, SB 203580, by alteration of one or more amino acids within the ATP binding pocket. J. Biol. Chem. 273:15605-15610. [DOI] [PubMed] [Google Scholar]
- 13.Gurnett, A. M., P. A. Liberator, P. M. Dulski, S. P. Salowe, R. G. K. Donald, J. W. Anderson, J. Wiltsie, C. A. Diaz-Saldana, G. S. Harris, B. C. Chang, S. J. Rattray, B. Nare, T. M. Crumley, P. S. Blum, A. S. Misura, T. Tamas, M. K. Sardana, T. Biftu, and D. M. Schmatz. Purification and characterization of cGMP dependent protein kinase from apicomplexan parasites: a novel chemotherapeutic target. J. Biol. Chem., in press. [DOI] [PubMed]
- 14.Haverkos, H. W. 1987. Assessment of therapy for toxoplasma encephalitis: the TE study group. Am. J. Med. 82:907-914. [DOI] [PubMed] [Google Scholar]
- 15.Isrealski, D. M., and J. S. Remington. 1993. Toxoplasmosis in patients with cancer. Clin. Infect. Dis. 17:423-435. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, L. N. 2001. Structural basis for substrate recognition and control in protein kinases. Ernst-Schering-Res-Found-Workshop 34:47-69. [DOI] [PubMed] [Google Scholar]
- 17.Kim, K., D. Soldati, and J. C. Boothroyd. 1993. Gene replacement in Toxoplasma gondii with chloramphenicol acetyltransferase as selectable marker. Science 262:911-914. [DOI] [PubMed] [Google Scholar]
- 18.Knighton, D. R., J. Zheng, L. F. Ten Eyck, V. A. Ashford, N.-H. Xuong, S. S. Taylor, and J. M. Sowadski. 1991. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253:407-414. [DOI] [PubMed] [Google Scholar]
- 19.Leport, C., F. Raffi, S. Matheron, C. Katlama, B. Regnier, A. G. Saimot, C. Marche, C. Vedrenne, and J. L. Vilde. 1988. Treatment of central nervous system toxoplasmosis with pyrimethamine/sulfadiazine combination in 35 patients with the acquired immunodeficiency syndrome. Efficacy of long-term continuous therapy. Am. J. Med. 84:94-100. [DOI] [PubMed] [Google Scholar]
- 20.Levitt, M. 1992. Accurate modeling of protein conformation by automatic segment matching. J. Mol. Biol. 226:507-533. [DOI] [PubMed] [Google Scholar]
- 21.Levitt, M. 1983. Protein folding by restrained energy minimization and molecular dynamics. J. Mol. Biol. 170:723-764. [DOI] [PubMed] [Google Scholar]
- 22.Lisnock, J., A. Tebben, B. Frantz, E. A. O'Neill, G. Croft, S. J. O'Keefe, B. Li, C. Hacker, S. de Laszlo, A. Smith, B. Libby, N. Liverton, J. Hermes, and P. LoGrasso. 1998. Molecular basis for p38 protein kinase inhibitor specificity. Biochemistry 37:16573-16581. [DOI] [PubMed] [Google Scholar]
- 23.Luft, B. J., and J. S. Remington. 1988. AIDS commentary: toxoplasmic encephalitis. J. Infect. Dis. 157:1-6. [DOI] [PubMed] [Google Scholar]
- 24.Luft, B. J., R. Hafner, A. H. Korzun, C. Leport, D. Antoniskis, E. M. Bosler, D. D. Bourland, R. Uttamchandani, J. Fuhrer, J. Jacobson, et al. 1993. Toxoplasmic encephalitis in patients with acquired immunodeficiency syndrome. N. Engl. J. Med. 329:995-1000. [DOI] [PubMed] [Google Scholar]
- 25.Luft, B. J., and J. S. Remington. 1992. Toxoplasmic encephalitis in AIDS. Clin. Infect. Dis. 15:211-222. [DOI] [PubMed] [Google Scholar]
- 26.Nare, B., J. Allocco, P. Liberator, and R. G. K. Donald. 2002. Evaluation of a cGMP-dependent protein kinase inhibitor in the treatment of murine toxoplasmosis: gamma-interferon is required for efficacy. Antimicrob. Agents Chemother. 46:300-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfefferkorn, E. R., and S. Borotz. 1994. Toxoplasma gondii: characterization of a mutant resistant to 6-thioxanthine. Exp. Parasitol. 79:374-382. [DOI] [PubMed] [Google Scholar]
- 28.Pfefferkorn, E. R., and L. C. Pfefferkorn. 1977. Specific labeling of intracellular Toxoplasma gondii with uracil. J. Protozool. 24:449-453. [DOI] [PubMed] [Google Scholar]
- 29.Pfeifer, A., P. Klatt, S. Massberg, L. Ny, M. Sausbier, C. Hirneiss, G.-X. Wang, M. Korth, A. Aszódi, K.-E. Andersson, F. Krombach, A. Mayerhofer, P. Ruth, R. Fässler, and F. Hofmann. 1988. Defective smooth muscle regulation in cGMP kinase I-deficient mice. EMBO J. 17:3045-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfeifer, A., A. Aszodi, U. Seidler, P. Ruth, F. Hofmann, and R. Fassler. 1996. Intestinal secretory defects and dwarfism in mice lacking cGMP-dependent protein kinase. Science 274:2082-2086. [DOI] [PubMed] [Google Scholar]
- 31.Reed, M. B., K. J. Saliba, S. R. Caruana, K. Kirk, and A. F. Cowman. 2000. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403:906-909. [DOI] [PubMed] [Google Scholar]
- 32.Remington, J. S., and G. Desmonts. 1990. Toxoplasmosis, p. 89-195. In J. S. Remington and J. O. Klein (ed.), Infectious diseases of the fetus and newborn infant. The W. B. Saunders Co., Philadelphia, Pa.
- 33.Reynolds, M., and D. S. Roos. 1998. A biochemical and genetic model for parasite resistance to antifolates: Toxoplasma gondii provides insights into pyrimethamine and cycloguanil resistance in Plasmodium falciparum. J. Biol. Chem. 273:3461-3469. [DOI] [PubMed] [Google Scholar]
- 34.Roos, D. S., R. G. K. Donald, N. S. Morrissette, and A. L. C. Moulton. 1994. Molecular tools for the genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol. 45:27-63. [DOI] [PubMed] [Google Scholar]
- 35.Ruth, P. 1999. Cyclic GMP-dependent protein kinases: understanding in vivo functions by gene targeting. Pharmacol. Ther. 82:355-372. [DOI] [PubMed] [Google Scholar]
- 35a.Salowe, S. P., J. Wiltsie, P. A. Liberator, and R. G. K. Donald. 2002. The role of a parasite-specific allosteric site in the distinctive activation behavior of Eimeria tenella cGMP-dependent protein kinase. Biochemistry 41:4385-4391. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J. F., D. W. Russell, and N. Irwin (ed.). 2000. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Schindler, T., W. Bornmann, P. Pellicena, W. T. Miller, B. Clarkson, and J. Kuriyan. 2000. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science 289:1938-1942. [DOI] [PubMed] [Google Scholar]
- 38.Soldati, D., and J. C. Boothroyd. 1993. Transient transfection and expression in the obligate intracellular parasite Toxoplasma gondii. Science 260:349-352. [DOI] [PubMed] [Google Scholar]
- 39.Sridhar, R., O. Hanson-Painton, and D. R. Cooper. 2000. Protein kinases as therapeutic targets. Pharmacol. Res. 17:1345-1353. [DOI] [PubMed] [Google Scholar]
- 40.Wang, Z., B. J. Canagarajah, J. C. Boehm, S. Kassisa, M. H. Cobb, P. R. Young, S. Abdel-Meguid, J. L. Adams, and E. J. Goldsmith. 1998. Structural basis of inhibitor selectivity in MAP kinases. Structure 6:1117-1128. [DOI] [PubMed] [Google Scholar]
- 41.van der Wel, A. M., A. M. Tomas, C. H. Kocken, P. Malhotra, C. J. Janse, A. P. Waters, and A. W. Thomas. 1997. Transfection of the primate malaria parasite Plasmodium knowlesi using entirely heterologous constructs. J. Exp. Med. 185:1499-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, Y., L. A. Kirkman, and T. E. Wellems. 1996. Transformation of Plasmodium falciparum malaria parasites by homologous integration of plasmids that confer resistance to pyrimethamine. Proc. Natl. Acad. Sci. USA 93:1130-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]