Abstract
Purpose
Liver cancers are among the deadliest malignancies due to a limited efficacy of early diagnostics, the lack of appropriate biomarkers and insufficient discrimination of different types of tumors by classic and molecular methods. In this study, we searched for novel long non-coding RNA (lncRNA) as well as validated several known candidates suitable as probable biomarkers for primary liver tumors of various etiology.
Methods
We described a novel lncRNA HELIS (aka “HEalthy LIver Specific”) and estimated its expression by RT-qPCR in 82 paired tissue samples from patients with hepatocellular carcinoma (HCC), cholangiocarcinoma (CCA), combined HCC-CCA, pediatric hepatoblastoma (HBL) and non-malignant hepatocellular adenoma (HCA) and focal nodular hyperplasia (FNH). Additionally, we examined expression of cancer-associated lncRNAs HULC, MALAT1, UCA1, CYTOR, LINC01093 and H19, which were previously studied mainly in HCC.
Results
We demonstrated that down-regulation of HELIS strongly correlates with carcinogenesis; whereas in tumors with non-hepatocyte origin (HBL, CCA) or in a number of poorly differentiated HCC, this lncRNA is not expressed. We showed that recently discovered LINC01093 is dramatically down-regulated in all malignant liver cancers; while in benign tumors LINC01093 expression is just twice decreased in comparison to adjacent samples.
Conclusion
Our study revealed that among all measured biomarkers only down-regulated HELIS and LINC01093, up-regulated CYTOR and dysregulated HULC are perspective for differential diagnostics of liver cancers; whereas others demonstrated discordant results and cannot be considered as potential universal biomarkers for this purpose.
Electronic supplementary material
The online version of this article (10.1007/s00432-020-03378-5) contains supplementary material, which is available to authorized users.
Keywords: LncRNA, Liver cancer, Biomarkers, Hepatocellular carcinoma, Cholangiocarcinoma, Hepatoblastoma
Introduction
Non-coding RNAs (ncRNAs) play important roles in various cellular processes, including cancer development and invasion (Slack et al. 2019; Anastasiadou et al. 2018). Many ncRNAs have oncogenic or tumor-suppressing functions and are often dysregulated during carcinogenesis. Therefore, some of them are considered as promising biomarkers and targets for cancer therapy (Matsui and Corey 2017; Ning et al. 2019). NcRNAs can circulate within exosomes in the body; so, ncRNA-based liquid biopsy (Liang et al. 2019; Pardini et al. 2019) or tissue biopsy after surgery (Slack et al. 2019) can be used for diagnostics and/or for prognosis of the treatment outcomes. Two main classes of ncRNAs are of the main interest for cancer diagnostics – miRNAs (Lan et al. 2015; Lujambio et al. 2012) and long non-coding RNAs (lncRNAs) with length more than 200 nt (Chi et al. 2019; Bohla et al. 2017) which participate in complex network regulation of gene expression. MALAT1, HOTTIP, NEAT1, ANRIL, etc. are the most well-known lncRNAs among hundreds of probable universal biomarkers dysregulated in different types of cancers (Wang et al. 2020). In contrast, only few lncRNAs are considered as tissue-specific ones. Among them are PCAT1 and PCA3 (prostate) (Misawa et al. 2017), HULC (liver) (Yu et al. 2017), BANCR (skin) (Zou et al. 2017) and SOX2-OT (brain) (Shahryari et al. 2015). Thus, identification and validation of novel lncRNA biomarkers with specificity for certain cancer type remain an unmet medical need.
In the present research, we focused on lncRNAs associated with cell transformation in the liver. Liver tumors are widespread malignancies with relatively low survival rates of patients that comes from insufficient early diagnostics (Dhanasekaran et al. 2012; Dimitroulis et al. 2017). Hepatocellular carcinoma (HCC) is the most common liver cancer and an often cause of death for patients with hepatitis and cirrhosis (Mak et al. 2018). Commonly used blood serum biomarker – alpha fetoprotein (AFP) – is not good enough for routine HCC prediction (Tsuchiya et al. 2015; De Stefano et al. 2018). At the same time, many lncRNAs are dysregulated in HCC (Klingenberg et al. 2017; Qiu et al. 2017; Wong et al. 2018) and can even play an important role in the development of HCC drug resistance (Ding et al. 2018; Wei et al. 2019). Some lncRNA can be found in circulating exosomes by liquid biopsy that demonstrate their high potential for non-invasive diagnostics of HCC (Yuan et al. 2017; Tan et al. 2019). Recently, two novel liver-specific lncRNAs were reported to be down-regulated in HCC – LINC01093 (He et al. 2019) and FAM99A (Esposti et al. 2016). Meanwhile, the best-known HCC-associated lncRNA HULC was shown to be also involved in the development of gastric and colorectal cancers, osteosarcoma, diffuse large B cell lymphoma and glioma (Ma et al. 2017).
Cholangiocarcinoma (CCA) is another widespread liver cancer (~ 10–20% of all liver malignancies) originated mainly from cholangiocytes/bile duct cells (Rizvi et al. 2018; Zhang et al. 2016b). In some cases, HCC and CCA can be hardly distinguished and even combined HCC-CCA cancers (cHCC-CCA) occur as well (Komuta and Yeh 2019). To date, specific biomarker to diagnose CCA is not known: AFP is not increased in the blood serum, while carbohydrate antigen (CA) 19-9 has low specificity. CA19-9 is also elevated in pancreatic, colorectal, gastric cancers and several non-malignant conditions (Khan et al. 2012). However, CCA development results in the upregulation of many lncRNAs—different ones in comparison to HCC (Li et al. 2019a).
Another highly malignant liver tumor is hepatoblastoma (HBL). HBL is common for infant children, preferentially within the first two years of life (Lim et al. 2018). Being an embryonal tumor HBL derives from liver precursor cells and possess heterogeneous features assigning to fetal, mixed embryonic/fetal, epithelial/mesenchymal and other origin (Ng and Mogul 2018). So far, lncRNAs with specific functions in HBL are not known, although several miRNAs modulate Wnt/β-catenin pathway in this cancer (Indersie et al. 2017). Clinical relevance of AFP monitoring in HBL patients is controversial (Ferraro et al. 2019).
Among non-malignant liver tumors, two major types are focal nodular hyperplasia (FNH) and hepatocellular adenoma (HCA) – benign liver tumors of hepatocellular origin with asymptomatic development (Roncalli et al. 2016). Despite their non-malignant nature, these lesions usually undergo surgical resection to avoid liver injuries and decrease risks of cancer. HCA has a strong potential for malignant transformation. Moreover, immunohistochemical analyses of biopsies using GPC3, HSP70, glutamine synthetase (GS) or β-catenin staining cannot completely attribute these heterogeneous tumors to HCA, FNH, or well-differentiated HCC (Cristiano et al. 2014). Thus, search for new biomarkers that may help to discriminate benign and malignant liver tumors is challenging. However, data on the expression of any HCC-related or other specific lncRNAs in benign liver tumors are not available to date.
Here, we aimed to find lncRNAs dysregulated in primary liver cancers with the option to discriminate different types of tumors. We found a novel lncRNA AC068535.3 (synonym LINC01831, NR_135589) significantly downregulated in cancer tissues. Due to its active expression almost in normal liver, we named AC068535.3 as HELIS aka “HEalthy LIver Specific”. To estimate probable potential of HELIS as a diagnostic biomarker, we analyzed its expression in various types of liver tumors. Additionally, we performed integrated analysis of these samples by quantification of other lncRNAs: LINC01093, H19, HULC, MALAT1, UCA1 and CYTOR (Lim et al. 2019), which were not investigated earlier in various types of primary liver tumors.
Materials and methods
Patient samples
In this study, tumor specimens and adjacent liver tissues were collected from 75 patients diagnosed with HCC, intrahepatic CCA, cHCC-CCA, FNH, HCA or unspecified tumor origin (Table 1) after tumor resection in FSBI “N.N. Blokhin National Medical Research Center of Oncology” of the Ministry of Health of the Russian Federation, as well as paired cancer/normal samples of kidney and pancreas used as controls in the present study (Table S1). HBL samples and adjacent tissues (7 pairs, Table 1) as well as five liver donor samples (Table S1) were collected in Petrovsky Russian Research Centre of Surgery. General demographical features of the samples are summarized in Table 1. Additional clinicopathological characteristics for HCC, CCA and cHCC-CCA cases are available in Supplementary Tables S5, S6 and S7, respectively. All samples were hepatitis negative except of the one cHCC-CCA case with HBV infection (Table S7). Diagnosis and origin of samples (tumor or normal) were confirmed by histopathological analysis performed by two independent expertise in each case. All samples were fresh frozen in liquid nitrogen after tumor resection and stored at − 80 °C.
Table 1.
Number of patients with different diagnoses enrolled in the present study and their general demographical characteristics
| Diagnosis | HCC | CCA | cHCC/CCA | HBL | FNH | HCA | Unspecified |
|---|---|---|---|---|---|---|---|
| Total (82) | 45 | 9 | 5 | 7 | 10 | 3 | 3 |
| Gender | |||||||
| Male | 27 | 6 | 2 | 3 | 5 | 1 | 3 |
| Female | 18 | 3 | 3 | 4 | 5 | 2 | 0 |
| Age, years | * | 32, 36, 56 years | 41, 52, 71 years | ||||
| ≤ 60 | 29 | 4 | 2 | 10 | |||
| >60 | 16 | 5 | 3 | 0 | |||
| Median | 54 years | 62 years | 61 years | 33 years |
*HBL samples were collected from children with age from 7 months to 4 years with median of 14 months
The study was approved by the Ethics Committee of the “N.N. Blokhin National Medical Research Center of Oncology” of the Ministry of Health of the Russian Federation and the Local Ethics Committee of the Petrovsky National Research Centre of Surgery. All procedures were performed in accordance with Declaration of Helsinki (1964) and its later amendments (World Medical Association 2013). Written informed consent was obtained from all individual participants included in the study or their legally authorized representatives (in case of HBL samples).
RNA isolation and RT-qPCR
Liver tissues of CCA, HBL, cHCC-CCA, donor liver samples and paired kidney and pancreas samples (~ 50–100 mg) were homogenized by Precellys Lysing kit CK14 (soft tissue homogenizing) in 1 mL of Trizol reagent (Invitrogen) followed by standard protocol for total RNA extraction and isopropanol precipitation. Resolved RNA was subjected to DNAse I treatment (Thermo Scientific). For extraction of total RNA from HCC, FNH and HCA samples, commercial PureLink RNA Mini Kit (Life Technologies) was used including on column DNAse I treatment (PureLink DNAse Kit, Life Technologies). RNA samples were subjected to reverse transcription reaction by Maxima H Minus First Strand cDNA Synthesis Kit (Thermo Scientific) according to the manufacture’s protocol. Obtained cDNA samples in total volume of 20 μL were diluted by RNAse-free H2O (Sigma) up to 100 μl. Five μL of these cDNA samples was mixed with 2 μL of 5 × qPCRmix-HS master mix (Evrogen, Russia), 0.6 μL of the primer mix (10 μM each) and 2.4 μL of H2O. Three-step cycling reactions were conducted in CFX96 Touch Real-Time PCR Detection System (Bio-Rad). RNA amounts were calculated by 2(−ΔCt) method using U6 snRNA for normalization. Primers used in the study are listed in Supplementary Table S2.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 7 software. All represented graphs are expressed as the median with the range from minimal to maximal meaning. The expression of all measured parameters in tumor and adjacent tissues was compared by a paired sample Student’s t-test. All p-values were two-sided, and p < 0.05 was accepted as statistically significant. Chi square test was performed to evaluate the association between lncRNA expression and clinicopathological parameters of each HCC case. Correlations between levels of HELIS and size of benign tumors was analyzed by Spearman’s rank correlation test.
Results
Identification of novel liver-specific lncRNA HELIS
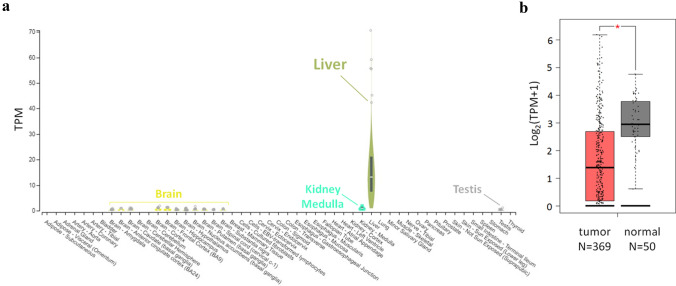
For bioinformatic analysis, we used public transcriptome data from The Cancer Genome Atlas (TCGA) to identify novel lncRNAs that are differentially expressed in HCC tissues and healthy liver. Among dozens of revealed lncRNAs, one located in 2q12.1 locus demonstrated unambiguous liver-specific expression according to GTEx data (Fig. 1a) and was down-regulated in HCC in comparison to healthy tissues (Fig. 1b). Previously, this lncRNA was just mentioned by Zhang et al. (2016a) among other 115 differentially expressed lncRNAs detected during integrative analysis of multiple RNA-Seq datasets for HCC.
Fig. 1.
High tissue-specificity of HELIS lncRNA and changes of its expression upon liver carcinogenesis. a GTEx data (www.gtexportal.org) for expression of HELIS in different healthy human tissues, average expression in liver represents 226 cases. b Decreased expression levels of HELIS lncRNA in HCC according to TCGA dataset visualized by Gene Expression Profiling Interactive Analysis (GEPIA, www.gepia.cancer-pku.cn)
Expression of HELIS and other markers in donor liver and control tissues
First, we confirmed HELIS expression in liver tissues from five healthy donors (Table S1) by RT-qPCR (Table S2). We confirmed tissue specificity of HELIS as no expression was found in paired (cancer/adjacent) samples of kidney and pancreas (Table S1, Fig. S1a). Similar results were obtained for HULC and LINC01093 that are known as liver-specific lncRNAs. In contrast, MALAT1 and CYTOR were detected in all liver and non-liver samples with relative up-regulation in cancer tissues (Fig. S1a). H19 was absent in kidney samples, while its expression was observed in liver and at low levels in pancreas. In case of kidney and pancreas, UCA1 was detected only in cancer tissues, but in the liver it was also expressed in healthy samples (Fig. S1a). We also quantified mRNA levels of liver- or HCC-specific proteins – GPC3, ALB, and ASGR1. As expected, they all showed high expression in the liver, and GPC3 mRNA demonstrated elevated levels in pancreas (Fig. S1b).
Expression of HELIS and other markers in HCC samples
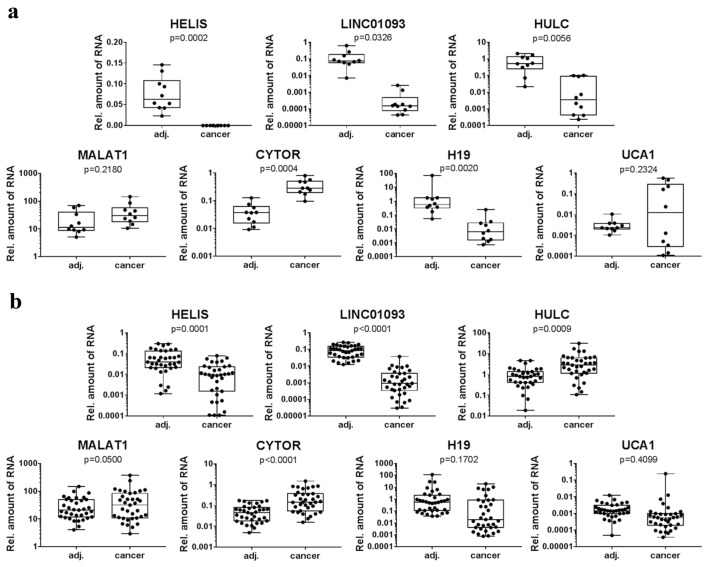
To validate dysregulation of HELIS expression in HCC, we analyzed 45 post-surgery liver tumor samples from patients with this diagnosis in comparison to adjacent normal tissues. Obtained data revealed dissection of samples in two subpopulations: in 10 HCC cases (Group 1) HELIS was not detected at all (Fig. 2a), and another 35 cases (Group 2) demonstrated significant down-regulation of HELIS expression (Fig. 2b). Notably, those 10 cancer tissues from Group 1 also expressed just residual amounts of HULC and LINC01093. In contrast, Group 2 demonstrated clear activation of HULC expression. Levels of MALAT1 and UCA1 were not markedly affected in all HCC population, and only up-regulation of CYTOR was statistically significant for both groups of samples. Relevant down-regulation of H19 was demonstrated only for Group 1. Notably, we have not observed any strong correlations between expression levels of these four lncRNAs (MALAT1, UCA1, H19, CYTOR) and clinicopathological features of patients. However, we revealed several trends of linked expression of certain pairs of biomarkers. For example, UCA1 and CYTOR were gradually up-regulated in HCC tissues, and H19 expression was positively correlated with levels of GPC3 and ALB mRNAs (Fig. S2).
Fig. 2.
Relative expression of HELIS and other lncRNAs in paired (adjacent/cancer) liver samples from patients diagnosed with HCC. Samples representing those with no expression of HELIS (a) and relatively low expression of HELIS (b) in cancer tissues
Two distinct groups of samples also differed in expression of GPC3, ALB, and ASGR1 mRNAs (Fig. S3), that was additionally approved by revealed correlations (Spearman’s rank test) between their levels and amount of HELIS (Table S8). Tumor samples with the lack of HELIS demonstrated strong transcription down-regulation of ALB and ASGR1 genes, while GPC3 changed ambiguously (Fig. S3a). In contrast, Group 2 showed less prominent down-regulation of ALB and ASGR1 mRNAs and significant up-regulation of GPC3 mRNA(Fig. S3b), that is common for liver carcinogenesis (Zhou et al. 2018). Reduced expression of ALB mRNA is also frequently observed in HCC as associated with cancer aggressiveness, larger tumor size and poor patients’ survival (Carr et al. 2017; Bagirsakci et al. 2017). In contrast, HCC-related down-regulation of ASGR1 mRNA is not a critical hallmark of tumorigenesis due to a wide range of variations in its expression levels (Shi et al. 2013).
Expression of HELIS and other markers in CCA and HBL samples
HELIS was detected neither in intrahepatic CCA nor in HBL tumor tissues in contrast to corresponding adjacent liver samples (Fig. 3). LINC01093 was not expressed in CCA and only residual amounts were observed in case of HBL tumors. Similar tendency was demonstrated for HULC. Other lncRNAs also changed uniformly in CCA and HBL, but only up-regulation of CYTOR was statistically significant (Fig. 3). In line with not-hepatocyte origin of CCA, these tumors expressed only residual amounts of ALB and ASGR1 mRNAs, and no changes in GPC3 transcription levels were detected (Fig. S4). Previously low levels of ASGR1 protein were detected in CCA tissues by immunostaining (Gu et al. 2016), and GPC3 was reported to be not affected in this type of cancer (Man et al. 2005; Hass et al. 2018). In case of HBL samples, expression of these three measured mRNAs was not informative (Fig. S4). No data are available about any diagnostic potential of ASGR1 or ALB in HBL, and GPC3 is not considered as a suitable biomarker for this liver malignancy (Zhou et al. 2017).
Fig. 3.
Relative expression of HELIS and other lncRNAs in paired (adjacent/cancer) liver samples from patients diagnosed with CCA (a) or HBL (b)
Expression of HELIS and other markers in cHCC-CCA samples
Considering the fact that at least some measured lncRNAs demonstrated different behavior in case of HCC and CCA, we also analyzed five extremely rare cases of cHCC-CCA tumors (Table S7). Interestingly, we have observed dissection of this small population into two parts. Three cases showed total lack of HELIS and LINC1093 and just residual expression of HULC in tumors; whereas, other two cases demonstrated either up-regulation or no changes in HULC levels in line with still active HELIS expression and detectable amounts of LINC1093 (Fig. S5). We have not found any correlation of these results with general clinicopathological features of the tumors (Table S7). However, cHCC-CCA tumors are known to be highly heterogenic and are divided into three so-called “Allen and Lisa subtypes” (according to Allen and Lisa, 1949) such as separate type (two histologically different lesions), combined type (defined HCC and CCA components in the same tumor) and mixed type (mixed HCC and CCA components with no boundaries). Genomic and transcriptomic analysis of cHCC-CCA tumors revealed significant differences between these subtypes including changes in expression levels of many oncomarkers. It was shown that combined type of cHCC-CCA has CCA-like features and mixed type is more similar to HCC (Xue et al. 2019). Discrimination of cHCC-CCA subtypes by routine histopathological analysis of tumor samples seems to be rather challenging and was beyond the scope of the present study. Thus, we can only hypothesize that those samples with lack of HELIS might belong to combined subtype of cHCC-CCA; whereas, two other cases showed features of mixed cHCC-CCA tumors. Notably, CYTOR was up-regulated in all cases and other measured parameters were not informative (Fig. S5).
Expression of HELIS and other markers in non-malignant tumors
To investigate changes of lncRNA levels in non-malignant tumors, we studied ten samples with FNH and three samples with HCA. As no distinctive features of HCA samples were observed for all measured markers, we decided to represent these benign tumors as one population. All tested samples expressed both HELIS and LINC01093 (Fig. 4). In benign tumors, HELIS expression was similar to the major part (Group 2) of HCC samples, while expression of LINC01093 in FNH and HCA was much higher than in HCC. Namely, just ~ twofold LINC01093 level decrease was demonstrated in comparison to several orders in case of HCC. Significant up-regulation of HULC in non-malignant tumors (Fig. 4) makes it doubtful whether it can be used as a discriminating biomarker for benign and cancer tissues. Similar pattern of expression was demonstrated for CYTOR, while other lncRNAs were not relevant in our study. Besides, all benign tumors showed significant increase of GPC3 mRNA, while ALB and ASGR1 mRNAs were not changed (Fig. S6). Despite HELIS expression in benign tumors did not deviate from that in HCC, we observed strong negative correlation between amount of HELIS and size of HCA/FNH tumors (Fig. S7). Thus, widely spread formations express less HELIS.
Fig. 4.
Relative expression of HELIS and other lncRNAs in paired (adjacent/benign tumor) liver samples from patients diagnosed with FNH or HCA
Expression of HELIS and other markers in three tumors of unspecified origin
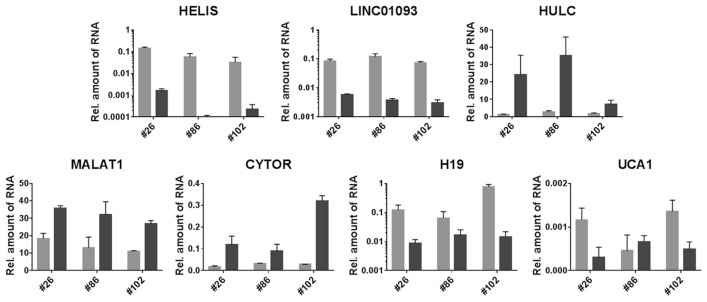
To assess potential diagnostic value of all studied biomarkers, we performed blind analysis of three tumor samples with unspecified origin. All samples showed significant down-regulation of HELIS and 15–30-fold decrease of LINC01093 expression, as well as reduction of H19 (Fig. 5) and ALB mRNA (Fig. S9). HULC, CYTOR and MALAT1 were up-regulated in all cases (Fig. 5); whereas, GPC3 mRNA increased only in case of samples #26 and #102 (Fig. S8). High expression of HULC together with the presence of HELIS exclude ranking these tumors to CCA and HBL, while significant down-regulation of HELIS and LINC01093 allow us to assume malignant HCC origin of the examined samples. This hypothesis appeared to match perfectly to the disclosed histological description of the tumors (Table S3). In particular, all three tumors were initially diagnosed as benign. However, post-surgery histology revealed their ambiguous features related to malignant HCC or at least to mixed HCC/HCA (HCC/FNH for case#102) genesis.
Fig. 5.
Relative expression of HELIS and other lncRNAs in three pairs of liver samples with unspecified origin of tumor (black bars – tumor, gray bars – adjacent liver tissues)
Discussion
Identification of new biomarkers for cancer diagnostics is an actual task of the biomedical research. In the present study, we revealed differential expression of novel lncRNA HELIS in various types of primary liver tumors and investigate its probable value for cancer diagnostics.
Upon estimation of HELIS expression in HCC, we observed a clear division of samples into two subgroups: Group 1 (10 cases, Fig. 2a) was characterized by total lack of HELIS and Group 2 (35 cases, Fig. 2b) demonstrated its significant down-regulation. We estimated probable relationship of HELIS loss in each HCC case characterized by clinical pathology (Tables S4, S5, S8) and revealed a certain link with tumor differentiation grade. In particular, lack of HELIS was a distinctive feature only for moderately or poorly differentiated tumors (G2 + G3); while all highly differentiated (G1) species still expressed low levels of HELIS. Besides, in all fibrolamellar tumors (for which Edmondson–Steiner differentiation grading is not applicable) HELIS demonstrated low but detectable levels of expression (Table S4). We also observed that low expression of HELIS clearly correlates with the reduction of HULC (Fig. S9a) and lack of HELIS is connected to the essential decrease of ALB, GPC3 and ASGR1 mRNA levels (Table S8). We could not observe any effect of HELIS lack on overall or recurrence-free survival of HCC patients (Tables S4, S5, S8). TCGA data also do not show any correlation between HELIS expression and survival rate of HCC patients (Fig. S10). Thus, we propose that HELIS has limited prognostic potential and should be used mainly in diagnostics.
We also demonstrated a complete loss of HELIS in all samples diagnosed as CCA and HBL and its down-regulation in benign tumors. Although expression rates of HELIS in FNH/HCA were comparable to those for majority of HCC, decrease of HELIS levels in these benign tissues strongly correlated with the increase of tumor size (Fig. S7). Notably, no such trend was observed for HCC tumors. In contrast, certain reduction of HELIS was observed for liver tissues adjacent to HCC tumors of a large size (Fig. S9b). We suggest that extended lesions intensively influence on surrounding tissues that might result into dysregulation of lncRNA expression as well. Crucial difference between adjacent tumor samples and non-HCC-derived (“super normal”) liver tissues was reported by Sonohara et al. in the context of MALAT1 and HULC expression levels (Sohohara et al. 2017).
Surprisingly, in our study MALAT1 and UCA1 were not up-regulated in the majority of HCC samples. Several groups reported the absence of expected activation of MALAT1 expression in HCC (in particular in case of comparison between tumor and adjacent non-tumor pair samples (Konoshi et al. 2016; Liu et al. 2014; Sohohara et al. 2017) as well as unchanged UCA1 and H19 levels (Yang et al. 2015). Although in our work H19 was down-regulated in most of HCC samples (Fig. 1). Statistical significance was poor, mainly due to the vast variability of expression between samples. We further observed no changes of expression of these three markers MALAT1, UCA1 and H19 in all other types of investigated liver tumors as well, in particular in case of CCA. In contrast to our data, up-regulation of H19 (Xu et al. 2017a) as well as enhanced expression of UCA1 and MALAT1 (Li et al. 2019b; Tan et al. 2017; Xu et al. 2017b) were reported for CCA previously. Among other tested lncRNAs, CYTOR (former LINC00152) was upregulated in all types of investigated liver cancers. Notably, in case of HCC the levels of CYTOR were much higher than for other types of tumors. Thus, for the first time, we have shown here that CYTOR increases in different types of liver tumors including non-malignant ones. This fact confirms that this lncRNA is a universal biomarker for different liver tumors, but does not allow distinguishing malignant specimens from benign ones.
Variable patterns of expression in the examined tumor samples were observed for liver-specific lncRNAs HULC and LINC01093. Up to date, there were no studies considering expression of these two lncRNAs in liver tumors except of HCC. For the first time, we showed loss of HULC in CCA, its up-regulation in benign liver tumors and no relevant changes in HBL. Thus, HULC levels are not informative for discrimination of HCC and benign liver tissues, but still can be used in diagnostics of CCA. Levels of LINC01093 significantly dropped down in HBL, and total loss of this lncRNA in case of CCA was demonstrated. In contrast, for FNH and HCA tumors, relatively high expression of LINC01093 was observed – only twofold decrease in comparison to adjacent tissues. Regarding this fact we proposed using LINC01093 expression level as an effective tool for analysis of histologically controversial cases with ambiguous morphological features and showed its probable potential for three unspecified liver tumors enrolled in the present research for blind analysis. Up-regulation of CYTOR and HULC (~ tenfold) together with the crucial decrease of both HELIS and LINC01093 (Fig. 5) apparently corresponded to malignant origin of the examined liver species as was further confirmed by rigorous histopathological description (Table S3). In particular, all three cases were initially determined as non-malignant followed by further recategorization to early-staged HCC or mixed benign/HCC. Thus, we illustrated potential of our lncRNA-based panel at least for additional diagnostic of difficult ambiguous cases.
In summary, we demonstrated that liver-specific lncRNA HELIS is highly expressed in healthy liver tissues and drops down during tumorigenesis. Until now, only lncRNA LINC01093 was known to have the similar expression pattern. For the first time, we showed distinct behavior of these two lncRNAs in malignant and benign tumors that indicates their capacity to distinguish different types of liver tumors in postoperative diagnosis or in biopsies. We also studied several lncRNAs upregulated in liver cancers and have selected HULC and CYTOR for improved tumor identification. We assume that simultaneous quantification of these four lncRNAs can allow to define a particular type of liver tumor (Table 2). This feature might be helpful in resolving issues for tumors with unspecified or mixed genesis. Undoubtedly, before going to practice a thorough validation of the proposed lncRNA, the diagnostic panel should be carried out using extended sets of liver cancer samples with different etiologies.
Table 2.
Expression levels of lncRNAs HELIS, HULC, LINC1093 and CYTOR in normal liver and under tumors development
| HELIS | LINC01093 | HULC | CYTOR | |
|---|---|---|---|---|
| Healthy liver | +++ | +++ | + | + |
| HCC | ‒/+ | + | +/++ | +++ |
| CCA | ‒ | ‒ | ‒ | ++ |
| HBL | ‒ | + | + | ++ |
| Benign FNH/HCA | + | ++ | ++ | ++ |
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
OYB: performed experiments, analyzed and interpreted the data and wrote initial draft; NLL: analyzed data and participated in the manuscript writing; IFK: performed some initial experiments; DASh, DASk.: collected and characterized samples; EAM: performed histopathological analysis of samples; NEK, YIP, AVM, EFK: performed surgery, collection of tissue samples and monitored outcome of patients; TSZ: provided conceptual advice and wrote the manuscript; MPR, OAD: designed research and wrote the manuscript. We confirm that the manuscript has been read and approved by all named authors, and the order of authors listed in the manuscript is accepted by all of us.
Funding
This study was funded by Russian Science Foundation (RSF) grant №18-74-00120 for O.Y.B.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest neither competing interests.
Ethical approval
We confirm that all experiments involving human liver samples were approved by medical ethics committee of our institutions. All procedures were performed in accordance with Declaration of Helsinki (1964) and its later amendments (World Medical Association, 2013).
Consent to participate
Written informed consent for the use of tissue samples for scientific purposes was obtained from all individual participants included in the study.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Allen RA, Lisa JR (1949) Combined liver cell and bile duct carcinoma. Am J Pathol 25:647–655 [PMC free article] [PubMed] [Google Scholar]
- Anastasiadou E, Jacob LS, Slack FJ (2018) Non-coding RNA networks in cancer. Nat Rev Cancer 18:5–18. 10.1038/nrc.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagirsakci E, Sahin E, Atabey N, Erdal E, Guerra V, Carr BI (2017) Role of albumin in growth inhibition in hepatocellular carcinoma. Oncology 93:136–142. 10.1159/000471807 [DOI] [PubMed] [Google Scholar]
- Bolha L, Ravnik-Glavac M, Glavac D (2017) Long noncoding RNAs as biomarkers in cancer. Dis Markers 2017:7243968. 10.1155/2017/7243968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr BI, Guerra V (2017) Serum albumin levels in relation to tumor parameters in hepatocellular carcinoma patients. Int J Biol Markers 32:391–396. 10.5301/ijbm.5000300 [DOI] [PubMed] [Google Scholar]
- Chi Y, Wang D, Wang J, Yu W, Yang J (2019) Long non-coding RNA in the pathogenesis of cancers. Cells 8:1015. 10.3390/cells8091015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristiano A, Dietrich A, Spina JC, Ardiles V, De Santibanes E (2014) Focal nodular hyperplasia and hepatic adenoma: current diagnosis and management. Updates Surg 66:9–21. 10.1007/s13304-013-0222-3 [DOI] [PubMed] [Google Scholar]
- Cristobal I, Sanz-Alvarez M, Luque M, Carames C, Rojo F, Garcia-Foncillas J (2019) The role of microRNAs in hepatoblastoma tumors. Cancers (Basel) 11:409. 10.3390/cancers11030409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stefano F, Chacon E, Turcios L, Marti F, Gedaly R (2018) Novel biomarkers in hepatocellular carcinoma. Dig Liver Dis 11:1115–1123. 10.1016/j.dld.2018.08.019 [DOI] [PubMed] [Google Scholar]
- Dhanasekaran R, Limaye A, Cabrera R (2012) Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis, and therapeutics. Hepat Med 4:19–37. 10.2147/HMER.S16316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitroulis D, Damaskos C, Valsami S, Davakis S, Garmpis N, Spartalis E et al (2017) From diagnosis to treatment of hepatocellular carcinoma: an epidemic problem for both developed and developing world. World J Gastroenterol 23:5282–5294. 10.3748/wjg.v23.i29.5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Lou W, Xu L, Fan W (2018) Non-coding RNA in drug resistance of hepatocellular carcinoma. Biosci Rep 38:BSR20180915. 10.1042/BSR20180915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposti DD, Hernandez-Vargas H, Voegele C, Fernandez-Jimenez N, Forey N, Bancel B et al (2016) Identification of novel long non-coding RNAs deregulated in hepatocellular carcinoma using RNA-sequencing. Oncotarget 7:31862–31877. 10.7717/peerj.3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro S, Panzeri A, Braga F, Panteghini M (2019) Serum α-fetoprotein in pediatric oncology: not a children’s tale. Clin Chem Lab Med 57:783–797. 10.1515/cclm-2018-0803 [DOI] [PubMed] [Google Scholar]
- Gu D, Jin H, Jin G, Wang C, Wang N, Hu F et al (2016) The asialoglycoprotein receptor suppresses the metastasis of hepatocellular carcinoma via LASS2-mediated inhibition of V-ATPase activity. Cancer Lett 379:107–116. 10.1016/j.canlet.2016.05.030 [DOI] [PubMed] [Google Scholar]
- Hass HG, Vogel U, Scheurlen M, Jobst J (2018) Subclassification and detection of new markers for the discrimination of primary liver tumors by gene expression analysis using oligonucleotide arrays. Gut Liver 12:306–315. 10.5009/gnl17277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Zuo Q, Hu B, Jin H, Wang C, Cheng Z et al (2019) A novel, liver-specific long noncoding RNA LINC01093 suppresses HCC progression by interaction with IGF2BP1 to facilitate decay of GLI1 mRNA. Cancer Lett 450:98–109. 10.1016/j.canlet.2019.02.033 [DOI] [PubMed] [Google Scholar]
- Indersie E, Lesjean S, Hooks KB, Sagliocco F, Ernault T, Cairo S et al (2017) MicroRNA therapy inhibits hepatoblastoma growth in vivo by targeting β-catenin and Wnt signaling. Hepatol Commun 1:168–183. 10.1002/hep4.1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Pan J, Fang S, Zhou C, Han Y, Chen J et al (2019) Liquid biopsy: circulating exosomal long noncoding RNAs in cancer. Clin Chim Acta 495:331–337. 10.1016/j.cca.2019.04.082 [DOI] [PubMed] [Google Scholar]
- Khan SA, Davidson BR, Goldin RD, Heaton N, Karani J, Pereira SP et al (2012) Guidelines for the diagnosis and treatment of cholangiocarcinoma: an update. Gut 61:1657–1669. 10.1136/gutjnl-2011-301748 [DOI] [PubMed] [Google Scholar]
- Klingenberg M, Matsuda A, Diederichs S, Patel T (2017) Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol 67:603–618. 10.1016/j.jhep.2017.04.009 [DOI] [PubMed] [Google Scholar]
- Komuta M, Yeh MM (2019) A review on the update of combined hepatocellular cholangiocarcinoma. Semin Liver Dis. 10.1055/s-0039-3402515 [DOI] [PubMed] [Google Scholar]
- Konishi H, Ichikawa D, Yamamoto Y, Arita T, Shoda K, Hiramoto HPL et al (2016) Plasma level of metastasis-associated lung adenocarcinoma transcript 1 is associated with liver damage and predicts development of hepatocellular carcinoma. Cancer Sci 107:149–154. 10.1111/cas.12854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan H, Lu H, Wang X, Jin H (2015) MicroRNAs as potential biomarkers in cancer: opportunities and challenges. Biomed Res Int 201:125094. 10.1155/2015/125094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Huang L, Li Z, Zhong X, Tai S, Jiang X et al (2019a) Functions and roles of long noncoding RNA in cholangiocarcinoma. J Cell Physiol 234:17113–17126. 10.1002/jcp.28470 [DOI] [PubMed] [Google Scholar]
- Li O, Yi W, Yang P, Guo C, Peng C (2019b) Long non-coding RNA UCA1 promotes proliferation and invasion of intrahepatic cholangiocarcinoma cells through targeting microRNA-122. Exp Ther Med 18:25–32. 10.3892/etm.2019.7564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim IIP, Bondoc AJ, Geller JI, Tiao GM (2018) Hepatoblastoma—the evolution of biology, surgery, and transplantation. Children (Basel) 6:1. 10.3390/children6010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L, Wong SYS, Huang F, Lim S, Chong SS, Ooi LL et al (2019) Roles and regulation of long noncoding RNAs in hepatocellular carcinoma. Cancer Res 79:5131–5139. 10.1158/0008-5472.CAN-19-0255 [DOI] [PubMed] [Google Scholar]
- Liu WT, Lu X, Tang GH, Ren JJ, Liao WJ, Ge PL et al (2014) LncRNAs expression signatures of hepatocellular carcinoma revealed by microarray. World J Gastroenterol 20:6314–6321. 10.3748/wjg.v20.i20.6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Lowe SW (2012) The microcosmos of cancer. Nature 482:347–355. 10.1038/nature10888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Huang H, Xu Y, He X, Wang J, Hui B et al (2017) Current advances of long non-coding RNA highly upregulated in liver cancer in human tumors. OncoTargets Ther 10:4711–4717. 10.2147/OTT.S136915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak LY, Cruz-Ramon V, Chinchilla-Lopez P, Torres HA, LoConte NK, Rice JP et al (2018) Global Epidemiology, prevention, and management of hepatocellular carcinoma. Am Soc Clin Oncol Educ Book 38:262–279. 10.1200/EDBK_200939 [DOI] [PubMed] [Google Scholar]
- Man XB, Tang L, Zhang BH, Li SJ, Qiu XH, Wu MC et al (2005) Upregulation of glypican-3 expression in hepatocellular carcinoma but downregulation in cholangiocarcinoma indicates its differential diagnosis value in primary liver cancers. Liver Int 25:962–966. 10.1111/j.1478-3231.2005.01100.x [DOI] [PubMed] [Google Scholar]
- Matsui M, Corey DR (2017) Non-coding RNAs as drug targets. Nat Rev Drug Discov 16:167–179. 10.1038/nrd.2016.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa A, Takayama KI, Inoue S (2017) Long non-coding RNAs and prostate cancer. Cancer Sci 108:2107–2114. 10.1111/cas.13352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K, Mogul DB (2018) Pediatric liver tumors. Clin Liver Dis 22:753–772. 10.1016/j.cld.2018.06.008 [DOI] [PubMed] [Google Scholar]
- Ning B, Yu D, Yu AM (2019) Advances and challenges in studying noncoding RNA regulation of drug metabolism and development of RNA therapeutics. Biochem Pharmacol 169:113638. 10.1016/j.bcp.2019.113638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardini B, Sabo AA, Birolo G, Calin GA (2019) Noncoding RNAs in extracellular fluids as cancer biomarkers: the new frontier of liquid biopsies. Cancers (Basel) 11:1170. 10.3390/cancers11081170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Tang Q, Li G, Chen K (2017) Long non-coding RNAs as biomarkers and therapeutic targets: recent insights into hepatocellular carcinoma. Life Sci 191:273–282. 10.1016/j.lfs.2017.10.007 [DOI] [PubMed] [Google Scholar]
- Rizvi S, Khan S, Hallemeier CL, Kelley RK, Gores GJ (2018) Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat Rev Clin Oncol 15:95–111. 10.1038/nrclinonc.2017.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncalli M, Sciarra A, Tommaso LD (2016) Benign hepatocellular nodules of healthy liver: focal nodular hyperplasia and hepatocellular adenoma. Clin Mol Hepatol 22:199–211. 10.3350/cmh.2016.0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahryari A, Jazi MS, Samaei NM, Mowla SJ (2015) Long non-coding RNA SOX2OT: expression signature, splicing patterns, and emerging roles in pluripotency and tumorigenesis. Front Genet 6:196. 10.3389/fgene.2015.00196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B, Abrams M, Sepp-Lorenzino L (2013) Expression of asialoglycoprotein receptor 1 in human hepatocellular carcinoma. J Histochem Cytochem 61:901–909. 10.1369/0022155413503662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ, Chinnaiyan AM (2019) The role of non-coding RNAs in oncology. Cell 179:1033–1055. 10.1016/j.cell.2019.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonohara F, Inokawa Y, Hayashi M, Yamada S, Sugimoto H Fujii et al (2017) Prognostic value of long non-coding RNA HULC and MALAT1 following the curative resection of hepatocellular carcinoma. Sci Rep 7:16142. 10.1038/s41598-017-16260-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Huang Z, Li X (2017) Long non-coding RNA MALAT1 interacts with miR-204 to modulate human hilar cholangiocarcinoma proliferation, migration, and invasion by targeting CXCR4. J Cell Biochem 11:3643–3653. 10.1002/jcb.25862 [DOI] [PubMed] [Google Scholar]
- Tan C, Cao J, Chen L, Xi X, Wang S, Zhu Y et al (2019) Noncoding RNAs serve as diagnosis and prognosis biomarkers for hepatocellular carcinoma. Clin Chem 65:905–915. 10.1373/clinchem.2018.301150 [DOI] [PubMed] [Google Scholar]
- Tsuchiya N, Sawada Y, Endo I, Saito K, Uemura Y, Nakatsura T (2015) Biomarkers for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol 21:10573–10583. 10.3748/wjg.v21.i37.10573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang X, Chen W, Hu X, Li J, Liu C (2020) Regulatory roles of long noncoding RNAs implicated in cancer hallmarks. Int J Cancer 146:906–916. 10.1002/ijc.32277 [DOI] [PubMed] [Google Scholar]
- Wei L, Wang X, Lv L, Liu J, Xing H, Song Y et al (2019) The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol Cancer 25:147. 10.1186/s12943-019-1086-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CM, Tsang FHC, Ng IOL (2018) Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol 15:137–151. 10.1038/nrgastro.2017.169 [DOI] [PubMed] [Google Scholar]
- Xu Y, Wang Z, Jiang X, Cui Y (2017a) Overexpression of long noncoding RNA H19 indicates a poor prognosis for cholangiocarcinoma and promotes cell migration and invasion by affecting epithelial-mesenchymal transition. Biomed Pharmacother 92:17–23. 10.1016/j.biopha.2017.05.061 [DOI] [PubMed] [Google Scholar]
- Xu Y, Yao Y, Leng K, Li Z, Qin W, Zhong X et al (2017b) Long non-coding RNA UCA1 indicates an unfavorable prognosis and promotes tumorigenesis via regulating AKT/GSK-3β signaling pathway in cholangiocarcinoma. Oncotarget 8:96203–96214. 10.18632/oncotarget.21884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue R, Chen L, Zhang C, Fujita M, Li R, Yan SM et al (2019) Genomic and transcriptomic profiling of combined hepatocellular and intrahepatic cholangiocarcinoma reveals distinct molecular subtypes. Cancer Cell 35:932–947. 10.1016/j.ccell.2019.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Lu Y, Xu Q, Tang B, Park CK, Chen X (2015) HULC and H19 played different roles in overall and disease-free survival from hepatocellular carcinoma after curative hepatectomy: a preliminary analysis from gene expression omnibus. Dis Markers 2015:191029. 10.1155/2015/191029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Zheng H, Chan MT, Wu WK (2017) HULC: an oncogenic long non-coding RNA in human cancer. Cell Mol Med 21:410–417. 10.1111/jcmm.12956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Sun Y, Liu L, Zhou B, Wang S, Gu D (2017) Circulating lncRNAs serve as diagnostic markers for hepatocellular carcinoma. Cell Physiol Biochem 44:125–132. 10.1159/000484589 [DOI] [PubMed] [Google Scholar]
- Zhang L, Liu X, Zhang X, Chen R (2016a) Identification of important long non-coding RNAs and highly recurrent aberrant alternative splicing events in hepatocellular carcinoma through integrative analysis of multiple RNA-Seq datasets. Mol Genet Genomics 291:1035–1051. 10.1007/s00438-015-1163-y [DOI] [PubMed] [Google Scholar]
- Zhang H, Yang T, Wu M, Shen F (2016b) Intrahepatic cholangiocarcinoma: epidemiology, risk factors, diagnosis and surgical management. Cancer Lett 379:198–205. 10.1016/j.canlet.2015.09.008 [DOI] [PubMed] [Google Scholar]
- Zhou S, O’Gorman MR, Yang F, Andresen K, Wang L (2017) Glypican 3 as a serum marker for hepatoblastoma. Sci Rep 7:45932. 10.1038/srep45932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Shang W, Yu X, Tian J (2018) Glypican-3: a promising biomarker for hepatocellular carcinoma diagnosis and treatment. Med Res Rev 38:741–767. 10.1002/med.21455 [DOI] [PubMed] [Google Scholar]
- Zou Y, Li J, Chen Y, Xiao H, Zhang F, Yu D et al (2017) BANCR: a novel oncogenic long non-coding RNA in human cancers. Oncotarget 8:94997–95004. 10.18632/oncotarget.22031 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.